Abstract
Epstein-Barr virus (EBV) encodes oncogenic information and, oftentimes concomitant with host immunosuppression, gives rise to malignancies in all major categories of lymphoma defined by the World Health Organization.1 Here, we conditionally evicted the viral extrachromosomal genome from tumor cells in vitro to examine the role of EBV in different lymphomas, including Burkitt lymphoma (BL) and posttransplant lymphoproliferative disorder. Cells derived from 2 canonical BLs were found to have the least dependence on the virus; some required EBV to prevent the inefficient induction of apoptosis. In contrast, cells derived from a subset of BL, Wp-restricted BL, required EBV to block a robust apoptotic program that involves the up-regulation of the proapoptotic protein Bim. Wp-restricted BL cells also relied on the virus to promote efficient proliferation, a distinction that highlights the multiple contributions EBV makes to affect proliferation of its host cells. Like Wp-BL cells, posttransplant lymphoproliferative disorder cells depended on the virus to inhibit apoptosis. They furthermore required the virus to drive them out of G1/G0. Together, these results reveal a graded dependence on EBV among tumor cells that directly correlates with the number of viral genes expressed in the tumor cell.
Introduction
Epstein-Barr virus (EBV) is a human γ-herpes virus that preferentially infects B lymphocytes and transforms them in vitro. EBV has been implicated in a subset of most types of lymphoma,1 principally in cases in which the host's immune system is compromised pathogenically2 or even clinically.3 The viral genes expressed in both transformed and tumor cells are termed “latent,” those used by the virus when it is not producing progeny. However, the viral genes expressed in tumors vary widely; even viral oncogenes necessary to maintain cells transformed in vitro are frequently not expressed in tumor cells.4-8 Indeed, the particular viral genes expressed differ dramatically not only between, but also among, classes of tumors. We asked whether EBV makes different genetic contributions to diverse lymphomas with which it is associated. Previous studies have categorized the expression of distinct sets of viral genes in different lymphomas as being the result of varied programs of latent infection and not necessarily a selection for specific viral functions.
To examine EBV's contributions to lymphoma, we selected a set of diverse malignancies, including endemic Burkitt lymphoma (BL) and posttransplant lymphoproliferative disorder (PTLD), which express overlapping but nonidentical sets of viral genes (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). BLs (endemic in sub-Saharan Africa) are found in young children after repeated malarial attacks. Most of these lymphomas (“canonical” BL) have been found to express only the viral protein EBNA1, sometimes the viral protein LMP2A, and some viral noncoding RNAs.4,8 Wp-restricted BLs (Wp-BLs), approximately 15% of endemic BLs, express additional viral proteins, EBNA3A, EBNA3B, EBNA3C, truncated EBNA-LP, and BHRF1, compared with canonical BL.9,10 PTLD, identified as an outgrowth of lymphoblasts, occurs in transplant recipients placed on immunosuppressive therapy. Early-onset PTLDs (within a year after transplantation) are considered to express still more viral proteins than Wp-BL, including the well-characterized viral oncogenes EBNA2 and LMP1.11 Late-onset PTLDs tend to express fewer viral genes. Taken together, canonical BL, Wp-BL, and PTLD form a collection of lymphomas that range in their expression of viral genes from minimal (canonical BL) to full (PTLD).
Some explanted tumor cells mirror the viral gene expression seen in the tumor in vivo, providing the opportunity not only to study the role of EBV in tumors generally, but also to study differences between tumors specifically. Accordingly, we have used cell cultures to identify the different genetic contributions EBV makes to different tumors by experimentally ridding the tumor cells of EBV. Indeed, assays in cell culture are particularly apt for our mechanistic studies given that, in current xenograft models for EBV-associated malignancies, a major determinant of tumor formation after xenotransplantation is the genetic makeup of the recipient animal.12,13 For example, the host genetic background dictates parameters including the strength of the immune response or the support provided by the microenvironment. Properties of tumor cells identified in cell culture under optimized conditions in which cell growth is unrestrained (logarithmic) are not subject to the genetic constraints of the recipient animal model.
To rid the tumor cells of EBV, we made use of its genome being an extrachromosomal DNA plasmid maintained in proliferating cells by the activities of the viral protein EBNA1. EBNA1 fosters the initiation of replication of viral DNA during S phase and ensures the nonrandom partitioning of sister plasmids into daughter cells during mitosis.14,15 A means was devised to inhibit EBNA1 by the conditional expression of its dominant negative derivative16 (dnEBNA1), which uncovered EBV's roles in sustaining different tumors. We found that the viral contributions to sustaining these lymphoma cells differ substantially between tumors, and hypothesize that this difference reflects an in vivo evolution from an addiction to viral oncogenes to being largely independent of them.
Methods
Cell lines and culture
The lymphoma cell lines BJAB, MutuIII, Oku-BL, Sav-BL, and BL-5 have previously been described.17 The PTLD cell lines PTLD1, PTLD3, and PTLD5 were a kind gift from Dr Cliona Rooney. The canonical BL cell lines Dante-BL and Kem-BL were a kind gift from Dr Alan Rickinson. All cell lines were cultured as previously described.17,18
Retroviral transduction
Retroviral vectors (listed in supplemental Table 1) were generated in 293T cells as previously described, with modifications.18 In most cases, one day after transfecting 293T cells with the retroviral components, the cells were irradiated (3000 cGy). Lymphoma cells were then cocultivated on top of the irradiated 293T cells in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum for 16 to 24 hours.
Generating inducible cell lines
Cells were transduced with TetR-KRAB internal ribosome entry site PURO (from p3329). Puromycin-resistant clones were isolated and screened for a functional TetR-KRAB by a transient-transfection assay in which luciferase expression is regulated by doxycycline. Healthy clones that possessed a functional TetR-KRAB were then transduced with dnEBNA1 internal ribosome entry site green fluorescent protein (from p3299). Transduced cells were subcloned and screened for efficient green fluorescent protein (GFP) regulation by doxycycline. For each cell line, 2 independent clones were selected.
FISH
Cells were stained for fluorescence in situ hybridization analysis (FISH) as previously described.15 To determine the percentage of EBV-negative cells, at least 200 cells per experiment were assayed at each time point. To determine the average number of plasmids per cell (at steady state, in the absence of dnEBNA1), the total number of plasmids was counted in 50 cells per experiment.
Growth curves and cell viability measurements
Cells were diluted to 3 to 5 × 104 cells/mL in culture medium and treated with either 10 ng/mL doxycycline or the vehicle, ethanol. Live cell concentrations were measured every 2 or 4 days with a hemocytometer. Cells stained with trypan blue (with a 1:10 dilution of 0.3% trypan blue dissolved in phosphate-buffered saline [PBS]) or exhibiting an aberrant morphology were considered nonviable. After counting, when necessary, cells were diluted in fresh medium back to approximately the starting concentration (3-5 × 104 cells/mL), and doxycycline or ethanol was added.
Limiting dilution assays
Cells were cultured at limiting dilution as previously described.19
Apologix apoptosis detection
Cells were stained with SR-VAD-FMK (SR100, Cell Technology) following the manufacturer's protocol, except that cells were washed in PBS rather than the manufacturer's supplied wash buffer (PBS worked equally well as the wash buffer). After staining, cells were concentrated by centrifugation and placed on slides. Cells were analyzed and counted on an inverted fluorescence microscope (Axiovert 200 M, Carl Zeiss) using the Texas red filter set Version 20.
Cell sorting
Single cells transduced with retroviral vectors were sorted on FACS-Vantage SE with the FACS-DIVA option (BD Biosciences). To obtain cells that efficiently expressed the particular transgene, cells were sorted for the highest fluorescence intensity (the top 10%-20%) of the marker protein (mRFP or CFP) because it has been established with these vectors that the intensity of the marker correlates with levels of the coexpressed proteins.19
Cell-cycle analysis
Cells were fixed in 95% ethanol on ice and treated with 10 μg/mL DNAse-free RNAse A (Fermentas) in PBS-TBE (0.5% Tween-20, 0.1% bovine serum albumin, 1mM ethylenediaminetetraacetic acid in PBS). They were then stained with 50 μg/mL propidium iodide in PBS-TBE overnight at 4°C. At least 20 000 cells were analyzed per condition on a FACSCalibur (BD Biosciences) equipped with 488-nm and 633-nm lasers. Data were processed using ModFit, Version 3.2.1.
Western blotting
Cells were resuspended in NET lysis buffer (20mM Tris, pH 7.5, 150mM NaCl, 1mM ethylenediaminetetraacetic acid, 0.5% Triton-X 100) and then lysed by adding an equal volume of sample buffer (20mM Tris, pH 6.5, 100mM NaCl, 10% glycerol, 6% sodium dodecyl sulfate, 5% β-mercaptoethanol, 0.04% bromophenol blue). Samples were briefly sonicated. Cell lysates were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred electrophoretically to nitrocellulose membranes. The blots were blocked in blotto (5% nonfat milk, 0.05% Tween-20 in PBS) and probed with primary antibodies followed by alkaline phosphatase–labeled secondary antibodies (Jackson ImmunoResearch). The blots were scanned, and the signals from raw (unmodified) images were quantified using ImageQuant software Version 5.2. Images in Figure 2 and supplemental Figure 2 were prepared for publication with Adobe Photoshop CS2 Version 9.0.2, by adjusting the brightness −15 and the contrast +55. These are linear adjustments that allowed the blots to be visible on printing. The following primary antibodies were used: rabbit polyclonal anti-poly(ADP-ribose) polymerase p85 fragment (G7341, Promega) at a 1:1000 dilution, anti-EBNA1 E68 (rabbit antiserum affinity purified against the C terminus of EBNA-1) or rat monoclonal1H420 at a 1:250 or 1:50 dilution, respectively, rabbit polyclonal anti-Bim/BOD (AAP-330, Stressgen) at 1:1000 dilution, rabbit polyclonal anti-Myc (sc-764, Santa Cruz Biotechnology), and mouse monoclonal anti–α-tubulin (Sigma-Aldrich) at 1:5000 or 1:10 000.
Total DNA isolation
Total DNA was purified as previously described.17
RNA isolation
Total RNA was purified with TRIzol (Invitrogen) as previously described.17 The isolated RNA was then treated with Turbo DNAse (Ambion) following the manufacturer's instructions and repurified with TRIzol.
RT-PCR analysis
Total RNA (1 μg) was mixed with a 15-mer or 20-mer oligo-dT (1 μL of 100μM stock) in a total volume of 20 μL, incubated at 70°C for 10 minutes, and then rapidly cooled on an ice slurry. AMV reverse transcriptase (10 U, Roche Diagnostics) was diluted in the manufacturer's supplied buffer supplemented with deoxyribonucleoside triphosphates (0.2mM final concentration) and added to the cooled RNA, in a total volume of 50 μL. The reaction was incubated at 42°C for one hour. Aliquots of this reaction were then used as templates (4 μL of a 1:4 dilution in water of the reverse transcription reaction) in PCR with primers (IDT) listed in supplemental Table 2, using Herculase (Stratagene) and supplied buffer supplemented with deoxyribonucleoside triphosphates (0.2mM final concentration). Cycling parameters included an initial incubation at 95°C for 5 minutes, followed by 20 cycles (for ACTB) or 25 cycles (all other amplicons) of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute. DNA amplicons were separated on a 1.3% agarose gel and visualized with ethidium bromide staining. Signals were quantified using ImageQuant Version 5.2 software.
Real-time PCR analysis
Total RNA was reverse transcribed as described for reverse-transcribed (RT)-PCR or with SuperScript II (Invitrogen) following the manufacturer's instructions. Real-time PCR was carried out in a 384-well plate with each 20-μL reaction containing 4 μL of the reverse transcription reaction diluted 1:4 or 1:6 in water, 0.5μM of both the forward or reverse primer, 0.2μM of the probe (sequences for primers and probes listed in supplemental Table 2), Rox reference dye (Invitrogen), and AmpliTaq Gold PCR Master Mix (Applied Biosystems). Probes were labeled with 5′-FAMRA and 3′-TAMRA. For detection of EBV DNA, 100 ng purified total DNA per reaction was used as template, with primers and probe specific for the EBER1 sequence (as an internal control, cellular DNA was also detected using primers and probe specific to glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). The reactions were incubated at 50°C for 2 minutes, then at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data were collected on a 7900HT real-time instrument (Applied Biosystems) and analyzed using SDS Version 2.2.2 or 2.2.4 software. Fold changes were based on the assumption that amplicon levels increase by 1.8 times every cycle.
Statistical analysis
The program Mstat, Version 5.10, was used for all statistical analyses (N. Drinkwater, McArdle Laboratory for Cancer Research, School of Medicine and Public Health, University of Wisconsin) and is available for downloading (http://www.mcardle.wisc.edu/mstat).
Results
EBV's role in tumor survival is revealed
We identified selective advantages EBV provides different lymphomas by examining tumor cells engineered to lose the virus and thus all viral genes on treatment with doxycycline. Multiple tumor cell lines were collected that were EBV-positive, of an early passage, and differed in their expression of viral genes (supplemental Figure 1). Eight lymphoma lines were used to express dnEBNA1 when induced with doxycycline. Half of these were engineered successfully. Independent clones of the PTLD cell line PTLD1, the Wp-restricted BL cell line Oku-BL, and the canonical BL cell lines Sav-BL and Dante-BL were successfully engineered to express dnEBNA1 when induced with doxycycline (supplemental Figure 2). The ability of dnEBNA1 to evict the virus was confirmed by fluorescence in situ hybridization analysis of cells cultured in the presence or absence of dnEBNA1 (Table 1). The other 4 lines could not be correctly engineered for different reasons, including profound sensitivity to puromycin (cell lines PTLD5 and PTLD3), evolving viral gene expression as determined by Western blot analysis (cell line BL-5), or the inability to efficiently relieve repression of dnEBNA1 expression in the presence of doxycycline (cell line Kem-BL; data not shown).
The induction of dnEBNA1 forces the loss of EBV from tumor cells over time
| Tumor line . | Clone . | Plasmid copies* . | dnEBNA1 . | Percentage of EBV-negative cells† . | |||
|---|---|---|---|---|---|---|---|
| Day 6 . | Day 10 . | Day 12 . | Day 20 . | ||||
| PTLD1 | P1-1 | 9 | Off | 1 ± 0 | 4 ± 2 | ND | ND |
| On | 28 ± 3 | 37 ± 3 | |||||
| P5-1 | 9 | Off | 2 ± 2 | 1 ± 1 | ND | ND | |
| On | 43 ± 3 | 46 ± 9 | |||||
| Oku-BL | O2-4 | 13 | Off | 2 ± 1 | 2 ± 0 | ND | ND |
| On | 29 ± 8 | 55 ± 12 | |||||
| O5-2 | 13 | Off | 2 ± 1 | 2 ± 0 | ND | ND | |
| On | 22 ± 4 | 45 ± 5 | |||||
| Sav-BL | S1-1 | 27 | Off | ND | ND | 4 ± 3 | 2 ± 1 |
| On | 50 ± 6 | 79 ± 4 | |||||
| S14-2 | 33 | Off | ND | ND | < 1 | 3 | |
| On | 67 | 80 | |||||
| Dante-BL | D7-1 | 29 | Off | ND | ND | 1 ± 0 | 0 ± 0 |
| On | 46 ± 6 | 48 ± 11 | |||||
| D8-1 | 20 | Off | ND | ND | 4 ± 2 | 3 ± 1 | |
| On | 63 ± 11 | 68 ± 7 | |||||
| Tumor line . | Clone . | Plasmid copies* . | dnEBNA1 . | Percentage of EBV-negative cells† . | |||
|---|---|---|---|---|---|---|---|
| Day 6 . | Day 10 . | Day 12 . | Day 20 . | ||||
| PTLD1 | P1-1 | 9 | Off | 1 ± 0 | 4 ± 2 | ND | ND |
| On | 28 ± 3 | 37 ± 3 | |||||
| P5-1 | 9 | Off | 2 ± 2 | 1 ± 1 | ND | ND | |
| On | 43 ± 3 | 46 ± 9 | |||||
| Oku-BL | O2-4 | 13 | Off | 2 ± 1 | 2 ± 0 | ND | ND |
| On | 29 ± 8 | 55 ± 12 | |||||
| O5-2 | 13 | Off | 2 ± 1 | 2 ± 0 | ND | ND | |
| On | 22 ± 4 | 45 ± 5 | |||||
| Sav-BL | S1-1 | 27 | Off | ND | ND | 4 ± 3 | 2 ± 1 |
| On | 50 ± 6 | 79 ± 4 | |||||
| S14-2 | 33 | Off | ND | ND | < 1 | 3 | |
| On | 67 | 80 | |||||
| Dante-BL | D7-1 | 29 | Off | ND | ND | 1 ± 0 | 0 ± 0 |
| On | 46 ± 6 | 48 ± 11 | |||||
| D8-1 | 20 | Off | ND | ND | 4 ± 2 | 3 ± 1 | |
| On | 63 ± 11 | 68 ± 7 | |||||
ND indicates not done.
Average number of plasmids per cell in the absence of dnEBNA1 from 50 to 150 cells.
Average percentage of EBV-negative cells ± SD (3 independent experiments, except clone S14–2, which was done once).
The number of viral genomes per cell varies considerably in a clonal population of EBV-transformed cells.15 Therefore, cells with few genomes are the first to lose EBV in the presence of dnEBNA1, whereas cells with high numbers will go through further divisions before they completely lose the virus. An additional layer of complexity in these experiments arises from EBNA1's not only maintaining viral genomes, but also enhancing the transcription of some viral genes,21 an activity inhibited by dnEBNA1.
Therefore, to capture the consequences of the loss of viral gene expression either from transcriptional repression and/or template depletion, the fate of cells was tracked over multiple days in culture. As PTLD1 and Oku-BL cells lost EBV, a marked inhibition of growth was observed (Figure 1A). This inhibition correlated with the induction of apoptosis, as determined by the activation of caspases (Figure 1B) and was confirmed by increased levels of cleaved poly(ADP-ribose) polymerase (PARP; supplemental Figure 3). In contrast, doxycycline did not affect the growth of parental cells (which did not contain the inducible dnEBNA1 vector) at limiting dilution, a highly sensitive assay for survival and growth (supplemental Figure 4). These observations indicate that EBV provides a survival advantage to both PTLD and Oku-BL tumor cells by blocking apoptosis.
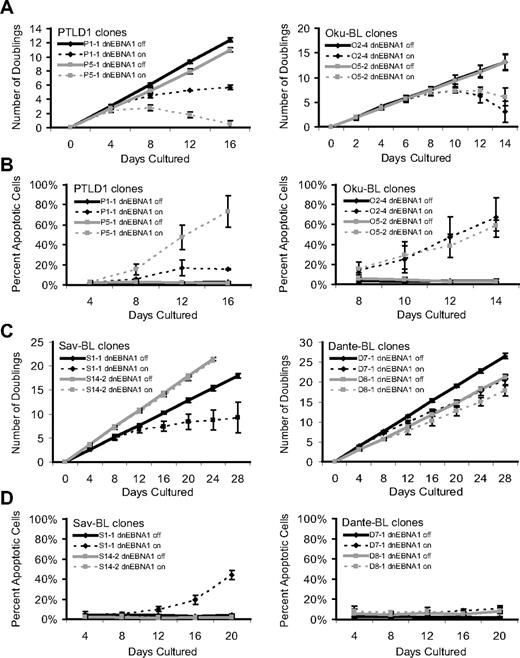
Phenotypes that emerge as EBV is evicted from tumor cells. The growth and survival of tumor cell populations were tracked over time as EBV was evicted. Dashed lines represent cells in which dnEBNA1 remains induced; and solid lines, cells in which dnEBNA1 is uninduced. (A) Growth curves of PTLD1 and Oku-BL clones. The average number of doublings from at least 3 independent experiments ± SD is shown. (B) PTLD1 and Oku-BL clones were assayed for the induction of apoptosis by staining cells with a fluorescently labeled peptide that is specifically bound by active caspases (APO LOGIX, Cell Technologies). For each time point, at least 200 cells were analyzed. The average percentage of apoptotic cells from at least 3 independent experiments plus or minus SD is shown. (C) Growth curves of Sav-BL and Dante-BL clones as measured in panel A. One of 3 S14-2 growth curves was halted at 20 days. (D) Apoptotic assays of Sav-BL and Dante-BL clones as measured in panel B. For clone S14-2, day 20 is the average of 2 independent experiments ± SD.
Phenotypes that emerge as EBV is evicted from tumor cells. The growth and survival of tumor cell populations were tracked over time as EBV was evicted. Dashed lines represent cells in which dnEBNA1 remains induced; and solid lines, cells in which dnEBNA1 is uninduced. (A) Growth curves of PTLD1 and Oku-BL clones. The average number of doublings from at least 3 independent experiments ± SD is shown. (B) PTLD1 and Oku-BL clones were assayed for the induction of apoptosis by staining cells with a fluorescently labeled peptide that is specifically bound by active caspases (APO LOGIX, Cell Technologies). For each time point, at least 200 cells were analyzed. The average percentage of apoptotic cells from at least 3 independent experiments plus or minus SD is shown. (C) Growth curves of Sav-BL and Dante-BL clones as measured in panel A. One of 3 S14-2 growth curves was halted at 20 days. (D) Apoptotic assays of Sav-BL and Dante-BL clones as measured in panel B. For clone S14-2, day 20 is the average of 2 independent experiments ± SD.
The PTLD clone P1-1 provided a contrast that indirectly confirmed these observations. In this clone, not only was dnEBNA1 inefficiently expressed, but viral gene expression from the Cp promoter was poorly inhibited by dnEBNA1, compared with that of PTLD clone P5-1 (supplemental Figures 2,5). Not surprisingly, clone P1-1 displayed the weaker phenotype (Figure 1A-B) in both growth inhibition and the induction of apoptosis. Thus, this clone fortuitously demonstrated that the phenotypes observed in cells losing EBV were dependent on the level of dnEBNA1 and additionally that the strength of the apoptotic signal was directly proportional to the strength of growth inhibition, indicating that apoptosis caused the demise of cell populations.
The phenotypes that emerged as EBV was lost from canonical BL cells not only differed from those observed in PTLD1 or Oku-BL cells, but also varied remarkably from clone to clone. Sav-BL clone S1-1 proliferated more slowly as EBV was lost, which correlated with the induction of apoptosis (Figure 1C-D). This apoptotic phenotype differed from that observed in PTLD1 or Oku-BL; S1-1 often persisted, with a large majority of the population becoming EBV-negative (Table 1). The inhibition of growth and a correlating induction of apoptosis were also observed in Dante-BL clone D7-1 (Figure 1C-D). This clone exhibited a more modest set of phenotypes than did Sav-BL clone S1-1, which correlated with inefficient loss of EBV on induction of dnEBNA1 (Table 1). Some clones of both Sav-BL and Dante-BL did not undergo apoptosis as EBV was lost (S14-2 and D8-1, respectively), even though EBV was being efficiently evicted from these cells (Figure 1C-D, Table 1). Furthermore, continued culturing of clone S14-2 in the absence of dnEBNA1 resulted in the spontaneous loss of the virus (41% of the cells were EBV-negative after ∼ 120 doublings). Together, these results indicate that, despite their clonality, Sav and Dante BLs are phenotypically heterogeneous tumor lines; some cells in these tumors rely on EBV to be protected from the inefficient induction of apoptosis, whereas others do not.
Efficient induction of apoptosis is associated with the expression of Bim
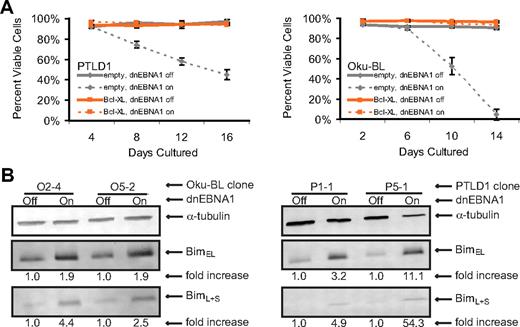
To study the properties of tumor cells in which apoptosis was induced efficiently after the loss of EBV (PTLD1 and Oku-BL), we complemented the antiapoptotic role of the virus by expressing the cellular oncogene Bcl-XL constitutively. Bcl-XL interacts with multiple proapoptotic genes and is important in B-cell development.22,23 Cellular death was examined by staining with trypan blue and monitoring cell morphology, allowing a comprehensive determination of the ability of Bcl-XL to complement EBV's antiapoptotic function. Populations of Bcl-XL-transduced cells strikingly lacked any detectable cellular death, even after several weeks of continuous expression of dnEBNA1 (Figure 2A, PTLD1 clone P5-1 and Oku-BL clone O2-4 only shown).
EBV's antiapoptotic function in both PTLD1 and Oku-BL cells is efficiently complemented by Bcl-XL and involves the repression of the proapoptotic protein Bim. (A) Cells were assayed for global cell death by staining cells with trypan blue. Dashed lines represent cells in which dnEBNA1 is induced; solid lines, cells in which dnEBNA1 remains uninduced. (Left graph) PTLD1 cells (clone P5-1 only shown) transduced with a control vector (gray lines) or a Bcl-XL expression vector (orange lines). (Right graph) Oku-BL cells (clone O2-4 only shown). For each time point, at least 200 cells were analyzed. The average of at least 3 independent experiments ± SD is reported. (B) The levels of endogenous Bim isoforms were analyzed by Western blotting in Oku-BL cells 14 days after induction of dnEBNA1 or in PTLD1 cells 12 days after induction. One of 2 independent experiments is shown. The levels of Bim were normalized to α-tubulin levels and then compared with control cells (dnEBNA1 off) whose normalized level was arbitrarily set to one. Bim isoforms L and S are not resolved on these blots.
EBV's antiapoptotic function in both PTLD1 and Oku-BL cells is efficiently complemented by Bcl-XL and involves the repression of the proapoptotic protein Bim. (A) Cells were assayed for global cell death by staining cells with trypan blue. Dashed lines represent cells in which dnEBNA1 is induced; solid lines, cells in which dnEBNA1 remains uninduced. (Left graph) PTLD1 cells (clone P5-1 only shown) transduced with a control vector (gray lines) or a Bcl-XL expression vector (orange lines). (Right graph) Oku-BL cells (clone O2-4 only shown). For each time point, at least 200 cells were analyzed. The average of at least 3 independent experiments ± SD is reported. (B) The levels of endogenous Bim isoforms were analyzed by Western blotting in Oku-BL cells 14 days after induction of dnEBNA1 or in PTLD1 cells 12 days after induction. One of 2 independent experiments is shown. The levels of Bim were normalized to α-tubulin levels and then compared with control cells (dnEBNA1 off) whose normalized level was arbitrarily set to one. Bim isoforms L and S are not resolved on these blots.
The presence of Bcl-XL imposes a specific block in the apoptotic pathway: anything downstream of the depolarization of the mitochondrial membrane is prevented.24 This block allows the identification of initiating, upstream events untangled with subsequent steps. Bcl-XL–protected PTLD1 and Oku-BL cells were examined for changes that might explain the induction of apoptosis. We found increases in the levels of all detected isoforms of the proapoptotic protein Bim in cells losing EBV (Figure 2B). In contrast, we failed to find consistent up-regulation of Bim in canonical BL cells (data not shown). These data provide a molecular distinction between the tumor cells that display the efficient induction of apoptosis (Bim is up-regulated) and those that display inefficient induction of apoptosis (Bim is not up-regulated) as the virus is lost and indicate that EBV blocks at least 2 distinct apoptotic pathways in tumor cells.
EBV's role in tumor proliferation is revealed
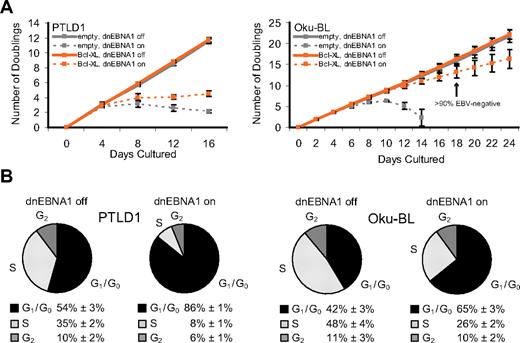
The ability of Bcl-XL to complement EBV's antiapoptotic function was used to unmask additional roles for the virus in PTLD1 and Oku-BL tumor cells. Bcl-XL–protected PTLD1 cells (specifically, clone P5-1) double in number every 33 hours, and the expression of Bcl-XL did not affect this rate (Figure 3A). In contrast, Bcl-XL–protected cells in which dnEBNA1 was induced and EBV was lost ceased to proliferate. To confirm this observation, cells were stained with propidium iodide, analyzed by fluorescence-activated cell sorter, and found to be arrested in G1/G0 (Figure 3B). Unlike PTLD1 cells, Bcl-XL–protected Oku-BL cells that had lost EBV continued to proliferate (Figure 3A, clone O2-4 only shown). However, as is apparent in Figure 3A, the growth rate of these cells was reduced from doubling every 26 hours (independent of Bcl-XL) to every 41 hours. Cell cycle analysis revealed a defect in proliferation. Compared with control EBV-positive cells, cells from which EBV had been evicted were apparently delayed in G1/G0 (Figure 3B). These results were confirmed in the second Oku-BL clone (data not shown). Bcl-XL–protected Oku-BL cells were also tested for growth at limiting dilution. In the presence of human fibroblast feeder layers, we were able to subclone EBV-negative cells (supplemental Figure 6A). The subclones were then tested for their ability to grow at limiting dilution in the absence of human fibroblast feeder layers (supplemental Figure 6B). Whereas EBV-positive subclones could form colonies from single cells, EBV-negative subclones could only form colonies from 10 000 cells or more. Taken together, these results demonstrate that EBV promotes the proliferation of Oku-BL cells in both density-dependent and density-independent assays. These data additionally indicate that both Oku-BL and PTLD1 cells rely on the virus to support their proliferation but differ in the strength of their reliance when apoptosis is inhibited experimentally. Oku-BL cells require the virus to “promote” proliferation but can grow independently of the virus. In contrast, PTLD1 cells require the virus to drive proliferation and cannot proliferate without it.
Oku-BL and PTLD1 tumor cells differ in their dependence on EBV for proliferation. (A) The growth of tumor cell populations was measured over time as EBV was evicted. Dashed lines represent cells in which dnEBNA1 remain induced; solid lines, cells in which dnEBNA1 is uninduced. (Left graph) PTLD1 cells (clone P5-1 only shown) transduced with a control vector (gray lines) or a Bcl-XL expression vector (orange lines). (Right graph) Oku-BL cells (clone O2-4 only shown). The percentage of EBV-negative Oku-BL cells was determined by fluorescence in situ hybridization analysis. The average number of doublings from at least 3 independent experiments plus or minus SD is shown. (B) The percentage of cells in each stage of the cell cycle was determined by propidium iodide staining and fluorescence-activated cell sorting analysis. Cells transduced with a Bcl-XL expression vector were analyzed 20 days (Oku-BL) or 12 days (PTLD1) after induction of dnEBNA1 (the cell populations marked in orange). The average of 3 independent experiments ± SD is shown.
Oku-BL and PTLD1 tumor cells differ in their dependence on EBV for proliferation. (A) The growth of tumor cell populations was measured over time as EBV was evicted. Dashed lines represent cells in which dnEBNA1 remain induced; solid lines, cells in which dnEBNA1 is uninduced. (Left graph) PTLD1 cells (clone P5-1 only shown) transduced with a control vector (gray lines) or a Bcl-XL expression vector (orange lines). (Right graph) Oku-BL cells (clone O2-4 only shown). The percentage of EBV-negative Oku-BL cells was determined by fluorescence in situ hybridization analysis. The average number of doublings from at least 3 independent experiments plus or minus SD is shown. (B) The percentage of cells in each stage of the cell cycle was determined by propidium iodide staining and fluorescence-activated cell sorting analysis. Cells transduced with a Bcl-XL expression vector were analyzed 20 days (Oku-BL) or 12 days (PTLD1) after induction of dnEBNA1 (the cell populations marked in orange). The average of 3 independent experiments ± SD is shown.
mycER cannot substitute for EBV's drive of proliferation
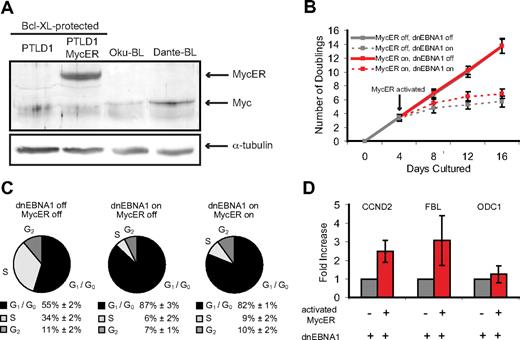
One explanation for the proliferative difference between PTLD1 and Oku-BL cells is that Oku-BL cells have acquired cellular mutation(s) that drive their proliferation, replacing the need for EBV to do so. An obvious candidate mutation is the myc translocation to the immunoglobulin locus, a hallmark of BL.25 If dysregulated myc substitutes for EBV as the driving force for proliferation in Oku-BL, then providing this oncogene to PTLD1 cells would convert their dependence on EBV to one on myc. To test this notion, the tamoxifen-inducible myc derivative Myc-estrogen receptor (mycER)26 was introduced into PTLD1 cells at a level comparable with the level of endogenous myc in BL cells (Figure 4A). The inducible nature of mycER was particularly attractive because it allows evaluation of the growth-promoting capacity of the dysregulation of this single oncogene (in this case, achieved by the addition of tamoxifen), in the absence of additional complementing mutations. PTLD1 cells that rely on EBV to drive proliferation but not block apoptosis (Bcl-XL–protected PTLD1 cells, clone P5-1) were used so that the proliferative phenotype could be studied in the absence of superseding cellular death. In the presence of dnEBNA1, cells in which mycER was active did proliferate slightly better than those in which it was inactive, but they clearly did not exhibit robust growth (Figure 4B). This finding was confirmed by cell cycle analysis (Figure 4C), which revealed a reproducible but quite modest decrease in quiescent cells (from 87% to 82%).
MycER, in the presence of Bcl-XL, is not sufficient to drive the proliferation of PTLD1 cells. (A) The level of MycER expression was analyzed by Western blot in Bcl-XL–protected PTLD1 cells and found to be comparable with the level of Myc in several BL cell lines (Oku-BL and Dante-BL) in which Myc is misregulated. (B) The growth of cell populations was measured over time in cells losing EBV but gaining MycER function. Dashed lines represent cells in which dnEBNA1 is induced; solid lines, cells in which dnEBNA1 is uninduced. Gray lines trace cells in which MycER is inactive, red lines cells in which MycER is activated (beginning 4 days after dnEBNA1 was induced). All cells were previously transduced with a Bcl-XL expression vector (Bcl-XL-protected). The average of 4 independent experiments ± SD is shown. The difference in the total number of doublings by day 16 between cells in which MycER is inactive or active is statistically significant (P = .04, one-sided Wilcoxon rank-sum test). (C) The percentage of cells in each stage of the cell cycle was determined by propidium iodide staining and fluorescence-activated cell sorting analysis. PTLD1 cells were analyzed 16 days after dnEBNA1 induction or mock induction and 12 days after activation or mock activation of MycER. The average of 3 independent experiments ± SD is shown. The reduction in the percentage of cells in G1/G0 in the presence of active MycER (from 87% to 82%) is statistically significant (P = .02, one-sided Wilcoxon rank-sum test). (D) RT-PCR of Myc target genes in cells in which EBV is evicted and MycER is active or inactive (day 16 from panel A). Target gene levels were normalized to β-actin levels and then compared with the control (inactive Myc) levels, which were arbitrarily set to one. The average of 3 independent experiments ± SD is shown.
MycER, in the presence of Bcl-XL, is not sufficient to drive the proliferation of PTLD1 cells. (A) The level of MycER expression was analyzed by Western blot in Bcl-XL–protected PTLD1 cells and found to be comparable with the level of Myc in several BL cell lines (Oku-BL and Dante-BL) in which Myc is misregulated. (B) The growth of cell populations was measured over time in cells losing EBV but gaining MycER function. Dashed lines represent cells in which dnEBNA1 is induced; solid lines, cells in which dnEBNA1 is uninduced. Gray lines trace cells in which MycER is inactive, red lines cells in which MycER is activated (beginning 4 days after dnEBNA1 was induced). All cells were previously transduced with a Bcl-XL expression vector (Bcl-XL-protected). The average of 4 independent experiments ± SD is shown. The difference in the total number of doublings by day 16 between cells in which MycER is inactive or active is statistically significant (P = .04, one-sided Wilcoxon rank-sum test). (C) The percentage of cells in each stage of the cell cycle was determined by propidium iodide staining and fluorescence-activated cell sorting analysis. PTLD1 cells were analyzed 16 days after dnEBNA1 induction or mock induction and 12 days after activation or mock activation of MycER. The average of 3 independent experiments ± SD is shown. The reduction in the percentage of cells in G1/G0 in the presence of active MycER (from 87% to 82%) is statistically significant (P = .02, one-sided Wilcoxon rank-sum test). (D) RT-PCR of Myc target genes in cells in which EBV is evicted and MycER is active or inactive (day 16 from panel A). Target gene levels were normalized to β-actin levels and then compared with the control (inactive Myc) levels, which were arbitrarily set to one. The average of 3 independent experiments ± SD is shown.
The ability of mycER to transactivate well-characterized target genes in PTLD1 cells was examined.27-29 The relative levels of cyclin D2, fibrillarin, and ornithine decarboxylase 1 (ODC1) were measured by RT-PCR. Although both cyclin D2 and fibrillarin levels were increased 2- to 3-fold in the presence of active mycER, we failed to detect significant up-regulation of ODC1 (Figure 4D). The ability of Myc to regulate a target gene is influenced by coregulators; for example, the Max-interacting factor Mxi1 can antagonize Myc's ability to transactivate ODC1.30 Thus, it appears that PTLD1 cells do not have the correct configuration of coregulators that would enable MycER to activate target genes like ODC1, a potential cause of the inability for MycER alone to replace EBV's ability to drive PTLD1 cells to proliferate.
Discussion
EBV is associated with subsets of many different forms of lymphomas, but the particular viral genes these different tumors express vary considerably (supplemental Figure 1). We found that the extent of viral gene expression correlated with the extent to which a tumor cell depends on the virus. This finding is counter to 2 notions regarding the role of EBV in lymphomas: (1) that the set of viral genes expressed is merely a reflection of the particular transcriptional controls within the cell and (2) that only the viral genes common to all lymphomas drive tumor maintenance.
We arrived at this insight by forcing the lymphoma cells to lose EBV. The forced loss of EBV led to the efficient induction of apoptosis in both PTLD1 and Oku-BL cells, which correlated with the up-regulation of Bim. In contrast, 2 canonical BL cells displayed an inefficient induction of apoptosis (which occurred in some but not all clones) as made apparent by the lack of consistent up-regulation of Bim and their accumulation of viable EBV-negative cells. Taken together, these results indicate that PTLD1 and Oku-BL cells rely more on EBV to inhibit apoptosis than do Sav-BL and Dante-BL cells. They further indicate that there probably are 2 distinct apoptotic pathways blocked by the virus: one that involves the up-regulation of Bim and another that does not.
PTLD1 and Oku-BL tumor cells differed in their dependence on EBV for proliferation. Oku-BL cells expressing Bcl-XL proliferated independently of EBV; however, because of their apparent prolongation in G1/G0, they proliferated more slowly. The ability of EBV to mediate efficient exiting from G1/G0 (defined here as the “promotion of proliferation”) is a newly recognized phenotype for EBV. This phenotype appears to be exacerbated in these EBV-negative Oku-BL cells grown under limiting dilution in the absence of feeder cells (EBV-negative cells require 104 as many cells as do EBV-positive cells to grow at limiting dilution). In contrast to Oku-BL cells, PTLD1 cells quiesced in G1/G0 as EBV was lost. Clearly, EBV influences cell proliferation in multiple ways, and these multiple activities distinguish the role of the virus in different tumor cells. Oku-BL cells require EBV to promote proliferation; PTLD1 cells require EBV to drive them out of G1/G0 into the proliferative cycle.
It is probable that multiple viral genes are required to confer a particular phenotype to tumor cells. For example, in other contexts, both EBNA3A and EBNA3C are required to down-regulate the proapoptotic protein Bim,31 or both LMP1 and EBNA2 appear to be needed to drive proliferation.32 Furthermore, different viral genes may be used in different tumor cells to carry out a similar function. For instance, EBNA3A and EBNA3C together, or BHRF1 alone, have been found to interfere with Bim.31,33 Either or both of these mechanisms may be used by PTLD1 or Oku-BL cells (which express these genes) to block Bim-associated apoptosis.
Taken together, our findings reveal a direct correlation. Among the lymphoma cell lines studied here, PTLD1 cells not only express the most viral genes (supplemental Figure 1) but also have the greatest reliance on EBV. They require the virus to both prevent the efficient induction of apoptosis and drive proliferation. Oku-BL cells express fewer viral genes than PTLD1 cells (for example, they do not express EBNA2 or LMP1, supplemental Figure 1) and although they share the requirement to block the efficient induction of apoptosis, they need the virus to promote but not drive proliferation. Sav-BL and Dante-BL, the 2 canonical BL lines, express the fewest viral genes (at the level of mRNA, they are best distinguished from Oku-BL cells by the absence of BHRF1, supplemental Figure 1) and rely least on EBV. Indeed, only half the clones examined were found to require the virus to prevent the inefficient induction of apoptosis. Thus, the degree to which these lymphoma cell lines depend on the virus correlates directly with the number of viral genes expressed within them.
In contrast, an inverse correlation emerges between the number of viral genes expressed in these tumor cells and their associated cellular mutations. Whereas canonical BL cells have been found to contain mutations in p53, Wp-BL cells appear to possess wild-type p53.31 Furthermore, although both Wp-BL and canonical BL cells (including Oku-BL and Sav-BL9 ) have previously been shown to contain clonal translocations of the Myc locus to an immunoglobulin locus, no clonal abnormalities were detected in the karyotype of PTLD1 cells (data not shown). Thus, cells with fewer expressed viral genes apparently contain a greater array of associated oncogenic mutations.
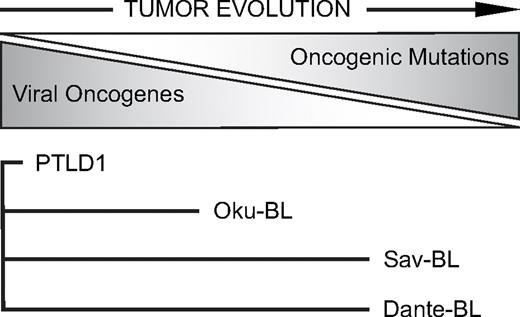
Our examination of 4 tumor cell lines from which EBV could be evicted (though undoubtedly a small sample size) is consistent with a hypothesis for EBV-induced lymphomagenesis (Figure 5). In the formation of this hypothesis, we have been significantly influenced by the work of others.9,11,34,35 EBV supplies a normal B-cell information to survive and grow that is potentially oncogenic. A healthy immune system removes these “proto” tumor cells because they express recognized viral antigens. Cells can avoid elimination by silencing targeted viral genes epigenetically, but infected cells with silenced viral genes are quiescent.36-38 Selection thus favors a proto tumor cell that evolves to reduce its dependence on the virus for proliferation and survival by gaining complementary cellular mutations. The increase of immunosurveillance in PTLD patients by infusion of CTLs directed against EBV proteins is a remarkably successful treatment for the disease, illustrating the need for these tumor cells to decrease expression of viral proteins to survive.39 Indeed, the tendency of early-onset PTLDs to express more viral genes than late-onset PTLDs is consistent with this selective pressure.11 For more evolved tumors, the viral genes that are expressed are those that are either poorly immunogenic (EBNA140,41 ) or nonimmunogenic (viral miRNAs and the EBERs). When these tumor cells are relieved of immunosurveillance (eg, by explanting canonical BL cells to culture), they can evolve to express the full battery of latent genes.42 Furthermore, some explanted tumor cells, importantly those with few viral genes expressed, can evolve in culture to spontaneously lose the virus altogether, as we (Sav-BL clone S14-2) and others43 have found. The perplexing heterogeneity between and among types of EBV-associated lymphomas can thus be explained by a dynamic process in which a selective pressure exerted by the immune system drives tumors to evolve to be independent of the virus.
A hypothesis for EBV-induced lymphomagenesis. EBV transforms B lymphocytes, providing cells with much potentially oncogenic information. However, the viral genes these EBV-positive “proto” tumor cells express are immunogenic, placing the cells under strong negative selection by the immune system. In response, tumor cells evolve to express fewer viral genes by gaining cellular mutations that replace the functions of viral oncogenes. Different tumor cells express distinct sets of latent viral genes reflecting their in vivo evolution away from dependence on the virus and toward dependence on cellular mutations. The lengths of the lines for each tumor cell line reflect the hypothesized extent of this evolution.
A hypothesis for EBV-induced lymphomagenesis. EBV transforms B lymphocytes, providing cells with much potentially oncogenic information. However, the viral genes these EBV-positive “proto” tumor cells express are immunogenic, placing the cells under strong negative selection by the immune system. In response, tumor cells evolve to express fewer viral genes by gaining cellular mutations that replace the functions of viral oncogenes. Different tumor cells express distinct sets of latent viral genes reflecting their in vivo evolution away from dependence on the virus and toward dependence on cellular mutations. The lengths of the lines for each tumor cell line reflect the hypothesized extent of this evolution.
The notion that EBV-positive tumors evolve to be independent of viral oncogenes may parallel findings in other contexts. For example, tumors treated with inhibitors of single oncogenes can develop resistance to this therapy by acquiring an independent means of survival and growth.44,45 In addition, studies in conditional mouse models reveal that tumors can evolve to be independent of the oncogene that induced them; when expression of the oncogene is inhibited, selection for growth can support tumorigenesis via presumably newly acquired cellular mutations.46,47
During the lymphomagenesis of EBV-induced tumors, there are probably genetic or epigenetic changes in addition to those selected for by the immune system that significantly contribute to tumor formation. For example, EBV-infected normal cells (lymphoblastoid cell lines) go through crisis on extended time in culture.48 In contrast, PTLD1 cells were not observed to go through crisis when cultured through at least 175 doublings (> 250 days, data not shown).
It is worth noting that our study, along with the studies of others, indicates that some lymphomas may evolve to be independent of the virus in vivo and thus present clinically as EBV-negative. Were a vaccine against EBV to become widely available, a significant decrease in the incidence of both EBV-negative and EBV-positive lymphomas in vaccinated populations would not only be evidence of this in vivo evolution but also reveal a much broader role for the virus in human lymphomagenesis.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Norman Drinkwater, Shannon Kenney, Ngan Lam, and Paul Lambert for critical evaluation of the manuscript.
This work was supported by the National Cancer Institute, National Institutes of Health (grants P01 CA022443, R01 CA133027, and R01 CA070723). D.T.V. was supported by the National Cancer Center (predoctoral fellowship). B.S. is an American Cancer Society Research Professor.
National Institutes of Health
Authorship
Contribution: D.T.V. designed and performed experiments, analyzed data, and wrote the manuscript; and B.S. designed experiments, performed experiments for supplemental Figure 6B, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bill Sugden, McArdle Laboratory for Cancer Research, 1400 University Ave, Rm 814, Madison, WI 53706; e-mail: sugden@oncology.wisc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal