Abstract
Prior genome-wide association (GWA) studies have identified 10 susceptibility loci for risk of chronic lymphocytic leukemia (CLL). To identify additional loci, we performed a GWA study in 407 CLL cases (of which 102 had a family history of CLL) and 296 controls. Moreover, given the strong familial risk of CLL, we further subset our GWA analysis to the CLL cases with a family history of CLL to identify loci specific to these familial CLL cases. Our top hits from these analyses were evaluated in an additional sample of 252 familial CLL cases and 965 controls. Using all available data, we identified and confirmed an independent association of 4 single-nucleotide polymorphisms (SNPs) that met genome-wide statistical significance within the IRF8 (interferon regulatory factor 8) gene (combined P values ≤ 3.37 × 10−8), located in the previously identified 16q24.1 locus. Subsetting to familial CLL cases, we identified and confirmed a new locus on chromosome 6p21.3 (combined P value = 6.92 × 10−9). This novel region harbors the HLA-DQA1 and HLA-DRB5 genes. Finally, we evaluated the 10 previously reported SNPs in the overall sample and replicated 8 of them. Our findings support the hypothesis that familial CLL cases have additional genetic variants not seen in sporadic CLL. Additional loci among familial CLL cases may be identified through larger studies.
Introduction
Chronic lymphocytic leukemia (CLL) is a hematologic malignancy, with ∼ 15 000 individuals diagnosed annually in the United States. Current evidence strongly supports a genetic component for CLL etiology.1 To date, 3 genome-wide association (GWA) studies of CLL have been conducted. The initial GWA study of 505 CLL cases and 1438 controls from the United Kingdom genotyped 299 983 single-nucleotide polymorphisms (SNPs) and identified 6 loci (2q13, 2q37.1, 6p25, 11q24, 15q23, and 19q13) associated with CLL risk.2 We replicated 5 of these 6 loci in 407 CLL cases and 296 controls.3 A follow-up analysis of the United Kingdom GWA study4 identified 4 additional susceptibility loci (2q37.3, 8q24.21, 15q21.3, and 16q24.1), bringing the total susceptibility loci to 10. The remaining 2 GWA studies were conducted in a sample from the San Francisco Bay area, and there was a 77% overlap of samples between the 2 studies: one used a pooled DNA genotyping strategy on 148 CLL cases and 592 controls5 and the other genotyped 339 528 SNPs on 211 CLL cases and 750 controls.6 While no novel CLL susceptibility loci were identified from these 2 studies, they provided additional support for the previously identified 6p25 and 11q24 regions.
Given that the previously identified loci account for ∼ 10% of the genetic risk of CLL and that CLL has one of the highest familial risks among hematologic malignancies (on the order of 8-fold increased risk7 ), we undertook a GWA study to identify additional CLL susceptibility loci using the Affymetrix 6.0 platform, which has greater genomic coverage than those previously used. Further, we enriched our case group with familial CLL cases to identify novel loci specific to familial CLL. We also evaluated the 10 recently reported CLL loci in our sample. Finally, as preliminary data, we evaluated the association of CLL susceptibility loci with risk of monoclonal B-cell lymphocytosis (MBL), a known precursor condition to CLL,8 using MBL samples ascertained from our CLL families.
Methods
GWA study sample
Peripheral blood samples were obtained from 2 ongoing studies: the Genetic Epidemiology of CLL (GEC) Consortium and the Mayo Clinic non-Hodgkin lymphoma (NHL)/CLL study. The GEC consortium is a collaboration of researchers from 7 institutions with the overall aim of investigating the genetic basis of CLL through the collection of CLL families (ie, families with 2 or more relatives with CLL). A total of 110 Caucasian CLL patients from 110 families were available at the time of genotyping. These families were found through Duke University, the Mayo Clinic, the University of Texas M. D. Anderson Cancer Center, the National Cancer Institute (NCI), the University of Minnesota/Minneapolis Veterans Administration Medical Center, the University of California-San Diego, and the University of Utah. The Mayo Clinic NHL/CLL case-control study is an ongoing, clinic-based study being conducted in Rochester, MN.9 Briefly, newly diagnosed NHL/CLL patients 20 years of age or older, HIV negative, and residents of the midwestern United States at the time of diagnosis are enrolled. Clinic-based controls are ascertained from patients visiting the general internal medicine clinic. Eligibility requirements include age 20 years or older and a resident of Minnesota, Iowa, or Wisconsin. Patients are excluded if they have prior diagnoses of lymphoma, leukemia, or HIV infection. From this study, genotype data were available from 328 Caucasian CLL cases and 328 controls. The diagnosis of all CLL cases across both studies were reviewed and confirmed by a hematopathologist and classified according to World Health Organization criteria.10
Replication study sample
In the replication stage, an additional 96 new Caucasian CLL families from the GEC Consortium and the University of Iowa were identified since the GWA study. From these, we selected 151 CLL cases, 28 MBL individuals, and 197 unaffected family members. Further, we selected relatives of the 102 CLL cases who were successfully genotyped in the GWA study. A total of 101 CLL cases, 32 MBL individuals, and 270 unaffected relatives were selected. In these families, relatives were screened for MBL in accordance with our previous work.11 From these 198 families, we had a total of 252 CLL cases, 60 MBLs, and 467 controls. We also included 500 age- and sex-frequency–matched independent Caucasian control samples collected from the Mayo Clinic Biobank, which is an institutional resource for biological specimens, risk-factor data, and clinical data on participants age 18 years or older. Participants are volunteers or patients prescheduled for a medical examination in the divisions of community internal medicine, family medicine, or general internal medicine (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Ethics
All data collection from study participants was approved by the respective institutional review boards of all participating centers, and all participants gave written informed consent in accordance with the Declaration of Helsinki.
Genotyping and quality control
For the GWA study, we genotyped 438 CLL cases (110 familial CLL and 328 sporadic CLL) and 328 controls using the Affymetrix 6.0 SNP array; all samples were also genotyped on an Illumina BeadXpress, and 84 SNPs overlapped with the Affymetrix 6.0 platform. Concordance of genotypes across these 2 platforms was > 99.7%. Within the Affymetrix 6.0 chip, there were 2906 duplicate SNPs, among which we observed > 99.7% concordance. Rigorous quality-control measures were implemented, such as excluding individuals with call rates < 95% (n = 30), who were related (n = 3), who had sex discrepancy issues (n = 2), or who had poor concordance among duplicate SNPs (n = 6). Multidimensional scaling within PLINK v1.07 software was used as an additional check for the presence of population stratification, and no evidence was observed. Cluster plots of SNPs that were top hits were reviewed. Twenty-two samples (14 controls and 8 cases) had no genotype calls. SNPs were dropped if call rates were < 95%, not mapped to a chromosome, had Hardy-Weinberg equilibrium P values < 10−10 in either the cases or the controls, or poor concordance among duplicates. We also excluded SNPs if call rates differed by 5% or more between cases and controls. For the replication study, we genotyped 252 CLL cases and 967 controls on a custom Illumina BeadXpress oligo pool assay as part of a larger genotyping project. SNPs and subjects were excluded if call rates were < 90%. Concordance among duplicate samples was > 99.99%.
Statistical analyses
Tests for Hardy-Weinberg equilibrium were done using either the Pearson goodness-of-fit test or the Fisher exact test. Tests for association were done using the Cochran-Armitage trend test and, where appropriate, familial relationships were accounted for in the statistical analyses by adjusting the variance of the test for the covariance of related subjects.12,13 We used unconditional logistic regression to estimate odds ratios and corresponding 95% confidence intervals for CLL risk. The analyses with independent samples consisted of unrelated cases and controls; 1 CLL case was selected from each family. Imputed genotypes and recombination rates were calculated using MACH 1.0 software14 and HapMap CEU (Utah residents with ancestry from northern and western Europe) samples as the reference data. Conditional analyses were conducted using the discovery sample and logistic regression. Tests for association between genotypes and mRNA expression were done using linear regression and publicly available expression and genotype data from the 60 unrelated CEU HapMap samples. Linkage disequilibrium (r2) values between SNPs were calculated by Haploview15 using genotypes from HapMap CEU data or from the unrelated controls from the GWA study.
Results
We genotyped 438 CLL cases from the United States, with 110 (25%) cases selected from high-risk CLL families (ie, families with confirmed multiple members with CLL) and the remaining 328 CLL cases and 328 controls drawn from the Mayo Clinic case-control study of non-Hodgkin lymphoma. Of the 766 samples selected for genotyping, 703 subjects (296 controls, 102 familial CLL cases, and 305 sporadic CLL cases) passed quality control. Of the 934 968 SNPs genotyped, 827 777 autosomal SNPs passed quality control. Mean call rates of the final 703 samples was 99%. Genotype concordance among duplicates was > 99.7%. The Cochran-Armitage trend test was used to compare genotype frequencies between cases and controls. There was no evidence of population stratification (inflation factor λ = 1.003 among the 90% least significant SNPs; supplemental Figure 1).
Among all 407 CLL cases and 296 controls, we observed evidence of association with CLL risk and 7 SNPs with P values < 10−5 (supplemental Table 2). Four of these SNPs were in strong linkage disequilibrium (LD) with each other (all pairwise r2 =0.99 based on our controls) and were located in the IRF8 (interferon regulatory factor 8) gene on 16q24, which has recently been identified as a CLL susceptibility locus.4 Given our hypothesis that familial cases have a stronger genetic component than sporadic cases, we then performed subset analyses comparing genotype frequencies between the 102 familial CLL cases and 296 controls. We observed an additional 39 SNPs with P values < 10−5 that were not identified in our full CLL sample analyses (supplemental Table 2). Ten of these SNPs reached the genome-wide significance threshold.
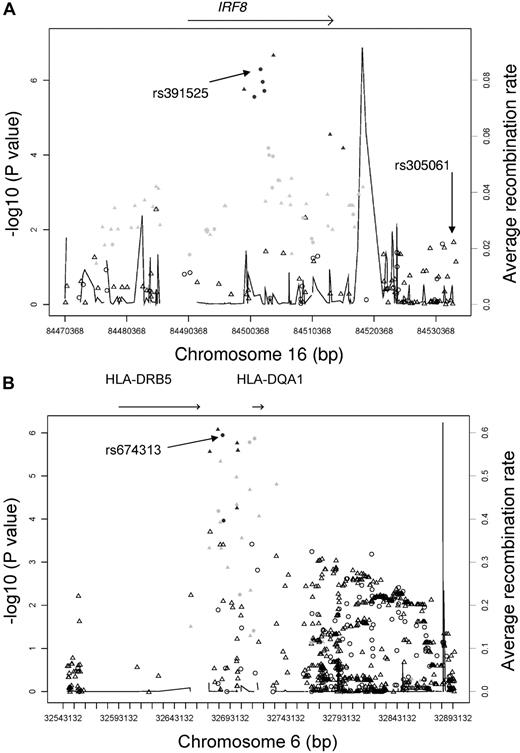
We genotyped these 46 top SNPs (supplemental Table 2) plus SNPs near these top hits with P values < 10−4 in a replication sample. The replication stage consisted of 252 familial CLL cases and 965 controls. We used the trend test to compare genotype frequencies between cases and controls; this test accounted for the familial relationship among related subjects.12,13 Of the 7 top hits identified from the full CLL sample GWA analyses, 3 did not replicate, whereas all 4 SNPs from IRF8 (rs305077, rs391525, rs2292982, and rs2292980) had clear evidence of replication, with P values < .0006 and effect sizes in the same direction as that in the discovery stage (Table 1). The combined analyses of CLL cases and controls from both stages reached significance, with P values = 3.16 × 10−9 to 3.37 × 10−8. These results also held if only independent samples (ie, only unrelated cases and control samples) were included in the combined analyses (Table 1). These SNPs are intronic within the IRF8 gene and are independent of the previously published4 SNP for CLL risk (rs305061, all pairwise r2 = 0 based on HapMap). Results of conditional analyses of our top IRF8 rs391525 SNP with rs305065 (a SNP typed in our GWA that was in high LD with rs305061) supported that these 2 SNPs independently tag different predisposing variants (adjusted P value < .0001). We imputed genotypes in our full discovery sample and evaluated those in or near the IRF8 gene, including the previously identified rs305061. One imputed intronic SNP (rs11649318) had greater association than that of our observed SNPs (Figure 1A) and was correlated (r2 = 0.8 based on HapMap) with our top IRF8 rs391525 SNP. We next evaluated the association of these IRF8 SNPs with IRF8 mRNA expression from lymphocytes using publicly available data. All 4 of the typed SNPs were significantly associated with mRNA expression (supplemental Figure 2); specifically, all showed increased IRF8 expression with 2 copies of the major allele, which we found to increase CLL risk (Table 1). Our results agree with a previous study reporting that IRF8 expression is associated with CLL.16
Associations of CLL risk with replicated SNPs among all CLL cases
| Locus . | Nearest gene . | SNP . | Position . | Risk allele . | Stage . | Number of subjects . | Minor allele frequency . | OR . | 95% CI . | P . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | Cases . | Controls . | ||||||||||
| 16q24.1 | IRF8 | rs305077 | 84500967 | C | Discovery | 407 | 296 | 0.26 | 0.38 | 0.57 | 0.45 | 0.73 | 2.75 × 10−6 |
| Replication | 252 | 965 | 0.25 | 0.33 | 0.68 | 0.55 | 0.85 | 5.62 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.26 | 0.34 | 0.66 | 0.57 | 0.77 | 3.37 × 10−8 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.57 | 0.48 | 0.69 | 5.91 × 10−10 | |||||
| rs391525 | 84501940 | G | Discovery | 407 | 296 | 0.25 | 0.38 | 0.54 | 0.43 | 0.69 | 5.00 × 10−7 | ||
| Replication | 252 | 965 | 0.25 | 0.33 | 0.67 | 0.53 | 0.84 | 2.80 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.25 | 0.34 | 0.64 | 0.55 | 0.74 | 3.16 × 10−9 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.55 | 0.46 | 0.66 | 6.94 × 10−11 | |||||
| rs2292982 | 84502324 | G | Discovery | 407 | 296 | 0.26 | 0.38 | 0.56 | 0.44 | 0.71 | 1.08 × 10−6 | ||
| Replication | 252 | 965 | 0.25 | 0.33 | 0.66 | 0.52 | 0.82 | 1.79 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.25 | 0.34 | 0.65 | 0.56 | 0.75 | 6.48 × 10−9 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.56 | 0.47 | 0.68 | 2.13 × 10−10 | |||||
| rs2292980 | 84502577 | C | Discovery | 407 | 296 | 0.26 | 0.38 | 0.57 | 0.45 | 0.72 | 1.90 × 10−6 | ||
| Replication | 252 | 965 | 0.25 | 0.33 | 0.67 | 0.53 | 0.84 | 3.02 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.25 | 0.34 | 0.66 | 0.56 | 0.76 | 1.89 × 10−8 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.57 | 0.48 | 0.69 | 6.34 × 10−10 | |||||
| Locus . | Nearest gene . | SNP . | Position . | Risk allele . | Stage . | Number of subjects . | Minor allele frequency . | OR . | 95% CI . | P . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | Cases . | Controls . | ||||||||||
| 16q24.1 | IRF8 | rs305077 | 84500967 | C | Discovery | 407 | 296 | 0.26 | 0.38 | 0.57 | 0.45 | 0.73 | 2.75 × 10−6 |
| Replication | 252 | 965 | 0.25 | 0.33 | 0.68 | 0.55 | 0.85 | 5.62 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.26 | 0.34 | 0.66 | 0.57 | 0.77 | 3.37 × 10−8 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.57 | 0.48 | 0.69 | 5.91 × 10−10 | |||||
| rs391525 | 84501940 | G | Discovery | 407 | 296 | 0.25 | 0.38 | 0.54 | 0.43 | 0.69 | 5.00 × 10−7 | ||
| Replication | 252 | 965 | 0.25 | 0.33 | 0.67 | 0.53 | 0.84 | 2.80 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.25 | 0.34 | 0.64 | 0.55 | 0.74 | 3.16 × 10−9 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.55 | 0.46 | 0.66 | 6.94 × 10−11 | |||||
| rs2292982 | 84502324 | G | Discovery | 407 | 296 | 0.26 | 0.38 | 0.56 | 0.44 | 0.71 | 1.08 × 10−6 | ||
| Replication | 252 | 965 | 0.25 | 0.33 | 0.66 | 0.52 | 0.82 | 1.79 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.25 | 0.34 | 0.65 | 0.56 | 0.75 | 6.48 × 10−9 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.56 | 0.47 | 0.68 | 2.13 × 10−10 | |||||
| rs2292980 | 84502577 | C | Discovery | 407 | 296 | 0.26 | 0.38 | 0.57 | 0.45 | 0.72 | 1.90 × 10−6 | ||
| Replication | 252 | 965 | 0.25 | 0.33 | 0.67 | 0.53 | 0.84 | 3.02 × 10−4 | |||||
| Combined* | 659 | 1261 | 0.25 | 0.34 | 0.66 | 0.56 | 0.76 | 1.89 × 10−8 | |||||
| Combined independent† | 503 | 794 | 0.25 | 0.37 | 0.57 | 0.48 | 0.69 | 6.34 × 10−10 | |||||
OR indicates odds ratio; and CI, confidence interval.
All cases and controls (related and unrelated) from both stages.
All unrelated cases and controls from both stages.
Trend test P values (as −log10values; leftyaxis) are shown for SNPs analyzed in GWA study. Recombination rate is shown across the region with the solid line (right y axis). Triangles indicate imputed SNPs and circles indicate observed SNPs. Coloring (black, light gray, white) shows the extent of LD between each SNP and rs391525. Black: r2 ≥ 0.75; light gray: 0.25 ≤ r2 < 0.75; white: r2 < 0.25. (A) Association results of the 16q24 locus across a 60-kb region between all discovery CLL cases and controls. (B) Association results of the 6q21.3 locus between the discovery familial CLL cases and controls.
Trend test P values (as −log10values; leftyaxis) are shown for SNPs analyzed in GWA study. Recombination rate is shown across the region with the solid line (right y axis). Triangles indicate imputed SNPs and circles indicate observed SNPs. Coloring (black, light gray, white) shows the extent of LD between each SNP and rs391525. Black: r2 ≥ 0.75; light gray: 0.25 ≤ r2 < 0.75; white: r2 < 0.25. (A) Association results of the 16q24 locus across a 60-kb region between all discovery CLL cases and controls. (B) Association results of the 6q21.3 locus between the discovery familial CLL cases and controls.
Of the top SNPs identified from the familial CLL GWA analyses, 3 SNPs (rs674313, rs9272219, and rs9272535) had clear evidence of replication, with P < .0009 and effect sizes in the same direction as that in the discovery sample (Table 2). Results from additional SNPs, rs615672 and rs502771, that are in LD (r2 > 0.6 based on our controls) with rs674313 also support these findings (Table 2). The combined analyses of all 354 familial CLL cases and 1261 controls from both stages for these 3 SNPs had significant associations (P = 6.92 × 10−9 to 1.84 × 10−7). Conditional analyses of these 5 SNPs showed that only our most significant SNP (rs674313) remained associated with CLL risk (adjusted P = 0.01), suggesting that these SNPs tag the same region. The effect of these SNPs was attenuated among our sporadic CLL cases versus controls (supplemental Table 3). These SNPs are located within the 6p21.32 region, which harbors the HLA-DQA1 and HLA-DRB5 genes. We evaluated the imputed SNPs in this region and found 1 imputed SNP (rs602875) with greater association (P = 8.1 × 10−7) than that observed (Figure 1B); this SNP was completely correlated (r2 = 1, based on HapMap) with rs674313, our top SNP in the region.
Associations of CLL risk with replicated SNPs among familial CLL cases
| Locus . | Nearest gene . | SNP . | Position . | Risk allele . | Stage . | Number of subjects . | Minor allele frequency . | OR . | 95% CI . | P . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | Cases . | Controls . | ||||||||||
| 6p21.3 | HLA-DRB5 | rs615672 | 32682149 | C | Discovery | 102 | 296 | 0.50 | 0.34 | 1.95 | 1.40 | 2.73 | 6.42 × 10−5 |
| Replication | 252 | 965 | 0.47 | 0.39 | 1.31 | 1.09 | 1.57 | 2.85 × 10−3 | |||||
| Combined* | 354 | 1261 | 0.48 | 0.38 | 1.42 | 1.22 | 1.67 | 1.29 × 10−5 | |||||
| Combined independent† | 198 | 794 | 0.47 | 0.36 | 1.46 | 1.19 | 1.81 | 3.40 × 10−4 | |||||
| rs674313 | 32686060 | T | Discovery | 102 | 296 | 0.44 | 0.26 | 2.40 | 1.67 | 3.45 | 1.12 × 10−6 | ||
| Replication | 252 | 965 | 0.35 | 0.26 | 1.50 | 1.22 | 1.84 | 6.90 × 10−5 | |||||
| Combined* | 354 | 1261 | 0.38 | 0.26 | 1.69 | 1.41 | 2.01 | 6.92 × 10−9 | |||||
| Combined independent† | 198 | 794 | 0.38 | 0.25 | 1.87 | 1.47 | 2.38 | 1.98 × 10−7 | |||||
| rs502771 | 32686948 | C | Discovery | 102 | 296 | 0.43 | 0.28 | 1.93 | 1.37 | 2.70 | 1.07 × 10−4 | ||
| Replication | 252 | 965 | 0.38 | 0.28 | 1.51 | 1.23 | 1.84 | 3.28 × 10−5 | |||||
| Combined* | 354 | 1261 | 0.40 | 0.28 | 1.61 | 1.36 | 1.91 | 5.58 × 10−8 | |||||
| Combined independent† | 198 | 794 | 0.39 | 0.27 | 1.68 | 1.33 | 2.11 | 8.17 × 10−6 | |||||
| HLA-DQA1 | rs9272219 | 32710247 | T | Discovery | 102 | 296 | 0.44 | 0.26 | 2.32 | 1.62 | 3.30 | 1.65 × 10−6 | |
| Replication | 252 | 965 | 0.35 | 0.27 | 1.40 | 1.14 | 1.71 | 9.46 × 10−4 | |||||
| Combined* | 354 | 1261 | 0.38 | 0.27 | 1.59 | 1.34 | 1.90 | 1.84 × 10−7 | |||||
| Combined independent† | 198 | 794 | 0.38 | 0.27 | 1.71 | 1.36 | 2.17 | 4.55 × 10−6 | |||||
| rs9272535 | 32714734 | A | Discovery | 102 | 296 | 0.44 | 0.26 | 2.33 | 1.64 | 3.32 | 1.33 × 10−6 | ||
| Replication | 252 | 965 | 0.35 | 0.27 | 1.42 | 1.15 | 1.74 | 5.96 × 10−4 | |||||
| Combined* | 354 | 1261 | 0.38 | 0.27 | 1.61 | 1.35 | 1.92 | 9.31 × 10−8 | |||||
| Combined independent† | 198 | 794 | 0.38 | 0.26 | 1.75 | 1.38 | 2.21 | 2.25 × 10−6 | |||||
| Locus . | Nearest gene . | SNP . | Position . | Risk allele . | Stage . | Number of subjects . | Minor allele frequency . | OR . | 95% CI . | P . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | Cases . | Controls . | ||||||||||
| 6p21.3 | HLA-DRB5 | rs615672 | 32682149 | C | Discovery | 102 | 296 | 0.50 | 0.34 | 1.95 | 1.40 | 2.73 | 6.42 × 10−5 |
| Replication | 252 | 965 | 0.47 | 0.39 | 1.31 | 1.09 | 1.57 | 2.85 × 10−3 | |||||
| Combined* | 354 | 1261 | 0.48 | 0.38 | 1.42 | 1.22 | 1.67 | 1.29 × 10−5 | |||||
| Combined independent† | 198 | 794 | 0.47 | 0.36 | 1.46 | 1.19 | 1.81 | 3.40 × 10−4 | |||||
| rs674313 | 32686060 | T | Discovery | 102 | 296 | 0.44 | 0.26 | 2.40 | 1.67 | 3.45 | 1.12 × 10−6 | ||
| Replication | 252 | 965 | 0.35 | 0.26 | 1.50 | 1.22 | 1.84 | 6.90 × 10−5 | |||||
| Combined* | 354 | 1261 | 0.38 | 0.26 | 1.69 | 1.41 | 2.01 | 6.92 × 10−9 | |||||
| Combined independent† | 198 | 794 | 0.38 | 0.25 | 1.87 | 1.47 | 2.38 | 1.98 × 10−7 | |||||
| rs502771 | 32686948 | C | Discovery | 102 | 296 | 0.43 | 0.28 | 1.93 | 1.37 | 2.70 | 1.07 × 10−4 | ||
| Replication | 252 | 965 | 0.38 | 0.28 | 1.51 | 1.23 | 1.84 | 3.28 × 10−5 | |||||
| Combined* | 354 | 1261 | 0.40 | 0.28 | 1.61 | 1.36 | 1.91 | 5.58 × 10−8 | |||||
| Combined independent† | 198 | 794 | 0.39 | 0.27 | 1.68 | 1.33 | 2.11 | 8.17 × 10−6 | |||||
| HLA-DQA1 | rs9272219 | 32710247 | T | Discovery | 102 | 296 | 0.44 | 0.26 | 2.32 | 1.62 | 3.30 | 1.65 × 10−6 | |
| Replication | 252 | 965 | 0.35 | 0.27 | 1.40 | 1.14 | 1.71 | 9.46 × 10−4 | |||||
| Combined* | 354 | 1261 | 0.38 | 0.27 | 1.59 | 1.34 | 1.90 | 1.84 × 10−7 | |||||
| Combined independent† | 198 | 794 | 0.38 | 0.27 | 1.71 | 1.36 | 2.17 | 4.55 × 10−6 | |||||
| rs9272535 | 32714734 | A | Discovery | 102 | 296 | 0.44 | 0.26 | 2.33 | 1.64 | 3.32 | 1.33 × 10−6 | ||
| Replication | 252 | 965 | 0.35 | 0.27 | 1.42 | 1.15 | 1.74 | 5.96 × 10−4 | |||||
| Combined* | 354 | 1261 | 0.38 | 0.27 | 1.61 | 1.35 | 1.92 | 9.31 × 10−8 | |||||
| Combined independent† | 198 | 794 | 0.38 | 0.26 | 1.75 | 1.38 | 2.21 | 2.25 × 10−6 | |||||
OR indicates odds ratio; and CI, confidence interval.
All available familial CLL cases from 198 CLL families and controls from both stages.
All unrelated familial CLL cases from 198 CLL families and controls from both stages.
Table 3 reports associations for the 10 previously reported CLL susceptibility loci.2,4 Earlier, we reported results on the first 6 discovered loci (2q13, 2q37.1, 6p25, 11q24, 15q23, and 19q13) using either observed or imputed data from our discovery sample.3 With the additional data from the replication stage, we still found that 5 of the 6 loci remained significant, with locus 19q13 still nonsignificant. For the 4 recently reported loci (2q37.3, 8q24.21, 15q21.3, and 16q24.1), we found all but locus 15q21.3 to be associated with CLL with either the exact SNP or the best tagged SNP based on data from our discovery sample.
OR and 95% CIs for previously reported CLL susceptibility loci
| Previously reported . | Current study SNP . | (r2)* . | Position . | Risk Allele . | Number of subjects . | Minor allele frequency . | OR . | 95% CI . | P . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | SNP . | Same or best tag . | Cases . | Controls . | Cases . | Controls . | |||||||
| 2q13 | rs17483466 | rs17483466 | 111513929 | G | 659 | 1261 | 0.26 | 0.22 | 1.27 | 1.09 | 1.48 | 2.03 × 10−3 | |
| 2q37.1 | rs13397985 | rs13397985 | 230799467 | G | 659 | 1261 | 0.26 | 0.20 | 1.33 | 1.13 | 1.55 | 2.79 × 10−4 | |
| 2q37.3 | rs757978† | rs757978 | 242019774 | T | 407 | 296 | 0.12 | 0.09 | 1.46 | 1.02 | 2.08 | 3.90 × 10−2 | |
| 6p25 | rs872071‡ | rs872071 | 356064 | G | 252 | 965 | 0.58 | 0.52 | 1.30 | 1.06 | 1.59 | 8.72 × 10−3 | |
| rs9378805 | rs9378805 | 362727 | C | 659 | 1261 | 0.57 | 0.49 | 1.38 | 1.20 | 1.58 | 1.59 × 10−6 | ||
| 8q24.21 | rs2456449† | rs1021955 | 0.87 | 128273155 | G | 407 | 296 | 0.45 | 0.37 | 1.37 | 1.10 | 1.70 | 5.24 × 10−3 |
| 11q24 | rs735665§ | rs735665 | 122866607 | T | 407 | 296 | 0.28 | 0.21 | 1.47 | 1.14 | 1.89 | 2.64 × 10−3 | |
| 15q21.3 | rs7169431† | rs7169431 | 54128188 | A | 407 | 296 | 0.12 | 0.09 | 1.33 | 0.94 | 1.87 | 1.07 × 10−1 | |
| 15q23 | rs7176508‡ | rs7176508 | 67806044 | A | 252 | 965 | 0.50 | 0.41 | 1.40 | 1.15 | 1.70 | 4.74 × 10−4 | |
| 16q24.1 | rs305061† | rs305065 | 0.93 | 84531367 | C | 407 | 296 | 0.29 | 0.35 | 0.77 | 0.61 | 0.97 | 2.37 × 10−2 |
| 19q13 | rs11083846‡ | rs11083846 | 51899494 | A | 252 | 965 | 0.21 | 0.22 | 0.92 | 0.72 | 1.17 | 4.98 × 10−1 | |
| Previously reported . | Current study SNP . | (r2)* . | Position . | Risk Allele . | Number of subjects . | Minor allele frequency . | OR . | 95% CI . | P . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | SNP . | Same or best tag . | Cases . | Controls . | Cases . | Controls . | |||||||
| 2q13 | rs17483466 | rs17483466 | 111513929 | G | 659 | 1261 | 0.26 | 0.22 | 1.27 | 1.09 | 1.48 | 2.03 × 10−3 | |
| 2q37.1 | rs13397985 | rs13397985 | 230799467 | G | 659 | 1261 | 0.26 | 0.20 | 1.33 | 1.13 | 1.55 | 2.79 × 10−4 | |
| 2q37.3 | rs757978† | rs757978 | 242019774 | T | 407 | 296 | 0.12 | 0.09 | 1.46 | 1.02 | 2.08 | 3.90 × 10−2 | |
| 6p25 | rs872071‡ | rs872071 | 356064 | G | 252 | 965 | 0.58 | 0.52 | 1.30 | 1.06 | 1.59 | 8.72 × 10−3 | |
| rs9378805 | rs9378805 | 362727 | C | 659 | 1261 | 0.57 | 0.49 | 1.38 | 1.20 | 1.58 | 1.59 × 10−6 | ||
| 8q24.21 | rs2456449† | rs1021955 | 0.87 | 128273155 | G | 407 | 296 | 0.45 | 0.37 | 1.37 | 1.10 | 1.70 | 5.24 × 10−3 |
| 11q24 | rs735665§ | rs735665 | 122866607 | T | 407 | 296 | 0.28 | 0.21 | 1.47 | 1.14 | 1.89 | 2.64 × 10−3 | |
| 15q21.3 | rs7169431† | rs7169431 | 54128188 | A | 407 | 296 | 0.12 | 0.09 | 1.33 | 0.94 | 1.87 | 1.07 × 10−1 | |
| 15q23 | rs7176508‡ | rs7176508 | 67806044 | A | 252 | 965 | 0.50 | 0.41 | 1.40 | 1.15 | 1.70 | 4.74 × 10−4 | |
| 16q24.1 | rs305061† | rs305065 | 0.93 | 84531367 | C | 407 | 296 | 0.29 | 0.35 | 0.77 | 0.61 | 0.97 | 2.37 × 10−2 |
| 19q13 | rs11083846‡ | rs11083846 | 51899494 | A | 252 | 965 | 0.21 | 0.22 | 0.92 | 0.72 | 1.17 | 4.98 × 10−1 | |
OR indicates odds ratio; and CI, confidence interval.
r2 is between published SNP and most correlated SNP in study based on HapMap CEU.
SNPs genotyped only in discovery cohort.
SNPs genotyped only in replication cohort.
SNP failed genotyping in replication cohort.
Given that MBL is a precursor to CLL, we analyzed the CLL-susceptibility SNPs with MBL risk. We genotyped 60 MBL individuals ascertained from our high-risk CLL families and then evaluated associations with the 6 initially reported susceptibility loci, as well as the HLA and IRF8 loci. We found significant associations (P < .05) within the 2q37.1 and 6p21.3 regions (Table 4) and suggestive associations within the 2q13, 15q23, and 16q24.1 regions. The effect sizes of these findings were comparable to and in the same direction as those from our CLL findings.
Associations of MBL risk with replicated SNPs in 60 familial MBL cases and 965 controls
| Locus . | Nearest gene . | SNP . | Position . | Allele frequency . | OR . | 95% CI . | P . | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | ||||||||
| 2q13 | ACOXL | rs17483466 | 111513929 | 0.30 | 0.22 | 1.49 | 0.99 | 2.24 | 0.074 |
| 2q37.1 | SP140 | rs13397985 | 230799467 | 0.30 | 0.21 | 1.56 | 1.05 | 2.31 | 0.041 |
| 6p25 | IRF4 | rs872071 | 356064 | 0.47 | 0.48 | 0.97 | 0.66 | 1.42 | 0.878 |
| IRF4 | rs9378805 | 362727 | 0.50 | 0.49 | 1.03 | 0.71 | 1.50 | 0.877 | |
| 6p21.3 | HLA-DRB5 | rs615672 | 32682149 | 0.46 | 0.39 | 1.27 | 0.91 | 1.78 | 0.198 |
| HLA-DRB5 | rs674313 | 32686060 | 0.37 | 0.26 | 1.57 | 1.08 | 2.29 | 0.028 | |
| HLA-DRB5 | rs502771 | 32686948 | 0.38 | 0.28 | 1.53 | 1.06 | 2.21 | 0.037 | |
| HLA-DQA1 | rs9272219 | 32710247 | 0.38 | 0.27 | 1.61 | 1.11 | 2.34 | 0.020 | |
| HLA-DQA1 | rs9272535 | 32714734 | 0.38 | 0.27 | 1.62 | 1.12 | 2.35 | 0.017 | |
| 11q24 | SF3A3P2 | rs12799226* | 122895435 | 0.28 | 0.25 | 1.15 | 0.77 | 1.72 | 0.533 |
| 15q23 | rs7176508 | 67806044 | 0.48 | 0.41 | 1.29 | 0.90 | 1.85 | 0.204 | |
| 16q24.1 | IRF8 | rs305077 | 84500967 | 0.25 | 0.33 | 0.68 | 0.44 | 1.03 | 0.094 |
| IRF8 | rs391525 | 84501940 | 0.25 | 0.33 | 0.67 | 0.44 | 1.03 | 0.090 | |
| IRF8 | rs2292982 | 84502324 | 0.25 | 0.33 | 0.67 | 0.44 | 1.03 | 0.091 | |
| IRF8 | rs2292980 | 84502577 | 0.25 | 0.33 | 0.68 | 0.45 | 1.04 | 0.100 | |
| 19q13 | rs11083846 | 51899494 | 0.24 | 0.22 | 1.10 | 0.71 | 1.69 | 0.696 | |
| Locus . | Nearest gene . | SNP . | Position . | Allele frequency . | OR . | 95% CI . | P . | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | ||||||||
| 2q13 | ACOXL | rs17483466 | 111513929 | 0.30 | 0.22 | 1.49 | 0.99 | 2.24 | 0.074 |
| 2q37.1 | SP140 | rs13397985 | 230799467 | 0.30 | 0.21 | 1.56 | 1.05 | 2.31 | 0.041 |
| 6p25 | IRF4 | rs872071 | 356064 | 0.47 | 0.48 | 0.97 | 0.66 | 1.42 | 0.878 |
| IRF4 | rs9378805 | 362727 | 0.50 | 0.49 | 1.03 | 0.71 | 1.50 | 0.877 | |
| 6p21.3 | HLA-DRB5 | rs615672 | 32682149 | 0.46 | 0.39 | 1.27 | 0.91 | 1.78 | 0.198 |
| HLA-DRB5 | rs674313 | 32686060 | 0.37 | 0.26 | 1.57 | 1.08 | 2.29 | 0.028 | |
| HLA-DRB5 | rs502771 | 32686948 | 0.38 | 0.28 | 1.53 | 1.06 | 2.21 | 0.037 | |
| HLA-DQA1 | rs9272219 | 32710247 | 0.38 | 0.27 | 1.61 | 1.11 | 2.34 | 0.020 | |
| HLA-DQA1 | rs9272535 | 32714734 | 0.38 | 0.27 | 1.62 | 1.12 | 2.35 | 0.017 | |
| 11q24 | SF3A3P2 | rs12799226* | 122895435 | 0.28 | 0.25 | 1.15 | 0.77 | 1.72 | 0.533 |
| 15q23 | rs7176508 | 67806044 | 0.48 | 0.41 | 1.29 | 0.90 | 1.85 | 0.204 | |
| 16q24.1 | IRF8 | rs305077 | 84500967 | 0.25 | 0.33 | 0.68 | 0.44 | 1.03 | 0.094 |
| IRF8 | rs391525 | 84501940 | 0.25 | 0.33 | 0.67 | 0.44 | 1.03 | 0.090 | |
| IRF8 | rs2292982 | 84502324 | 0.25 | 0.33 | 0.67 | 0.44 | 1.03 | 0.091 | |
| IRF8 | rs2292980 | 84502577 | 0.25 | 0.33 | 0.68 | 0.45 | 1.04 | 0.100 | |
| 19q13 | rs11083846 | 51899494 | 0.24 | 0.22 | 1.10 | 0.71 | 1.69 | 0.696 | |
OR indicates odds ratio; and CI, confidence interval.
rs12799226 SNP is correlated with rs735665 (r2 = 0.72); rs735665 failed genotyping in replication cohort.
Discussion
It is clear that there is an inherited genetic contribution to CLL etiology. Our findings herein provide an additional independent locus at the 6p21.32 region to the 10 previously reported loci. The estimated effect sizes of the SNPs within the region are modest (odds ratios ∼ 1.3-1.8) with common allele frequencies (MAF ∼0.25-0.40). The 6p21.32 region is a strong candidate region for harboring predisposing variants for CLL. The HLA-DQA1 and HLA-DRB5 genes belong to the HLA class II α and β chain paralogs, respectively, and play a central role in the immune system by presenting peptides derived from extracellular proteins. Further, this region has been recently identified to harbor variants associated with other B-cell malignancies (follicular lymphoma and diffuse large B-cell lymphoma).5,6,17 It is of interest that this locus was identified and validated only through our familial CLL cases and showed no evidence of association among our sporadic CLL cases. The initial GWA study2 also had 155 CLL cases with a family history of CLL or other related lymphoproliferative disorders included, but did not identify this locus. This may be most likely because they did not perform a GWA analysis stratified by family history status, but only reported stratified analyses among their significant findings. Further, our study limited the family history to CLL, so all of our familial CLL cases had a family history of CLL. It is unclear how many of the 155 CLL cases from the initial GWA study had a family history of CLL specifically and whether this matters. The underlying mechanism of this locus in familial CLL will need further study.
Our study also provided evidence that the IRF8 gene within the 16q24.1 locus is strongly associated with CLL risk. This association is seen in all CLL cases regardless of family history of CLL. IRF8 is also a strong candidate to be implicated in the pathogenesis of CLL. It is a transcription factor that regulates downstream target genes in response to interferons and is nearly exclusively expressed in hematopoietic cells.17
Our study replicated 8 of the 10 previously implicated susceptibility SNPs for CLL risk. We were unable to replicate the association between CLL risk and rs7169431 on 15q21.3 and rs11083846 on 19q13. However, for rs7169431, we estimated an effect size of 1.33, which is comparable to that of the pooled estimate of 1.36 reported by Crowther-Swanepoel et al,4 suggesting that statistical power might be a factor for the lack of statistical significance of this SNP. We have ∼ 60%-70% power to find an effect size of 1.36 with allele frequency between 0.10 and 0.15, given our sample size of 407 cases and 296 controls. In contrast, for rs11083846, we found little evidence of an association. Within our replication sample, our reported odds ratio was 0.92 (95% CI: 0.72, 1.17). Likewise, with our discovery sample, we previously reported imputation results for this SNP and found an odds ratio estimate of 1.11 (95% CI: 0.86, 1.43).3 The reported estimate by Crowther-Swanepoel et al was 1.35 (95% CI: 1.22, 1.49).4 This difference in findings may be due to heterogeneity between the populations.
Finally, we evaluated the CLL-susceptibility SNPs in our sample of familial MBL cases ascertained from our CLL families. Although our sample size was small, we found evidence that some of these susceptibility SNPs were also associated with MBL risk. It would be of interest to see if these SNPs identify those individuals who progress to CLL. These MBL individuals already have an 8-fold increase risk of CLL given the fact that they are relatives of CLL patients, yet half of our MBL individuals had an absolute lymphocyte count < 2.6 × 109 cells/L (range = 1.0-8.8), suggesting that these are the low-count MBL samples.11
The strength of our study includes the well-characterized CLL cases and controls, the large number of familial CLL cases with validated family history of CLL, and stringent quality-control measures. A limitation of our study is the small number of familial CLL cases in the discovery stage of our study. As a result, we were more likely to have a large type II error rate and miss genetic variants.
In summary, we identified a novel CLL susceptibility locus at 6p21.3 among our familial CLL cases and controls and provide strong support for IRF8 as a candidate gene. These data support the importance of evaluating familial cases as a separate group when evaluating the genetic associations for CLL. It is likely that additional loci among familial CLL cases can be identified through larger studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the study participants for their time and effort in study participation and the study coordinators for all of their hard work in recruitment.
This work was supported by National Institutes of Health (NIH) grants CA118444 and CA92153; the Intramural Research Program of the NIH, NCI; the Veterans Affairs Research Service, and the Chronic Lymphocytic Leukemia Research Consortium. Additional support was provided by the National Center for Research Resources, a component of NIH and the NIH Roadmap for Medical Research (1 UL1 RR024150) and by the NCI (CA15083). Data collection in Utah was made possible by the Utah Population Database and the Utah Registry. Partial support for all data in the Utah Population Database was provided by the University of Utah Huntsman Cancer Institute. The Utah Cancer Registry is funded by contract N01-PC-35 141 from the NCI Surveillance Epidemiology and End Results program with additional support from the Utah State Department of Health and the University of Utah. Sample collection at Duke University was supported by a Leukemia & Lymphoma Society Career Development Award (to M.C.L.) and by the Bernstein Family Fund for Leukemia and Lymphoma Research.
National Institutes of Health
Authorship
Contribution: S.L.S. directed the overall study and wrote the manuscript; K.G.R. and S.J.A. conducted the data analyses; and J.M.C. conducted the genotyping. L.R.G., N.E.C, and G.E.M. are the primary investigators (PIs) of the NCI site for chronic lymphocytic leukemia (CLL) family collection; S.S.S. is the PI of the M. D. Anderson site for CLL family collection; M.C.L. and J.B.W. are the PIs of the Duke University site for CLL family collection; L.G.S. and V.A.M. are the PIs of the University of Minnesota/Minneapolis Veteran Affairs Medical Center site for CLL family collection; B.K.L. is the PI of the University of Iowa site for CLL family collection; L.Z.R. is the PI of the University of California-San Diego site for CLL family collection; J.F.L., T.G.C., N.E.K., C.A.H., J.R.C., and C.M.V. contributed to CLL family recruitment at Mayo Clinic; N.J.C. and M.G. are the PIs of the University of Utah study for CLL family recruitment; and J.R.C. is PI of the Mayo Clinic non-Hodgkin lymphoma/chronic lymphocytic leukemia case-control study. All authors contributed to the study design and reviewed and provided revisions to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Susan L. Slager, Mayo Clinic College of Medicine, 200 1st St SW, Rochester, MN 55905; e-mail: slager@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal