Abstract
Effective hemostasis relies on the timely formation of α-thrombin via prothrombinase, a Ca2+-dependent complex of factors Va and Xa assembled on the activated platelet surface, which cleaves prothrombin at Arg271 and Arg320. Whereas initial cleavage at Arg271 generates the inactive intermediate prethrombin-2, initial cleavage at Arg320 generates the enzymatically active intermediate meizothrombin. To determine which of these intermediates is formed when prothrombin is processed on the activated platelet surface, the cleavage of prothrombin, and prothrombin mutants lacking either one of the cleavage sites, was monitored on the surface of either thrombin- or collagen-activated platelets. Regardless of the agonist used, prothrombin was initially cleaved at Arg271 generating prethrombin-2, with α-thrombin formation quickly after via cleavage at Arg320. The pathway used was independent of the source of factor Va (plasma- or platelet-derived) and was unaffected by soluble components of the platelet releasate. When both cleavage sites are presented within the same substrate molecule, Arg271 effectively competes against Arg320 (with an apparent IC50 = 0.3μM), such that more than 90% to 95% of the initial cleavage occurs at Arg271. We hypothesize that use of the prethrombin-2 pathway serves to optimize the procoagulant activity expressed by activated platelets, by limiting the anticoagulant functions of the alternate intermediate, meizothrombin.
Introduction
The activation of prothrombin to α-thrombin is a critical step in the response to vascular injury. The generation of α-thrombin is achieved through the action of prothrombinase, which is composed of the serine protease factor Xa and its nonenzymatic cofactor, factor Va, assembled on an appropriate membrane surface in the presence of Ca2+ ions.1 In the physiologic setting, this surface is provided by the activated platelet.2 Relative to the activity of factor Xa alone, incorporation of factor Xa into prothrombinase accelerates the rate of prothrombin cleavage by 5 orders of magnitude,3 and both factor Va and the membrane surface are critical in this rate amplification, as removal of either component results in a substantial decrease in the rate of prothrombin cleavage.3 Indeed, deficiencies or disorders of any component of this complex result in severe bleeding diatheses.4-6
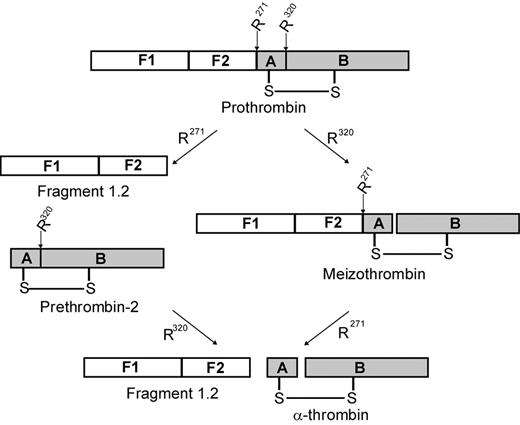
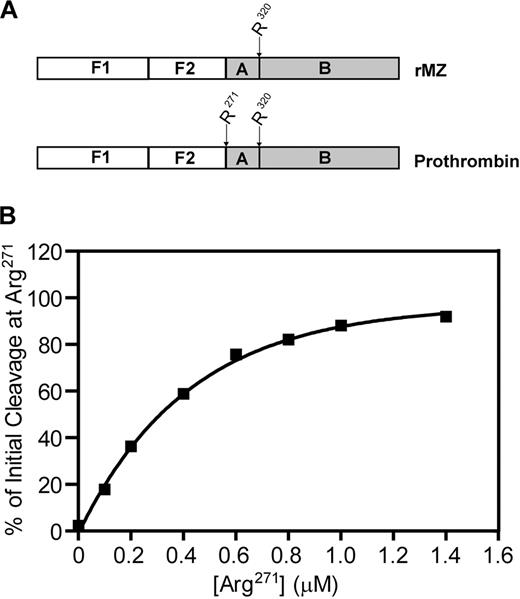
Prothrombin, the substrate for prothrombinase, consists of 4 domains: fragment 1, fragment 2, and the A and B chains of α-thrombin (Figure 1).1 Prothrombin is proteolytically activated to α-thrombin by cleavage on the C-terminal side of 2 specific residues: Arg271 (located between fragment 2 and the A chain) and Arg320 (located between the A and B chains). Initial cleavage at Arg271 results in the generation of prethrombin-2, an inactive intermediate, and the release of fragment 1.2 (F1.2). Subsequent cleavage at Arg320 converts prethrombin-2 to α-thrombin.7 Alternatively, cleavage may occur first at Arg320, leading to the generation of the enzymatically active intermediate meizothrombin, followed by cleavage at Arg271, releasing F1.2 and generating α-thrombin.8 Both of these activation pathways have been observed in systems using purified protein components and are dependent on the specific conditions applied. In the absence of factor Va, factor Xa cleaves prothrombin via the prethrombin-2 pathway.7,9,10 In contrast, when the entire, multicomponent prothrombinase complex is assembled on the surface of synthetic phosphatidylcholine/phosphatidylserine (PC/PS) vesicles, meizothrombin is generated with little, if any, prethrombin-2 formed.10,11 It is not clear which of these 2 pathways predominates in vivo.
Schematic of prothrombin activation. Human prothrombin consists of fragment 1 (F1), fragment 2 (F2), and the A and B chains of α-thrombin. Prothrombin is activated to α-thrombin by cleavage at Arg271 (R271) and Arg320 (R320). Initial cleavage at Arg271 (left pathway) generates fragment 1.2 and the inactive intermediate prethrombin-2. Subsequent cleavage at Arg320 generates α-thrombin. Alternatively, the active intermediate meizothrombin is produced when initial cleavage occurs at Arg320 (right pathway). Subsequent cleavage at Arg271 produces both α-thrombin and fragment 1.2.
Schematic of prothrombin activation. Human prothrombin consists of fragment 1 (F1), fragment 2 (F2), and the A and B chains of α-thrombin. Prothrombin is activated to α-thrombin by cleavage at Arg271 (R271) and Arg320 (R320). Initial cleavage at Arg271 (left pathway) generates fragment 1.2 and the inactive intermediate prethrombin-2. Subsequent cleavage at Arg320 generates α-thrombin. Alternatively, the active intermediate meizothrombin is produced when initial cleavage occurs at Arg320 (right pathway). Subsequent cleavage at Arg271 produces both α-thrombin and fragment 1.2.
In vivo, activated platelets2 provide both the membrane surface for assembly of prothrombinase and a key component of that complex, factor Va. Approximately 18% to 25% of the total factor V in blood is stored as a mixture of factor V and activated factor Va within the α-granules of platelets.12-14 Platelet-derived factor V/Va displays many unique properties compared with its plasma-derived counterpart, including resistance to inactivation by either activated protein C15 or plasmin16 and an inability to be phosphorylated by either casein kinase II17 or a platelet-associated kinase,18 both of which readily phosphorylate plasma-derived factor Va under the same conditions. Although platelets contain a significant store of factor Va, it is not released at a high enough concentration to saturate all of the factor Va binding sites on the platelet surface.19 Factor V in the plasma, when activated to factor Va, will also bind to the platelet surface, providing a second source of cofactor for prothrombinase assembly.19
Assembly of prothrombinase is also highly regulated by the activated platelet membrane. Activated platelets express specific, saturable binding sites for both factors Va and Xa,19,20 yet factor Xa binding to activated platelets is absolutely dependent on membrane-bound factor Va.21 Further, the agonist concentrations that are sufficient to induce maximal exposure of factor Va binding sites (≥ 1nM) are significantly lower than those required to effect factor Xa binding (≥ 20nM), suggesting that different components of the platelet surface are required for these binding interactions.19,22 In addition, a subpopulation of thrombin-activated platelets has been identified that binds factor Va but not factor Xa.22 As prothrombin activation occurs physiologically on the highly regulated, activated platelet membrane, knowledge of which intermediate is generated initially is critical in understanding how α-thrombin formation is regulated in vivo.
Methods
Materials
All of the protocols used in this research were approved by the University of Vermont's Institutional Review Board and Committee on Human Subjects. The thrombin inhibitor dansylarginine N-(3-ethyl-1,5-pentanediyl)amide (DAPA) was purchased from Haematologic Technologies. The peptides Arg-Gly Asp-Ser (RGDS) and Gly-Pro-Arg-Pro were synthesized and characterized by the Protein Core Facility, University of Vermont (Burlington, VT). Bovine serum albumin was purchased from MP Biomedicals, and hirudin was from Calbiochem. Iodine-125 and Western Lightning chemiluminescence reagents were purchased from PerkinElmer Life and Analytical Sciences, and IODO-GEN tubes were from Thermo Scientific. Collagen Type I, from rat tail, was obtained from BD Biosciences. PC/PS vesicles (75% PC/25% PS) were generously provided by Dr Kenneth Mann (University of Vermont). The PC/PS vesicles were prepared using synthetic phospholipids (Avanti Polar Lipids) according to the method of Higgins and Mann.23 Burro antiprethrombin-124 was provided by the Antibody Core Facility at the University of Vermont, and goat anti–horse IgG, conjugated to horseradish peroxidase was purchased from Vector Laboratories.
Preparation of coagulation proteins
Human prothrombin, thrombin, and factor Xa were purchased from Haematologic Technologies. The prothrombin mutants, R155/284/320A (rP2) and R155/271/284A (rMZ), were generously provided by Dr Michael Nesheim (Queen's University, Kingston, ON) and were expressed, purified, and characterized as described previously.25 These mutants have been used previously in studies assessing the mechanism of prothrombin activation by prothrombinase assembled on PC/PS vesicles.25-27 Prothrombin, rP2, and rMZ were dialyzed into 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), 150mM NaCl, 5mM CaCl2, 0.1% PEG-8000, pH 7.4 (HEPES-buffered saline/Ca2+/PEG) before use. Factor V was purified from pooled normal human plasma by immunoaffinity chromatography as described.28 Factor Va was prepared by incubation of factor V (1μM in HEPES-buffered saline/Ca2+/PEG), with α-thrombin (20nM) for 10 minutes at 37°C. After activation, hirudin (30nM) was added to inactivate the α-thrombin. Protein concentrations were determined by absorbance at 280 nm, using the following molecular weights and extinction coefficients (E1%280nm): factor V, 330 000, 9.629 ; prothrombin and prothrombin mutants, 72 000, 13.830 ; thrombin, 36 700, 18.331 ; and factor Xa, 46 000, 11.6.32
Iodination of prothrombin and prothrombin mutants
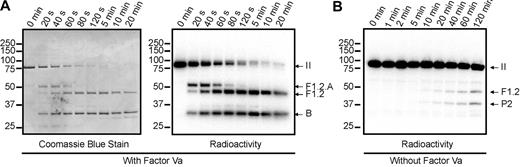
Prothrombin, rP2, and rMZ were radioactively labeled with iodine-125, using a method previously described for the labeling of factor V.33 Subsequent to labeling, protein integrity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli,34 after cystine reduction, followed by phosphorimaging, The ability of the radiolabeled proteins to be activated by prothrombinase was assessed using PC/PS vesicles (Figure 2A), as described in “Prothrombin activation time courses.” Consistent with previous reports,10,11 prothrombin was cleaved initially at Arg320, as evidenced by the transient generation of the F1.2.A chain of meizothrombin and subsequent cleavage at Arg271 to generate fragment 1.2 and α-thrombin. In the gel system used in these studies, the A chain of α-thrombin is not visible. Identical cleavage patterns were observed by both protein staining (Figure 2A left panel) and phosphorimaging (Figure 2A right panel), demonstrating that the labeled and unlabeled prothrombin were activated identically. To ensure that the alternative pathway of activation, with initial cleavage at Arg271, was not compromised, similar experiments were performed in the absence of factor Va (Figure 2B). As has been described previously,9,10 under these conditions, the initial cleavage occurred at Arg271 to generate fragment 1.2 and prothrombin-2, and the overall rate of cleavage was significantly slower.
Cleavage of prothrombin by prothrombinase assembled on PC/PS vesicles. Human prothrombin was radioactively labeled with 125I and assessed for its ability to be cleaved either by human prothrombinase (5nM factor Xa and 0.5nM factor Va) (A) or by factor Xa alone (B) assembled on PC/PS vesicles as detailed in “Prothrombin activation time courses.” Aliquots of the reaction mixtures were quenched at timed intervals and subjected to SDS-PAGE. The gels were analyzed by staining with Coomassie blue (A, left panel) to visualize unlabeled protein, and by phosphorimaging (A, right panel, B) to visualize 125I-prothrombin and fragments generated by prothrombinase. Time points are indicated at the top of each panel. Molecular weight markers are indicated on the left, and the locations of prothrombin (II), the fragment 1.2.A chain of meizothrombin (F1.2.A), fragment 1.2 (F1.2), prethrombin-2 (P2), and the B chain, are indicated on the right.
Cleavage of prothrombin by prothrombinase assembled on PC/PS vesicles. Human prothrombin was radioactively labeled with 125I and assessed for its ability to be cleaved either by human prothrombinase (5nM factor Xa and 0.5nM factor Va) (A) or by factor Xa alone (B) assembled on PC/PS vesicles as detailed in “Prothrombin activation time courses.” Aliquots of the reaction mixtures were quenched at timed intervals and subjected to SDS-PAGE. The gels were analyzed by staining with Coomassie blue (A, left panel) to visualize unlabeled protein, and by phosphorimaging (A, right panel, B) to visualize 125I-prothrombin and fragments generated by prothrombinase. Time points are indicated at the top of each panel. Molecular weight markers are indicated on the left, and the locations of prothrombin (II), the fragment 1.2.A chain of meizothrombin (F1.2.A), fragment 1.2 (F1.2), prethrombin-2 (P2), and the B chain, are indicated on the right.
Preparation of activated platelet suspensions
Blood was drawn by venipuncture from nonmedicated, consenting adults into 71mM citric acid, 85mM trisodium citrate, 2% dextrose, pH 4.5 (acid citrate dextrose) at a 6:1 ratio. Platelets were isolated and washed according to the procedure of Mustard et al,35 with minor modifications, as described previously.15 After washing, platelets were resuspended in HEPES-Tyrode buffer (5mM HEPES, 137mM NaCl, 2.68mM KCl, 11.9mM NaHCO3, 0.42mM NaH2PO4, 1mM MgCl2, 5mM CaCl2, 0.2% dextrose, pH 7.4; HT). Platelet concentration was determined using a Coulter Z1 particle counter (Beckman Coulter). Washed platelets were diluted to 3 × 108 platelets/mL in HT containing 1mM RGDS and were activated by incubation with α-thrombin (50nM) for 5 minutes at 25°C (conditions that allow for maximum exposure of factor Va and Xa binding sites19 ), followed by addition of 75nM hirudin to inactivate the α-thrombin. For experiments in which platelets were washed extensively subsequent to thrombin-catalyzed activation, the activation buffer contained 3mM RGDS and 2mM Gly Pro-Arg-Pro to prevent platelet aggregation. Control experiments demonstrated that the presence of these 2 peptides had no effect on either platelet activation or the rate of thrombin generation at the activated platelet surface (data not shown). Platelets were pelleted by centrifugation (3 minutes, 660g) and gently resuspended in HT containing 5mM RGDS. Washing was repeated 3 times, and, after the final wash, the platelets were resuspended in HT containing 1mM RGDS. Platelet concentration after washing was determined.
Prothrombin activation time courses
Reaction mixtures contained 1 × 108 activated platelets/mL, 1.4μM prothrombin, rP2, or rMZ, 3μM DAPA, 1mM RGDS, and trace amounts of 125I-prothrombin, 125I-rP2, or 125I-rMZ, respectively (500 cpm/μL). In some experiments, 5nM plasma-derived factor Va was added to saturate all of the factor Va binding sites on the activated platelet surface. Saturation of these sites results in the formation of approximately 0.5nM membrane-bound prothrombinase, based on an average of 2700 (± 1000) factor Xa binding sites per platelet.21 To best approximate these conditions, control experiments were performed using PC/PS vesicles (20μM) to provide the membrane surface, 0.5nM factor Va, 1.4μM prothrombin, rP2, or rMZ, 3μM DAPA, and trace amounts of 125I-prothrombin, 125I-rP2, or 125I-rMZ (500 cpm/μL). Under either condition, reactions were initiated by adding factor Xa to a final concentration of 5nM in the reaction and incubated at 25°C. Aliquots were removed at timed intervals and quenched by addition of 5 × sample preparation buffer (312.5mM Tris, 25mM ethylenediaminetetraacetic acid, 10% SDS, 50% glycerol, 0.05% bromphenol blue, pH 6.8; 5 × sample preparation buffer) to a final concentration of 1 × sample preparation buffer. Radioactivity in all samples was determined using a Packard Cobra II Auto-Gamma Counter (PerkinElmer Life and Analytical Sciences). The contribution of free factor Xa to prothrombin cleavage on activated platelets was assessed previously. Addition of saturating concentrations of factor Xa to thrombin-activated platelets from a person completely devoid of factor V yielded no detectable thrombin generation over a 20-minute time course,21 consistent with the reported approximately 10 000-fold decrease in catalytic activity of free factor Xa in the absence of factor Va.3
For experiments assessing the activation of prothrombin on the surface of collagen-activated platelets, reaction mixtures contained 3 × 108 platelets/mL, 1.4μM prothrombin, and 1mM RGDS in HT. Reactions were initiated by simultaneous addition of collagen (0.1 mg/mL final concentration) and factor Xa (5nM final concentration) at 37°C, in the absence of DAPA. Prothrombin activation was monitored as detailed in the preceding paragraph.
Determination of the kinetic constants governing cleavage at Arg271 and Arg320
In experiments using PC/PS vesicles, reaction mixtures were prepared and subsequently analyzed as described in “Prothrombin activation time courses”; however, the factor Va concentration was decreased to 0.1nM to reduce the reaction rate, and various amounts of unlabeled rP2 or rMZ, containing trace 125I-rP2 or 125I-rMZ, were added. Activated platelet mixtures contained 2 × 107 thrombin-activated platelets per milliliter to reduce the reaction rate, and 5nM plasma-derived factor Va to saturate all factor Va binding sites. Reactions were initiated, and aliquots removed at timed intervals. In experiments assessing competition for cleavage at the Arg271 and Arg320 sites within prothrombin, reaction mixtures were prepared using thrombin-activated platelets, with the following modification. Concentrations of prothrombin and rMZ, containing trace 125I-prothrombin or 125I-rMZ, were varied to maintain the total concentration of substrate at 1.4μM in every reaction. Reactions were initiated and processed.
SDS-PAGE, phosphorimaging, and immunoblotting
Radioiodinated protein samples (5000 cpm/lane) were reduced with 5% β-mercaptoethanol and separated by SDS-PAGE on 10% polyacrylamide gels (Invitrogen), according to the method of Laemmli.34 Gels were treated with 9% acetic acid, 18% methanol (30 minutes), followed by 5% glycerol (30 minutes), then dried under vacuum at 65°C and exposed overnight to a phosphorimaging screen (Bio-Rad). The imaging screen was scanned using a Molecular Imager FX running Quantity One v.4.6.3 (Bio-Rad). For experiments using collagen-activated platelets, reduced samples (100 ng prothrombin/lane) were separated by SDS-PAGE on 10% polyacrylamide gels, transferred to nitrocellulose membranes according to the method of Towbin et al,36 and analyzed by immunoblotting with a burro antiprethrombin-1 polyclonal antibody (7.5 μg/mL). Bound antibody was detected using horseradish peroxidase-conjugated goat antihorse IgG (0.2 μg/mL) and chemiluminescence detection.
Data analyses
Densitometric analyses were performed using the Quantity One v.4.6.3. Data obtained from the rP2 and rMZ time courses and the Arg271 competition experiments were fit to single exponential equations, whereas data from experiments determining the kinetic constants for cleavage of rP2 and rMZ were fit to the Michaelis-Menten equation, all using GraphPad Prism v.5.0 (GraphPad Software).
Results
Cleavage of 125I-prothrombin on the activated platelet surface
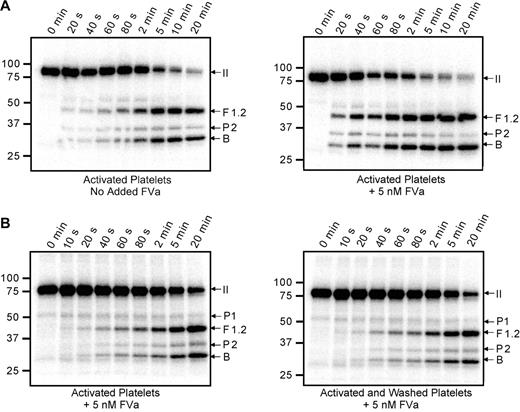
Cleavage of 125I-prothrombin was monitored using activated platelets (1 × 108/mL) as the membrane surface (Figure 3A). Substantial generation of the prethrombin-2 intermediate was observed, suggesting that initial cleavage of prothrombin occurred at Arg271. However, little prethrombin-2 accumulated during the time course, suggesting that it was rapidly converted to α-thrombin by cleavage at Arg320. Initial formation of prethrombin-2 was observed whether the reaction was performed exclusively with platelet-associated factor Va (Figure 3A left panel) or with the addition of plasma-derived factor Va to saturate all prothrombinase binding sites on the activated platelet surface (Figure 3A right panel). As expected, saturation of these sites resulted in a substantial increase in the rate of prothrombin cleavage. In these analyses, only trace amounts of meizothrombin were apparent, as evidenced by a lack of the F1.2.A chain.
Cleavage of prothrombin by prothrombinase assembled on the activated platelet surface. Thrombin-activated platelets (1 × 108/mL) were incubated with 1.4μM human prothrombin containing trace 125I-prothrombin and 3μM DAPA. Prothrombinase assays were initiated by addition of 5nM factor Xa; aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by phosphorimaging. (A) The assays were performed either relying on the platelet-released factor Va (left panel) or adding 5nM plasma-derived factor Va to saturate all factor Va binding sites on the platelet surface (right panel). (B) Prothrombin activation was similarly followed on the surface of either thrombin-activated platelets (left panel) or thrombin-activated platelets that had been washed 3 times after activation to remove any soluble materials released by the platelets (right panel). Under both conditions, plasma-derived factor Va (5nM) was added to saturate the activated platelet surface. All panels are labeled as described in Figure 2.
Cleavage of prothrombin by prothrombinase assembled on the activated platelet surface. Thrombin-activated platelets (1 × 108/mL) were incubated with 1.4μM human prothrombin containing trace 125I-prothrombin and 3μM DAPA. Prothrombinase assays were initiated by addition of 5nM factor Xa; aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by phosphorimaging. (A) The assays were performed either relying on the platelet-released factor Va (left panel) or adding 5nM plasma-derived factor Va to saturate all factor Va binding sites on the platelet surface (right panel). (B) Prothrombin activation was similarly followed on the surface of either thrombin-activated platelets (left panel) or thrombin-activated platelets that had been washed 3 times after activation to remove any soluble materials released by the platelets (right panel). Under both conditions, plasma-derived factor Va (5nM) was added to saturate the activated platelet surface. All panels are labeled as described in Figure 2.
When prothrombin is cleaved by prothrombinase on the surface of activated platelets, it is reasonable to hypothesize that the pathway of cleavage may be dictated by multiple components of the activated platelet membrane, such as the unique lipid content, expression of lipid rafts, and/or the expression of receptor proteins for factors Va, Xa, and/or II. Alternatively, soluble components released on platelet activation (the platelet releasate) may play a significant role. To address this latter possibility, the prothrombinase-catalyzed cleavage of prothrombin was monitored on activated platelets both before (Figure 3B left panel) and after (Figure 3B right panel) extensive platelet washing to remove soluble components contributed by the platelet releasate. These assays were performed using saturating concentrations of factors Va and Xa; and in both instances, prothrombin activation proceeded through the prethrombin-2 intermediate. In addition, the reaction rates were nearly identical under both conditions, indicating that no significant alteration of the platelet surface had occurred during the washing procedure and that the removal of soluble components did not alter the pathway of activation. Although some prethrombin-1 was visible in the 125I-prothrombin preparation used in this experiment, its concentration did not increase with time, indicating that the α-thrombin generated during the reaction was being inhibited by the presence of DAPA and therefore was incapable of cleaving the prothrombin substrate at Arg155.
Cleavage of rP2 and rMZ by prothrombinase
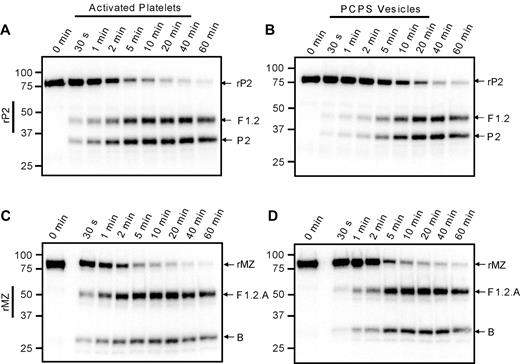
The apparent preference for initial cleavage of prothrombin at Arg271 versus Arg320 on the activated platelet surface could be the result of a faster rate of cleavage at Arg271, a greater affinity of prothrombinase for that site, or both. To investigate these possibilities, 2 prothrombin mutants were used: rP2, in which Arg320 has been substituted with Ala and only cleavage at Arg271 is possible; and rMZ, which can only be cleaved at Arg320 resulting from substitution of Arg271 with Ala.25,37 The prothrombinase-catalyzed cleavage of 1.4μM 125I-rP2 or 125I-rMZ was assessed under conditions in which the activated platelet membrane was saturated with both factors Va and Xa (Figure 4A,C). Both rP2 and rMZ were cleaved in these assays, resulting in the time-dependent generation of the anticipated product bands, F1.2 and prethrombin-2 from rP2, and the F1.2.A and F1.2.B chains of meizothrombin from rMZ. Densitometric quantification of the radioactive protein bands was used to generate progress curves of prothrombin consumption; and because initial rates could not be determined directly from these data because of the rapid reaction rates, the data were fit to single exponential decay equations to obtain reasonable estimates of the initial rates of cleavage. The experiments were performed using platelets isolated from 2 individual donors. Surprisingly, rMZ was cleaved 1.3- to 1.5-fold faster than rP2 on the thrombin-activated platelet surface (7.0nM/s and 5.4nM/s, respectively, for donor 1; and 11.6nM/s and 7.7nM/s, respectively, for donor 2), inconsistent with the preferred initial cleavage of plasma-derived prothrombin at Arg271. In contrast, when both mutants were cleaved via prothrombinase assembled on PC/PS vesicles (Figure 4B,D), the rate of cleavage of rMZ (8.4 ± 3.4nM/s) was approximately 4-fold greater than the rate of cleavage of rP2 (1.7 ± 0.2nM/s), consistent with the preferred initial cleavage at Arg320 when prothrombin is activated in systems using PC/PS vesicles.10,11
Cleavage of prothrombin variants by prothrombinase assembled on activated platelets or PC/PS vesicles. Prothrombinase assays were performed, as described in the legend to Figure 3, using either the prothrombin variant rP2 (A-B), which may only be cleaved at Arg271 to generate fragment 1.2 (F1.2) and prethrombin-2 (P2), or rMZ (C-D), which may only be cleaved at Arg320 to generate meizothrombin consisting of F1.2.A and B chains. Reactions were performed using either: (A,C) 1 × 108/mL thrombin-activated platelets, 5nM factor Xa, and 5nM factor Va; or (B,D) 20μM PC/PS vesicles, 5nM factor Xa, and 0.5nM factor Va. Aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by phosphorimaging. The locations of rP2, rMZ, and their cleavage products are indicated at the right of each panel. Time points are indicated at the top, and molecular weight markers are indicated on the left.
Cleavage of prothrombin variants by prothrombinase assembled on activated platelets or PC/PS vesicles. Prothrombinase assays were performed, as described in the legend to Figure 3, using either the prothrombin variant rP2 (A-B), which may only be cleaved at Arg271 to generate fragment 1.2 (F1.2) and prethrombin-2 (P2), or rMZ (C-D), which may only be cleaved at Arg320 to generate meizothrombin consisting of F1.2.A and B chains. Reactions were performed using either: (A,C) 1 × 108/mL thrombin-activated platelets, 5nM factor Xa, and 5nM factor Va; or (B,D) 20μM PC/PS vesicles, 5nM factor Xa, and 0.5nM factor Va. Aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by phosphorimaging. The locations of rP2, rMZ, and their cleavage products are indicated at the right of each panel. Time points are indicated at the top, and molecular weight markers are indicated on the left.
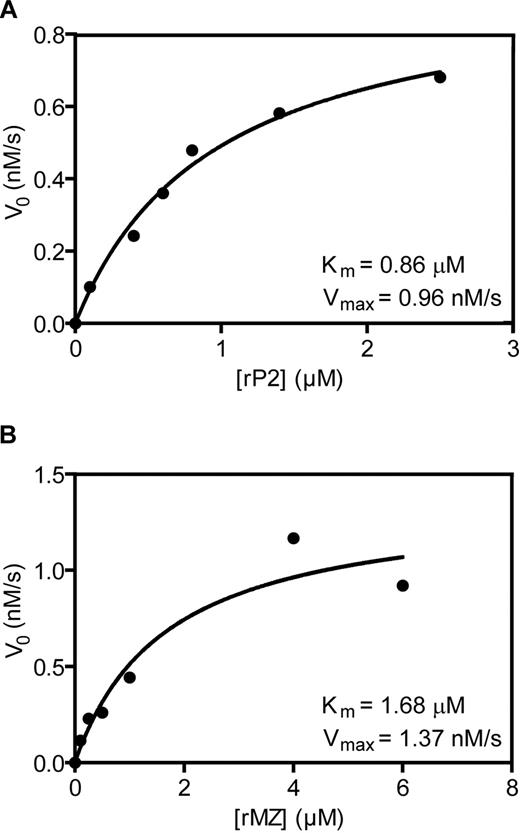
When assayed at concentrations equivalent to the physiologic concentration of prothrombin, the rates at which the 2 prothrombin variants were cleaved do not support, and cannot explain, the preference for cleavage at Arg271 when prothrombin is cleaved on thrombin-activated platelets. Therefore, the kinetic constants governing the cleavage reactions at each site were derived from substrate versus velocity curves, obtained by titration of the rP2 and rMZ mutants on thrombin-activated platelets isolated from 2 different platelet donors, as well as on PC/PS vesicles to serve as a control (Figure 5; Table 1). As expected, the Km values determined for cleavage of rP2 (0.34μM) and rMZ (0.32μM) by prothrombinase in experiments using PC/PS vesicles were consistent with previously published values25,37 yet are in marked contrast to kinetic parameters obtained for prothrombinase assembled on activated platelets from the 2 different donors. The average Km for rMZ (1.91 ± 0.24μM) was approximately 2.4-fold greater than that for rP2 (0.82 ± 0.05μM). Whereas the Km values were consistent between the 2 donors, the Vmax values for cleavage of either rP2 or rMZ varied approximately 4-fold (4.43nM/s and 4.50nM/s, respectively, for donor 1; and 0.96nM/s and 1.37nM/s, respectively, for donor 2), indicating that the concentration of prothrombinase binding sites differed between the individual platelet donors, consistent with previous reports of interdonor variability.19,21 However, these experimentally determined Km and Vmax values were consistent with the rates of cleavage observed in the extended time courses using rP2 and rMZ as substrates (Figure 4) and indicated that, at a physiologic concentration of prothrombin (1.4μM), rP2 and rMZ would be expected to be cleaved at similar rates on the activated platelet surface. However, although these data suggest a preferential binding of prothrombin to facilitate cleavage at Arg271 versus Arg320 when the 2 cleavage sites are present in separate prothrombin variants, they do not adequately explain the almost exclusive predomination of the prethrombin-2 pathway seen when plasma-derived prothrombin is cleaved on activated platelets (Figure 3).
Determination of kinetic constants governing the cleavage of rP2 and rMZ by prothrombinase assembled on the activated platelet membrane. Prothrombinase assays were performed as described in Figure 3. Reaction mixtures contained 2 × 107/mL thrombin-activated platelets, equimolar concentrations of added plasma-derived factors Va and Xa (5nM), and various concentrations of rP2 (A) or rMZ (B). Subsequent to initiation of the reaction, aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by densitometric analyses of protein bands visualized by phosphorimaging. The initial rates of cleavage of the variants were plotted as a function of the substrate concentration. Substrate consumption was less than 10% under all conditions. The data were fit to the Michaelis-Menten equation using GraphPad Prism software v5.0. The data shown, including the calculated Km and Vmax values, are from one of 2 platelet donors and are representative of experiments performed using platelets from both persons.
Determination of kinetic constants governing the cleavage of rP2 and rMZ by prothrombinase assembled on the activated platelet membrane. Prothrombinase assays were performed as described in Figure 3. Reaction mixtures contained 2 × 107/mL thrombin-activated platelets, equimolar concentrations of added plasma-derived factors Va and Xa (5nM), and various concentrations of rP2 (A) or rMZ (B). Subsequent to initiation of the reaction, aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by densitometric analyses of protein bands visualized by phosphorimaging. The initial rates of cleavage of the variants were plotted as a function of the substrate concentration. Substrate consumption was less than 10% under all conditions. The data were fit to the Michaelis-Menten equation using GraphPad Prism software v5.0. The data shown, including the calculated Km and Vmax values, are from one of 2 platelet donors and are representative of experiments performed using platelets from both persons.
Kinetic parameters defining cleavage of prothrombin variants on membrane surfaces*
| Prothrombin variant . | Available cleavage site . | Donor . | Activated platelets† . | PCPS vesicles . | ||||
|---|---|---|---|---|---|---|---|---|
| Km, μM . | Vmax, nM/s . | R2 . | Km, μM . | Vmax, nM/s . | R2 . | |||
| rP2 | Arg271 | 1 | 0.77 | 4.43 | 0.970 | 0.34 | 2.27 | 0.935 |
| 2 | 0.86 | 0.96 | 0.990 | |||||
| Mean ± SD | 0.82 ± 0.05 | |||||||
| rMZ | Arg320 | 1 | 2.15 | 4.50 | 0.987 | 0.32 | 4.72 | 0.988 |
| 2 | 1.68 | 1.37 | 0.934 | |||||
| Mean ± SD | 1.91 ± 0.24 | |||||||
| Prothrombin variant . | Available cleavage site . | Donor . | Activated platelets† . | PCPS vesicles . | ||||
|---|---|---|---|---|---|---|---|---|
| Km, μM . | Vmax, nM/s . | R2 . | Km, μM . | Vmax, nM/s . | R2 . | |||
| rP2 | Arg271 | 1 | 0.77 | 4.43 | 0.970 | 0.34 | 2.27 | 0.935 |
| 2 | 0.86 | 0.96 | 0.990 | |||||
| Mean ± SD | 0.82 ± 0.05 | |||||||
| rMZ | Arg320 | 1 | 2.15 | 4.50 | 0.987 | 0.32 | 4.72 | 0.988 |
| 2 | 1.68 | 1.37 | 0.934 | |||||
| Mean ± SD | 1.91 ± 0.24 | |||||||
Km and Vmax values were determined by titration of rP2 or rMZ and fitting to the Michaelis-Menten equation, as described in “Determination of the kinetic constants governing cleavage at Arg271 and Arg320” and “Data analyses.” The R2 values for these fits are indicated. Substrate consumption did not exceed 10% in these experiments. Experiments on PCPS vesicles were performed once.
Experiments with activated platelets were performed using platelets from 2 individual donors. Shown are the fit values of Km and Vmax for each individual donor, as well as mean values for Km.
Competition between Arg271 and Arg320 within the same prothrombin molecule for cleavage by prothrombinase
Because plasma-derived prothrombin contains both cleavage sites within the same molecule, experiments were designed to determine the extent to which the Arg271 cleavage site is able to compete with the Arg320 cleavage site when both are available within the same molecule of prothrombin. Titrations were performed using the rMZ mutant, which contains only the Arg320 site, and prothrombin, which contains both cleavage sites (Figure 6A). The total concentration of the 2 substrates (rMZ and prothrombin) in these reactions was fixed at 1.4μM, but the proportion of each species varied. Thus, the Arg320 cleavage site remained constant at 1.4μM, whereas the Arg271 cleavage site was varied from 0 to 1.4μM. For each reaction mixture, the initial rate of cleavage at either Arg271 or Arg320 was determined from densitometric analyses of their product bands, the F1.2 fragment for cleavage at Arg271, and the F1.2.A fragment of meizothrombin for cleavage at Arg320, and the percentage of initial cleavage at Arg271 was calculated from these rates (Figure 6B). As expected, when only rMZ was present and the Arg271 cleavage site was not available, 100% of the initial cleavage occurred at Arg320. As the concentration of Arg271 in the reaction mixture was increased, it quickly became the preferred cleavage site. At 0.1μM Arg271 and 1.4μM Arg320, approximately 20% of the initial cleavage occurred at the Arg271 site. Both sites were cleaved equivalently (50%) when only 0.3μM Arg271 was present. Little, if any, cleavage occurred at Arg320 once the concentration of Arg271 surpassed 0.8μM, even though the concentration of the Arg320 site remained at 1.4μM. These data clearly demonstrate that Arg271 is the preferred site of initial cleavage when prothrombinase is assembled on thrombin-activated platelets. Availability of Arg271 as a cleavage site within prothrombin inhibits cleavage at the alternative Arg320 site, with an apparent IC50 of 0.3μM, demonstrating a substantially tighter binding for normal prothrombin (containing both cleavage sites) over rMZ (containing only Arg320). When both sites are present at 1.4μM within the same molecule, as they are in prothrombin, more than 90% of initial cleavage occurs at Arg271.
Competition between Arg271 and Arg320 within the same prothrombin molecule for cleavage by prothrombinase assembled on the activated platelet membrane. (A) To titrate the Arg271 cleavage site within the same molecule as the Arg320 cleavage site, prothrombinase assays were performed using mixtures of prothrombin (containing both Arg271 and Arg320 as potential cleavage sites) and rMZ (containing only Arg320). (B) Reactions were performed, and data were analyzed as described in Figure 5, to determine the rate of generation of fragment 1.2.A as an indication of initial cleavage at Arg320, and of fragment 1.2 as an indication of initial cleavage at Arg271. Data are represented as the concentration of Arg271 versus the percentage of initial cleavage at Arg271.
Competition between Arg271 and Arg320 within the same prothrombin molecule for cleavage by prothrombinase assembled on the activated platelet membrane. (A) To titrate the Arg271 cleavage site within the same molecule as the Arg320 cleavage site, prothrombinase assays were performed using mixtures of prothrombin (containing both Arg271 and Arg320 as potential cleavage sites) and rMZ (containing only Arg320). (B) Reactions were performed, and data were analyzed as described in Figure 5, to determine the rate of generation of fragment 1.2.A as an indication of initial cleavage at Arg320, and of fragment 1.2 as an indication of initial cleavage at Arg271. Data are represented as the concentration of Arg271 versus the percentage of initial cleavage at Arg271.
Prothrombin activation on the surface of collagen-activated platelets
In an effort to more closely approximate how platelets might be activated subsequent to vascular injury, experiments were performed by incubating platelets with prothrombin and simultaneously adding both collagen and factor Xa (Figure 7). Under these conditions, as the platelets activate and express platelet-derived factor Va at their surface, factor Xa binds, forming active prothrombinase, and prothrombin is activated to thrombin. In the absence of inhibition by DAPA, the generated thrombin may feedback, activating additional platelet-released factor V to factor Va. Using these collagen-stimulated platelets, the same pattern of prothrombin cleavage was observed (Figure 7) as with thrombin-stimulated platelets (Figure 3A). Initial cleavage at Arg271 was again evident, with fragment 1.2 becoming apparent earlier than the B chain, indicating that the order of prothrombin cleavage was independent of the agonist used to effect platelet activation. As DAPA was not present, thrombin-catalyzed cleavages at Arg155 also occurred, generating prethrombin-1 from prothrombin and fragment 2 from fragment 1.2. (Fragment 1 could not be detected with the antibody used in the analysis.) Interestingly, the generated prethrombin-1 was efficiently processed to produce α-thrombin.
Prothrombin activation on the surface of collagen-activated platelets. Isolated platelets (3 × 108/mL) were incubated with 1.4μM human prothrombin. Platelet activation and prothrombinase assembly and function were simultaneously initiated by addition of 0.1 mg/mL collagen and 5nM factor Xa. Aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by Western blotting using a polyclonal antibody against prethrombin-1, which recognizes prothrombin (II), prethrombin-1 (P1), fragment 1.2.A, fragment 1.2 (F1.2), prethrombin-2 (P2), the B chain, fragment 2.A (F2.A), and fragment 2 (F2). The panel is labeled as described in Figure 2. The vertical line was inserted to indicate samples run on 2 separate gels.
Prothrombin activation on the surface of collagen-activated platelets. Isolated platelets (3 × 108/mL) were incubated with 1.4μM human prothrombin. Platelet activation and prothrombinase assembly and function were simultaneously initiated by addition of 0.1 mg/mL collagen and 5nM factor Xa. Aliquots were removed at timed intervals and analyzed by SDS-PAGE followed by Western blotting using a polyclonal antibody against prethrombin-1, which recognizes prothrombin (II), prethrombin-1 (P1), fragment 1.2.A, fragment 1.2 (F1.2), prethrombin-2 (P2), the B chain, fragment 2.A (F2.A), and fragment 2 (F2). The panel is labeled as described in Figure 2. The vertical line was inserted to indicate samples run on 2 separate gels.
Discussion
Activation of prothrombin by prothrombinase assembled on the surface of thrombin-activated platelets proceeded predominantly through initial cleavage at Arg271, generating prethrombin-2, with subsequent cleavage at Arg320 to generate α-thrombin. The rate of prothrombin activation was dependent on factor Va, but the order of cleavage was independent of the source of factor Va (ie, platelet- or plasma-derived). The prothrombin activation pathway was also independent of the agonist used to activate the platelets, as prothrombinase assembled on either thrombin- or collagen-activated platelets activated prothrombin via the prethrombin-2 intermediate. Furthermore, experiments in which the soluble components of the platelets were removed before prothrombinase assembly suggested that the pathway of prothrombin activation was dependent solely on components of the activated platelet membrane.
The data from the studies monitoring proteolysis of rP2 and rMZ by prothrombinase offer an explanation for preferential initial cleavage at Arg271. Whereas the rates of cleavage of rP2 and rMZ by prothrombinase assembled on activated platelets were approximately equal, the Km for cleavage at Arg271 in rP2 was approximately 2.4-fold lower than the Km for cleavage at Arg320 in rMZ. Interestingly, the Km determined for rP2 cleavage by prothrombinase on activated platelets (0.82 ± 0.05μM) is very similar to the previously reported Km for prothrombin cleavage by prothrombinase on activated platelets (0.82μM),21 suggesting that prothrombin and rP2 bind similarly to prothrombinase assembled on activated platelets. Although the Km data suggest a slight preference for binding at Arg271, they do not explain the cleavage pattern observed when prothrombin is the substrate, as at physiologic concentrations, substantial initial cleavage would still be anticipated at Arg320. The conclusion that prothrombinase assembled on activated platelets preferentially binds prothrombin to facilitate initial cleavage mainly at Arg271 was most dramatically supported by the Arg271 titration data, as only 50% of initial cleavage occurred at Arg320 when the ratio of the available sites was as high as 5:1 (Arg320/Arg271). At a cleavage site ratio of 1:1, approximately 90% to 95% of the initial cleavage of prothrombin by prothrombinase occurs at Arg271. The trace levels of meizothrombin observed in the prothrombin activation time courses in Figure 3 are consistent with these relative rates of Arg271 and Arg320 cleavage. These data strongly demonstrate a preferential binding to allow cleavage at Arg271 when both cleavage sites are available within the same molecule of prothrombin. Together with the data obtained from independent titrations of rP2 and rMZ, they also imply that this situation is not mimicked precisely when only one cleavage site is available. Not only is cleavage at Arg271 the first step in prothrombin activation on the activated platelet surface, but it also appears to be the rate-limiting step, as the prethrombin-2 that is generated is rapidly converted to α-thrombin with little of the intermediate accumulating at any stage of the reaction.
As the identification of prethrombin-2 in these experiments was based on its electrophoretic mobility, one must consider that the α-thrombin produced during the reactions may cleave prothrombin at Arg284 to produce prethrombin-2 des 1-13,38 which is similar in size to prethrombin-2. To obviate this potential cleavage, DAPA, an inhibitor of α-thrombin, was included in all reaction mixtures. Under these conditions, the α-thrombin-catalyzed cleavage of prothrombin at Arg155 to form prethrombin-1 was not observed, indicating that the DAPA was effectively inhibiting the α-thrombin generated during the reactions. Therefore, it seems unlikely that the band assigned as prethrombin-2 is the result of α-thrombin-catalyzed cleavage of prothrombin at Arg284.
The pathway of prothrombin cleavage on the activated platelet surface is in marked contrast to that observed with PC/PS vesicles containing 25% PS. These vesicles are commonly used as a model phospholipid surface, as this concentration of PS has been shown to support maximal prothrombinase activity.39 However, thrombin-activated platelets appear to express less than 8% PS on their surface, when quantified by lactadherin and annexin V binding to exposed PS.40 Substantial data also indicate that assembly of prothrombinase on the surface of activated platelets versus PC/PS vesicles is quite different. Unique, saturable binding sites exist on platelets for both factors Va and Xa.19 Although factor Xa will bind to PC/PS vesicles independently of factor Va,41,42 it will not bind to platelets in the absence of platelet-bound factor Va.21 The mutational studies of Larson et al,43 which used phospholipid vesicles that more closely resembled the total lipid content of platelets, suggest that the binding of factor Xa to these 2 membrane surfaces involves different interactions, as γ-carboxyglutamic acid residues 16, 29, and 32 of factor Xa appear to have significantly less involvement in binding to activated platelets than to phospholipid vesicles. In addition, data have suggested that a component of the platelet membrane, other than exposed PS, is necessary for binding of factor Xa to activated platelets because factor Va and factor Xa binding is differentially regulated by the level of platelet activation.19 More compelling perhaps is the observation of distinct platelet subpopulations, one that binds both factors Va and Xa and another that supports only factor Va binding.19,22
Although it is not yet clear what structural aspects of prothrombinase assembled on the surface of activated platelets promote initial cleavage at Arg271, the generation of prethrombin-2 as an intermediate is consistent with data obtained from studies assessing prothrombin activation in whole blood or platelet-rich plasma. Using an active site-blotting detection system, Tans et al44 observed little or no meizothrombin generation when clotting was initiated in plasma containing collagen-stimulated platelets and thus proposed that prothrombin activation might occur through the prethrombin-2 pathway. Rand et al45 observed substantial prethrombin-2 accumulation in a whole blood clotting system, and Undas et al observed similar levels of this intermediate in their microvascular injury model.46 In both studies, much higher concentrations of prethrombin-2 accumulated (> 0.4μM45) than were observed in our study, although the difference may be the result of the increased complexity of the whole blood systems. As those systems contain other cells capable of supporting prothrombinase assembly and function (ie, monocytes, lymphocytes, and neutrophils),21 the prethrombin-2 accumulation observed may be the result of a reduced rate of its conversion to α-thrombin on one or more of these cell surfaces. Consistent with this possibility, prothrombinase bound to lymphocytes has been found to be significantly less active than that bound to platelets.21 Alternatively, when nonanticoagulated whole blood was clotted by exposure to glass, Bovill et al47 demonstrated generation of meizothrombin, using an active site-labeling approach, which precluded identification of prethrombin-2. However, the low levels of meizothrombin generated may suggest that formation of meizothrombin as a prothrombin activation intermediate in their system is minor.
As prethrombin-2 is an inactive precursor of α-thrombin,7 the physiologic significance of its generation may lie in comparing the substrate specificity of meizothrombin with that of α-thrombin. Meizothrombin is significantly less efficient (∼ 1%) than α-thrombin in effecting platelet activation, fibrinogen cleavage, and factor V activation.48 Rather, meizothrombin expresses greater anticoagulant activity than α-thrombin, as meizothrombin effects a 6-fold to 7-fold increase in the rate of protein C activation in the presence of thrombomodulin and phospholipid.49 Interestingly, studies have suggested that meizothrombin is formed as a prothrombin activation intermediate on the surface of endothelial cells,49 expressing thrombomodulin.
It is interesting to hypothesize that in the initial stages of clot formation, when activated platelets are the site of prothrombin activation, promotion of the procoagulant response would be preferred and activation through prethrombin-2, which does not share the anticoagulant properties of meizothrombin, would be preferred. When sufficient thrombin, or platelet-released cytokines, accumulate to activate endothelial cells, down-regulation of the procoagulant response becomes more important and prothrombinase assembled on the activated endothelium generates meizothrombin, which interacts with thrombomodulin to activate protein C.
In conclusion, the studies detailed here are the first to demonstrate that prothrombin activation on the surface of activated platelets occurs through the prethrombin-2 pathway. We hypothesize that use of the prethrombin-2 pathway serves to optimize the procoagulant activity expressed by activated platelets, by limiting the anticoagulant functions of the alternate intermediate meizothrombin.
Presented in part in abstract form at the International Society on Thrombosis and Haemostasis XXII Congress, Boston, MA, July 13, 2009.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Paul Kim and Michael Nesheim for their generous gift of the rP2 and rMZ mutant proteins, Dr Kenneth Mann for his donation of PC/PS vesicles, Dr Beth Bouchard for critical review of the manuscript, and Shirlee Wohl for technical assistance.
This work was supported by the National Institutes of Health (grant P01HL046703, Project 3) (P.B.T.). J.P.W. and L.M.H. were supported by Hemostasis and Thrombosis Program for Academic Trainees (grant T32HL07594; Kenneth Mann). N.M.M. was supported by a Hughes Endeavor for Life Science Excellence and an Undergraduate Research Endeavors Competitive Award (University of Vermont).
J.P.W. is a PhD candidate at the University of Vermont; this work is submitted in partial fulfillment of the requirement for that degree.
National Institutes of Health
Authorship
Contribution: J.P.W. designed and performed experiments, analyzed the results, and wrote the manuscript; J.R.S. performed experiments and wrote the manuscript; N.M.M. and L.M.H. designed and performed experiments and analyzed the results; and P.B.T. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paula B. Tracy, University of Vermont College of Medicine, Biochemistry Department, 89 Beaumont Ave, Burlington, VT 05405; e-mail: ptracy@uvm.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal