Abstract
We have recently shown that approximately half of primary multiple myeloma (MM) samples display constitutive Akt activity, which disposes them for sensitivity to Akt inhibition. The Akt pathway counts among the signaling conduits for oncogenic RAS and activating mutations of K- and N-RAS frequently occur in MM. We therefore analyzed the relation between RAS mutation and Akt dependency in biopsies and CD138-purified cells from MM patients (n = 65) and the function of oncogenic RAS for MM cell survival in a range of MM cell lines with differing RAS status. Whereas RAS mutations do not predict Akt dependency, oncogenic RAS retains an important role for MM cell survival. Knockdown of either K- or N-RAS strongly decreased the viability of MM cells that harbored the respective oncogenic isoform, whereas ablation of wild-type RAS isoforms had little or no effect. Silencing of oncogenic RAS did not affect the Akt pathway, again indicating lack of a direct link. Combined inhibition of RAS and Akt strongly enhanced MM cell death. These data suggest that oncogenic RAS and Akt may independently contribute to MM cell survival. Targeting of both pathways could provide an attractive therapeutic strategy for patients with oncogenic RAS and dysregulated Akt signaling.
Introduction
Multiple myeloma (MM) is an incurable neoplasia of the terminally differentiated plasma cell and accounts for approximately 10% of all hematologic cancers.1 Intrinsic genetic lesions as well as the bone marrow microenvironment contribute to the activation of proliferation and survival pathways, impairment of cell death mechanisms, and drug resistance.2-5 Two pathways commonly dysregulated in MM involve activation of the serine/threonine kinase Akt (Akt pathway) and of the guanine nucleotide exchange factor RAS (RAS/MAPK pathway). The Akt pathway has repeatedly been found to be important for MM cell survival,6,7 and we have recently shown that approximately 50% of primary MM samples display an Akt-dependent, phospho-Akt-positive phenotype.7 Little is known about the mechanisms that lead to constitutive Akt activation in MM, but suitable genetic lesions, such as deletion of PTEN8,9 or activating mutations of PIK3CA,10,11 appear to be relatively rare in this disease. Conversely, the reported prevalence of activating mutations of K- and N-RAS in MM ranges from approximately 30% to 50% of patient samples.12-17 The occurrence of RAS mutation appears independent of clinical stage,13,17 but oncogenic RAS is associated with disease progression, aggressive phenotype, and shorter survival.12,17-19 It is also implied in the transition to MM because RAS mutations are rarely found in monoclonal gammopathy of undetermined significance.13,16 However, analyses of the functional role of oncogenic RAS in MM are rare, mainly because of the entire lack of specific inhibitors. Prenylation blockers, such as farnesyltransferase inhibitors, had initially been developed to curb RAS signaling. Encouraging preclinical data, however, did not translate into clinical efficiency, and it has subsequently been shown that the observed effects were largely independent of oncogenic RAS.20-23 It has been reported that ectopic overexpression of oncogenic RAS induces MM cell proliferation24 and lowers drug efficacy.25 However, it is unknown whether endogenous oncogenic RAS still contributes to the malignant phenotype of clinically overt MM and whether it therefore constitutes a potential therapeutic target in its own right. Because oncogenic RAS is a potential activator of the PI3K/Akt signaling axis,26-29 we investigated the correlation between RAS mutation and presence of/dependence on activated Akt in primary MM samples. We also used knockdown approaches in MM cell lines to assess the effects of oncogenic RAS depletion on MM cells. We show that isoform-specific knockdown of RAS reduces the viability of MM cells harboring the respective oncogenic K- or N-RAS mutation, whereas little or no effects are seen when RAS isoforms are lost against a wild-type background. Because RAS mutation was not predictive for sensitivity to Akt blockade and knockdown of oncogenic RAS did not affect Akt activation, constitutively activated RAS and Akt appear to contribute independently to MM cell survival. These observations might have direct implications for the development of targeted therapies in MM.
Methods
Cell culture and primary MM cells
The human MM cell lines AMO-1, JJN-3, and U266 were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). MM.1S cells were purchased from LGC Biolabs (ATCC-CRL-2974). Cell line INA-6 was a gift from Prof Martin Gramatzki (Kiel, Germany). MM cell culture conditions and acquisition of primary MM cells are described in detail by Stühmer et al.30 All cell cultures were regularly tested for mycoplasma. Bone marrow aspirates from myeloma patients were obtained after informed consent. Permission was granted by the Ethics Committee of the Medical Faculty of the University of Würzburg, Würzburg, Germany (reference no. 73/05).
Application of drugs
MM cells were seeded in 100 μL full medium in 96-well-plates. An equal amount of medium was added containing the respective drug in double concentration. Control incubations with dimethyl sulfoxide were always included. Akti-1,2 was obtained from Merck, and PD184352 was from Axon Medchem.
Cell viability assay
The fraction of apoptotic cells was identified by flow cytometry as described before.31 Briefly, cells were washed in phosphate-buffered saline, stained with propidium iodide and annexin V-fluorescein isothiocyanate or annexin V-allophycocyanin (Bender MedSystems), and analyzed using a FACSCalibur with CellQuest software Version 5.2 (BD Biosciences).
RAS mutation analysis
Primary MM samples and cell lines were screened for K- and N-RAS mutations at codons 12, 13 (both exon 2), and 61 (exon 3). A total of 105 cells from freshly isolated (CD138+ selected) patient samples or from cultured MM cell lines were washed twice with ice-cold phosphate-buffered saline and either genomic DNA (using digestion buffer, pH 8, containing 5M NaCl, 1M Tris, 0.5M ethylenediaminetetraacetic acid, 10% sodium dodecyl sulfate, 0.1 mg/mL proteinase K, followed by phenol-chloroform extraction) or total RNA (using the RNeasy Mini Kit, QIAGEN) were prepared. cDNA was synthesized using the RevertAid First-Strand cDNA Synthesis Kit (Fermentas). Polymerase chain reaction was performed for 35 cycles, with 1 minute each for denaturation (94°C), annealing (61°C), and extension (72°C). Polymerase chain reaction products were isolated from agarose gels and sequenced at LGC Genomics. The following primers for amplification off of genomic DNA were used: 5′-ACCTTATGTGTGACATGTTC-3′ (K-RAS forward for exon 2), 5′-AATGGTCCTGCACCAGTAAT-3′ (K-RAS reverse for exon 2), 5′-GCACTGTAATAATCCAGACT-3′ (K-RAS forward for exon 3), 5′-ATTACTCCTTAATGTCAGCT-3′ (K-RAS reverse for exon 3), 5′-AGTACTGTAGATGTGGCTCG-3′ (N-RAS forward for exon 2), 5′-TGATCCGACAAGTGAGAGAC-3′ (N-RAS reverse for exon 2), 5′-CCTTGGCAATAGCATTGCAT-3′ (N-RAS forward for exon 3), and 5′-AATGCTCCTAGTACCTGTAG-3′ (N-RAS reverse for exon 3). For amplification off of first-strand cDNA, the primers 5′-GCCTGCTGAAAATGACTGAA-3′ (K-RAS forward), 5′-CTTGCTAAGTCCTGAGCCTG-3′ (K-RAS reverse), 5′-TCTGTCCAAAGCAGAGGCAG-3′ (N-RAS forward), and 5′-GTGTCAGTGCAGCTTGAAAG-3′ (N-RAS reverse) were used.
Construction of shRNA and HA-tagged RAS expression vectors
shRNA expression constructs were based on pSUPER.32 The following target sequences were used: 5′-GTTGGAGCTGGTGGCGTAG-3′ (human K-RAS wild-type33 ), 5′-GTTGGAGCTGCTGGCGTAG-3′ (K-RAS as mutated in cell line MM.1S), 5′-GTTGGAGCAGGTGGTGTTG-3′ (human N-RAS wild-type), 5′-GTTGGAGCAGATGGTGTTG-3′ (N-RAS as mutated in cell line INA-6), and 5′-ACGAGGGGAGTACATCAAGAC-3′ (human Akt17 ).
A pcDNA3.1-based expression vector for N-terminally triple-hemagglutinin (HA)-tagged human K-RAS was purchased from the University of Missouri-Rolla cDNA Resource Center (no. RASK20TN00). The insert was excised by KpnI/XhoI digest and subcloned into pBluescript (SK) (yielding plasmid pBluescript-HA-K-RAS wt), from which the insert was transferred via KpnI/NotI digest into a modified pCAGGS protein expression vector34 (pCAGGS/SE-HA-K-RAS wt). This construct produced considerably better expression levels of HA-tagged K-RAS in MM cells.
To generate HA-tagged N- and H-RAS expression vectors, the Quik Change II Site-Directed Mutagenesis Kit (Agilent Technologies) was used to introduce an in-frame Bst1107I restriction site between the triple-HA-tag and the K-RAS coding sequences in pBluescript-HA-K-RAS wt. First-strand cDNA generated from U266 cells was used to amplify N- and H-RAS genes with primers 5′-AGTCGTATACTGAGTACAAACTGGTGG-3′ (N-RAS forward), 5′-AGTCCTCGAGTTACATCACCACACATG-3′ (N-RAS reverse), 5′-AGTCGTATACGGAATATAAGCTGGTGG-3′ (H-RAS forward), and 5′-AGTCCTCGAGTCAGGAGAGCACACACT-3′ (H-RAS reverse). The primers introduce Bst1107I and XhoI restriction sites (boldface type) to immediately flank the 5′- and 3′-ends of the coding regions, and they lead to deletion of the native start codons. The genes were cloned to replace K-RAS in pBluescript-HA-K-RAS wt and again transferred to pCAGGS/SE.
Point mutated forms of K-RAS (representing the mutation in MM.1S cells) and N-RAS (as mutated in INA-6 cells; supplemental Table 1B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were generated with the Quik Change II kit in the respective pBluescript constructs described above, and subcloned into pCAGGS/SE.
Transfection of MM cells by electroporation
AMO-1, INA-6, JJN-3, MM.1S, and U266 cells were washed and resuspended in fresh, pure RPMI 1640 medium at a density of up to 1.5 × 107 cells/500 μL. Transient transfection of shRNA and protein expression vectors was performed at 950 μF and 280 V (310 V for MM.1S cells; Gene Pulser 2, Bio-Rad; in 4-mm cuvettes, PeqLab). AMO-1, JJN-3, MM.1S, and U266 cells were cotransfected with an expression plasmid for enhanced green fluorescent protein (pcDNA3.1-EGFP), INA-6 cells were cotransfected with an expression plasmid for truncated CD4.31 Cells were kept for one day in full RPMI 1640 medium with 10% fetal bovine serum, and purification of strongly transfected cells was achieved either by CD4 microbead selection31 or by cell sorting of EGFP+ cells (MoFlo; Beckman Coulter). For electroporation of control cells, shRNA-expressing pSUPER constructs were substituted for the same amount of empty pSUPER vector. For Western analyses, cells were harvested 2 days after electroporation. After one wash in ice-cold phosphate-buffered saline, cell pellets were snap frozen in liquid nitrogen and stored at −80°C until further use.
Western analysis
Western blotting was performed essentially as described before.35 Briefly, cell pellets were dissolved in 20 μL of lysis buffer, protein concentrations determined, and equal amounts mixed with Laemmli buffer run on sodium dodecyl sulfate 10%-polyacrylamide gels before blotting on nitrocellulose membranes. The antiHA-tag antibody (ab9110) was purchased from Abcam, antiK-RAS (sc-30, lot A2309) and antiN-RAS antibodies (sc-31, lot K1709) were from Santa Cruz Biotechnology. Antibodies detecting pan-Akt (no. 9272), phospho-Akt (Ser473, no. 4058), phospho-Akt (Thr308, no. 2965), ERK1/2 (no. 9102), phospho-ERK1/2 (no. 9101), phospho-FOXO1/3a (no. 9464), and phospho-GSK-3β (no. 9336) were purchased from Cell Signaling Technology. Antiβ-actin antibody (A5316) was from Sigma-Aldrich, antiα-tubulin (no. 03568) was from Biozol. Secondary antibodies specific for rabbit (no. 111–036-045), mouse (no. 115036-003), or rat (no. 112036-062) were from Jackson ImmunoResearch Laboratories.
Immunofluorescent histochemical stainings of bone marrow biopsies
To analyze the expression of phospho-Akt (Ser473) and of phospho-Akt (Thr308) in MM cells, immunofluorescent stainings were performed for either antigen in combination with detection of CD138 in formalin-fixed, paraffin-embedded bone marrow biopsies heavily infiltrated with MM cells. For both the CD138/phospho-Akt (Ser473) and the CD138/phospho-Akt (Thr308) double stainings, the primary antibodies were derived from different species. The following antibodies were used: anti-CD138 (M7728, 1:50 dilution; Dako Germany), anti-phospho-Akt (Ser473, no. 4060), anti–phospho-Akt (Thr308, no. 4056; both in 1:100 dilution; Cell Signaling Technology), donkey anti–mouse CY2 (no. 715–225-151), and donkey antirabbit CY3 (no. 711–165-152; both in 1:1000 dilution; Dako Germany). A typical staining protocol used antibody diluent as blocking reagent for 15 minutes, followed by incubation with primary antibodies for 1 hour and with fluorescent dye-marked secondary antibodies for another hour. Between steps, slides were washed 3 times for 5 minutes in Tris-buffered saline. After staining, slides were mounted with antifading medium (Fluormount G; Biozol) and kept at 4°C in the dark. The slides were reviewed and scored by an experienced hematopathologist. The percentage of positive tumor cells (ie, cells carrying a signal for phospho-Akt within the CD138+ tumor cell population) varied from 0% to 80% between cases. An arbitrary cutoff of 20% positive tumor cells for at least one of the 2 phospho-Akt antibodies defined a case as positive or negative. Of the 34 positive cases, 15 showed positivity for both phospho-Akt (Ser473) and phospho-Akt (Thr308), 9 showed reactivity for phospho-Akt (Ser473) only, and 10 for phospho-Akt (Thr308) only. The average percentage of positive tumor cells was 42% for all phospho-Akt (Ser473)-positive cases and 46% for all phospho-Akt (Thr308)-positive cases. The entire immunohistochemical evaluation of primary MM samples was conducted without any knowledge of their RAS mutation status. Images were taken with a confocal laser scanning system (Leica TCS SP2; Leica) connected to a Leica DMRE microscope (original magnification ×400, objective HCX PL APO 40×/1.25-0.75 NA OIL CS; Leica), and imported into the Leica Confocal Software Version 2.61, Build 1537. For each antibody combination, multiple images were taken from different areas of the bone marrow trephine.
Statistical analysis
A 2-tailed Student t test was applied to perform statistical analysis of cell viability assays. For categorical data, Fisher exact test was used. Results were considered significant at P < .05. Calculations were performed with IBM SPSS Statistics 18.
Results
RAS mutation status does not predict dependency on Akt in primary MM
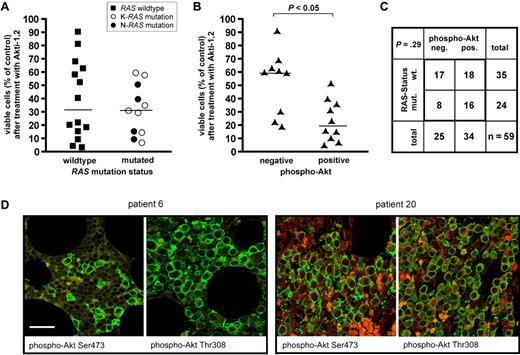
We have previously shown that the phospho-Akt status in MM cells is largely predictive of cell death induction through inhibition of Akt, and that for primary MM samples approximately 50% displayed a phospho-Akt-positive/Akt inhibition-sensitive phenotype.7 Because a similar number has been reported for activating mutations of RAS and oncogenic RAS is a potential upstream activator of the PI3K/Akt pathway, we decided to analyze the RAS/phospho-Akt status of primary MM samples (n = 65) and to investigate the functional role of mutant RAS in MM cell lines. Freshly isolated primary MM cells (CD138+ selection; purity > 95%) were treated with Akt inhibitor Akti-1,2 (10μM), and survival relative to dimethyl sulfoxide-treated controls was determined after 5 days in coculture with bone marrow stromal cells. Simultaneously, the mutation status of K- and N-RAS genes was determined by polymerase chain reaction off of genomic DNA isolated from these samples and, where available, bone marrow biopsies were retrospectively analyzed for the in situ presence of phospho-Akt at positions Ser473 and/or Thr308 (Figure 1).
Analysis of RAS mutation and Akt activation in primary MM samples. (A) RAS mutation status and sensitivity to Akt blockade in freshly isolated primary MM cells (n = 25). Viability measurements of MM cells cocultured with bone marrow stromal cells were performed after 5-day treatment with 10μM Akti-1,2. Sensitivity to Akt inhibition shows no obvious correlation with the presence or absence of RAS mutation. Medians are indicated. (B) Correlation between Akti-1,2 (10μM) sensitivity of freshly isolated, cocultured primary MM cells and the presence or absence of phospho-Akt in immunohistochemical stains of matched bone marrow biopsies (n = 19). Phospho-Akt positivity in the biopsies was significantly (P < .05) correlated with sensitivity to Akt inhibition of the respective MM cells. (C) RAS mutation in primary MM cells and phospho-Akt status in corresponding bone marrow biopsies (n = 59). RAS mutations were present in 16 of 34 phospho-Akt-positive and in 8 of 25 phospho-Akt-negative samples. (D) Two examples of MM patient biopsies with immunofluorescent double staining for plasma-cell marker CD138 (green) and either phospho-Akt Ser473 or phospho-Akt Thr308 (both in red). A phospho-Akt-negative (patient 6) and a phospho-Akt-positive (patient 20) sample are shown. Bar represents 75 μm.
Analysis of RAS mutation and Akt activation in primary MM samples. (A) RAS mutation status and sensitivity to Akt blockade in freshly isolated primary MM cells (n = 25). Viability measurements of MM cells cocultured with bone marrow stromal cells were performed after 5-day treatment with 10μM Akti-1,2. Sensitivity to Akt inhibition shows no obvious correlation with the presence or absence of RAS mutation. Medians are indicated. (B) Correlation between Akti-1,2 (10μM) sensitivity of freshly isolated, cocultured primary MM cells and the presence or absence of phospho-Akt in immunohistochemical stains of matched bone marrow biopsies (n = 19). Phospho-Akt positivity in the biopsies was significantly (P < .05) correlated with sensitivity to Akt inhibition of the respective MM cells. (C) RAS mutation in primary MM cells and phospho-Akt status in corresponding bone marrow biopsies (n = 59). RAS mutations were present in 16 of 34 phospho-Akt-positive and in 8 of 25 phospho-Akt-negative samples. (D) Two examples of MM patient biopsies with immunofluorescent double staining for plasma-cell marker CD138 (green) and either phospho-Akt Ser473 or phospho-Akt Thr308 (both in red). A phospho-Akt-negative (patient 6) and a phospho-Akt-positive (patient 20) sample are shown. Bar represents 75 μm.
For all primary MM samples (n = 65), the genetic status of K- and N-RAS was determined, and 25 samples (38.5%) were found to harbor oncogenic point mutations, thus matching the consensus based on published literature.12 Both genes were found mutated with about the same frequency (12 K-RAS and 13 N-RAS mutated samples). Twenty-five of these samples, of which 11 contained RAS mutations, could be evaluated for their sensitivity against Akti-1,2 (Figure 1A). Comparing the distribution of the survival rates between the RAS wild-type and RAS mutated groups, we found no statistical correlation (P = .47, Student t test; Figure 1A).
Correlation of these results with immunohistochemical detection of phosphorylated Akt (Ser473 and/or Thr308) in the matching bone marrow biopsies, however, largely confirmed our previous observations in that, of the 19 MM samples for which both pharmacologic and immunohistochemical data were available, the large majority of phospho-Akt-positive samples was sensitive to Akt inhibition, whereas phospho-Akt-negative samples were mostly resistant (P = .005, Student t test; Figure 1B). For the correlation between phospho-Akt positivity in biopsies and the presence of oncogenic RAS in the cognate MM samples, we found that mutant RAS was present in 16 of 34 (47%) phospho-Akt-positive cases, as well as in 8 of 25 (32%) phospho-Akt-negative samples (Figure 1C). These numbers do not imply a statistically significant correlation between the presence of oncogenic RAS and phospho-Akt status (P = .29, Fisher exact test), although with the qualification that at n = 59 the statistical power for these conclusions is still limited. Collectively, these data suggest that oncogenic RAS is not a primary driver of intrinsic Akt activity in MM.
Validation of isoform-specific shRNA expression constructs against K- and N-RAS
Because of the lack of suitable inhibitors, it remains unclear whether oncogenic RAS retains a functional role in MM. We therefore decided to establish an isoform-specific knockdown approach (ie, to investigate the loss-of-function consequences in these gain-of-function phenotypes). Genetic analyses of 5 MM cell lines suitable for transfection by electroporation showed that AMO-1 and U266 cells are RAS wild-type, INA-6, and JJN-3 cells display an activating N-RAS mutation, and MM.1S cells harbor oncogenic K-RAS (supplemental Table 1B and supplemental Figure 1; see legend to supplemental Figure 1 for comments on discrepancies in the published RAS status for some of these cell lines). Because of the high sequence homology of RAS isoforms at protein as well as at DNA level, and to assess potential selectivity for wild-type and mutant RAS alleles, it was mandatory to control the specificity of the reagents. Complicating matters, isoform-specific RAS antibodies can display some cross-reactivity (supplemental Figure 2), and RAS proteins may be present in different levels in different MM cells. We therefore cotransfected expression vectors for HA-tagged wild-type and mutant RAS proteins to assess the suitability of the allele- and isoform-specific shRNA expression vectors. Both the K-RAS and the N-RAS-specific knockdown constructs led to strong and selective depletion of their respective target (Figure 2A). However, the constructs did not distinguish between the wild-type and mutant alleles of either the K-RAS (Figure 2B) or N-RAS isoform (data not shown), showing that the single-base differences between target sequences were insufficient to confer effective selectivity. The constitutively active K- and N-RAS proteins are therefore of necessity always depleted in concert with their respective wild-type twin.
Verification of the isoform-specific shRNA expression vectors. MM.1S cells were transiently transfected with expression vectors for HA-tagged K-, N-, or H-RAS (1 μg/mL, 10 μg/mL, and 2 μg/mL, respectively) in combination with shRNA expression vectors against either K- or N-RAS, or empty pSUPER vector (15 μg/mL). (A) Exemplarily shown are Western blots for wild-type RAS proteins in combination with shRNA expression constructs based on the wild-type RAS alleles. Transfected cell fractions were purified and harvested for Western blotting 2 days after electroporation. The vertical white lines indicate that lanes representing a nonfunctional H-RAS shRNA expression vector have been deleted. (B) Expression of wild-type or mutant (G12A; the mutation present in MM.1S) K-RAS in MM.1S cells. Although allele-specific shRNA expression vectors may show slightly better efficiency with their perfect match, both lead to effective knockdown of mutant or wild-type K-RAS, rendering mutation specific knockdown impossible. β-Actin served as loading control.
Verification of the isoform-specific shRNA expression vectors. MM.1S cells were transiently transfected with expression vectors for HA-tagged K-, N-, or H-RAS (1 μg/mL, 10 μg/mL, and 2 μg/mL, respectively) in combination with shRNA expression vectors against either K- or N-RAS, or empty pSUPER vector (15 μg/mL). (A) Exemplarily shown are Western blots for wild-type RAS proteins in combination with shRNA expression constructs based on the wild-type RAS alleles. Transfected cell fractions were purified and harvested for Western blotting 2 days after electroporation. The vertical white lines indicate that lanes representing a nonfunctional H-RAS shRNA expression vector have been deleted. (B) Expression of wild-type or mutant (G12A; the mutation present in MM.1S) K-RAS in MM.1S cells. Although allele-specific shRNA expression vectors may show slightly better efficiency with their perfect match, both lead to effective knockdown of mutant or wild-type K-RAS, rendering mutation specific knockdown impossible. β-Actin served as loading control.
Silencing of oncogenic RAS induces MM cell death
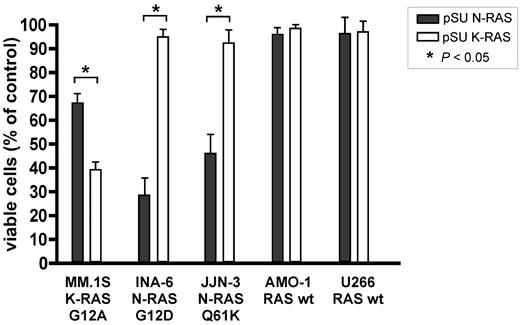
To assess the dependency of MM cells on oncogenic RAS, we transfected each of the 5 MM cell lines (AMO-1, INA-6, JJN-3, MM.1S, and U266) with K- or N-RAS knockdown constructs and measured cell viability over the course of 5 days in culture. Whereas the 2 RAS wild-type cell lines (AMO-1, U266) remained virtually unaffected by isoform-specific RAS depletion (Figure 3) or by combined knockdown of K- and N-RAS (supplemental Figure 3), the shRNA expression vectors led to decreased viability specifically in those MM cells that harbor the respective oncogenic RAS isoform (Figure 3). Thus, knockdown of N-RAS entailed significant cell death specifically in N-RAS mutated INA-6 (29% viability relative to empty vector treated controls) and JJN-3 (46% viability) cells. Conversely, MM.1S cells, which harbor oncogenic K-RAS, were most sensitive to knockdown of K-RAS (39% viability), although in this cell line a lesser effect was observed on N-RAS depletion (67% viability), too (Figure 3). Of note, both the respective mutation- and wild-type–directed shRNA expression constructs produced largely similar results, as would be expected from the fact that they do not discriminate between alleles (supplemental Figure 4).
Isoform-specific shRNA-mediated RAS knockdown reduces cell viability specifically in MM cell lines harboring the respective oncogenic isoform. MM cells were transiently transfected with shRNA expression vectors (20 μg/mL) targeting K- or N-RAS and cell viability was measured after 5 days in culture. N-RAS mutated cell lines INA-6 (G12D) and JJN-3 (Q61K) were specifically sensitive to N-RAS knockdown, whereas K-RAS mutated MM.1S cells (G12A) were most sensitive to K-RAS knockdown. The viability of MM cell lines AMO-1 and U266 (wild-type RAS) remained unaffected by knockdown of either RAS isoform.
Isoform-specific shRNA-mediated RAS knockdown reduces cell viability specifically in MM cell lines harboring the respective oncogenic isoform. MM cells were transiently transfected with shRNA expression vectors (20 μg/mL) targeting K- or N-RAS and cell viability was measured after 5 days in culture. N-RAS mutated cell lines INA-6 (G12D) and JJN-3 (Q61K) were specifically sensitive to N-RAS knockdown, whereas K-RAS mutated MM.1S cells (G12A) were most sensitive to K-RAS knockdown. The viability of MM cell lines AMO-1 and U266 (wild-type RAS) remained unaffected by knockdown of either RAS isoform.
Knockdown of oncogenic RAS in MM.1S and JJN-3 cells down-regulates signaling via the MAPK pathway but does not affect levels of constitutively activated Akt
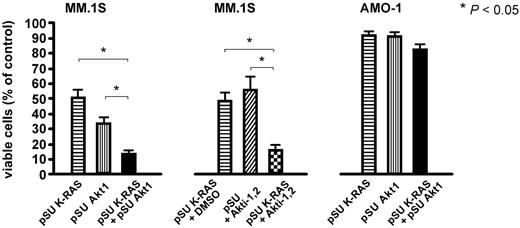
We analyzed the consequences of oncogenic K- or N-RAS depletion on the activity of downstream signaling pathways in MM.1S and JJN-3 cells, which display constitutive phosphorylation of ERK1/2 and of Akt. K-RAS knockdown in MM.1S and N-RAS knockdown in JJN-3 cells markedly reduced the levels of phosphorylated ERK1/2 but did not lead to notable decreases of Akt phosphorylation at either Ser473 or Thr308 (Figure 4), implying that oncogenic RAS may not signal via Akt and that in turn the intrinsic Akt activity in MM.1S and JJN-3 cells is not dependent on oncogenic RAS. Accordingly, the phosphorylation levels of Akt substrates GSK-3β and FOXO1/3a were not affected by K-RAS knockdown in MM.1S cells, either (Figure 4). Because oncogenic RAS and Akt may therefore represent components of 2 independent survival pathways, we tested the effects of simultaneous inhibition of these proteins. An shRNA expression vector against Akt1 (shown to be effectively inducing cell death in MM.1S cells7 ) or the small molecule Akt1&2 inhibitor Akti-1,2 were combined with the K-RAS knockdown vector (Figure 5). The K-RAS mutant MM.1S cells, which are dependent on both K-RAS and Akt, displayed significantly enhanced levels of cell death when these proteins were down-regulated together (Figure 5 left). This effect was also very prominent when K-RAS knockdown was complemented with pharmacologic inhibition of Akt (Figure 5 middle). Contrarily, the viability of RAS wild-type AMO-1 cells, which neither depend on RAS nor display constitutive activity of Akt, was unaffected by combined knockdown of K-RAS and Akt1 (Figure 5 right).
Western analysis of oncogenic RAS signaling. Knockdown of K-RAS in MM.1S cells (harboring an activating G12A K-RAS mutation) or N-RAS in JJN-3 cells (harboring a Q61K N-RAS mutation) led to strongly decreased levels of phosphorylated ERK1/2, indicative of involvement of the RAS/MAPK pathway in oncogenic RAS signaling. Phosphorylation levels of Akt and of its substrates FOXO1/3a and GSK-3β were not significantly affected (shown for MM.1S cells only). Cells were transfected with shRNA expression vectors (20 μg/mL) designed against either the mutant K-RAS allele (but which also leads to knockdown of the wild-type protein), or against both N-RAS alleles (the targeted sequence does not match the position of the mutation at codon 61), purified and harvested for Western analysis 2 days after transfection. β-actin served as loading control.
Western analysis of oncogenic RAS signaling. Knockdown of K-RAS in MM.1S cells (harboring an activating G12A K-RAS mutation) or N-RAS in JJN-3 cells (harboring a Q61K N-RAS mutation) led to strongly decreased levels of phosphorylated ERK1/2, indicative of involvement of the RAS/MAPK pathway in oncogenic RAS signaling. Phosphorylation levels of Akt and of its substrates FOXO1/3a and GSK-3β were not significantly affected (shown for MM.1S cells only). Cells were transfected with shRNA expression vectors (20 μg/mL) designed against either the mutant K-RAS allele (but which also leads to knockdown of the wild-type protein), or against both N-RAS alleles (the targeted sequence does not match the position of the mutation at codon 61), purified and harvested for Western analysis 2 days after transfection. β-actin served as loading control.
Combined knockdown of oncogenic RAS and Akt enhances cell death in MM.1S cells. Simultaneous depletion of oncogenic K-RAS and attenuation of Akt activity, either by shRNA-mediated knockdown (left) or by pharmacologic inhibition of Akt1 and 2 with Akti-1,2 (middle), significantly enhanced cell death in K-RAS mutated MM.1S cells. The concentrations chosen for the expression vectors for K-RAS shRNA (15 μg/mL) and Akt1 shRNA (10 μg/mL) and for Akti-1,2 (2.5μM) were such that an approximately medium size effect on the viability after 5 days in culture could be expected. RAS wild-type AMO-1 cells, which are insensitive to Akt inhibition, were not significantly affected by either single or combined knockdown of K-RAS and Akt1 (right).
Combined knockdown of oncogenic RAS and Akt enhances cell death in MM.1S cells. Simultaneous depletion of oncogenic K-RAS and attenuation of Akt activity, either by shRNA-mediated knockdown (left) or by pharmacologic inhibition of Akt1 and 2 with Akti-1,2 (middle), significantly enhanced cell death in K-RAS mutated MM.1S cells. The concentrations chosen for the expression vectors for K-RAS shRNA (15 μg/mL) and Akt1 shRNA (10 μg/mL) and for Akti-1,2 (2.5μM) were such that an approximately medium size effect on the viability after 5 days in culture could be expected. RAS wild-type AMO-1 cells, which are insensitive to Akt inhibition, were not significantly affected by either single or combined knockdown of K-RAS and Akt1 (right).
Inhibition of MEK/MAPK alone does not recapitulate the effects of RAS knockdown
The observation that knockdown of oncogenic RAS in MM.1S and JJN-3 cells strongly decreased the levels of phospho-ERK1/2 led us to examine to what extent oncogenic RAS might exert its prosurvival effect via the MAPK pathway. Treatment of MM cells with 10μM of the MEK inhibitor PD184352 did not lead to significant decreases in viability, even though at this concentration the phosphorylation of the MEK substrates ERK1 and ERK2 is completely and permanently blocked (supplemental Figure 5). The prosurvival effect of constitutively active RAS in MM cells is therefore not primarily mediated via the MEK/ERK module, again underpinning that RAS represents a potentially useful therapeutic target in its own right.
Discussion
Both the RAS/MAPK and the PI3K/Akt pathway have garnered considerable attention for their presumed role in the pathogenesis and potential suitability for therapeutic intervention in MM.36-38 We have recently shown that Akt contributes to tumor cell survival in approximately 50% of primary MM cases and that sensitivity to Akt inhibition is predicted by the presence of constitutive Akt phosphorylation.7 Because genetic lesions within the Akt pathway are relatively rare in MM, we decided to investigate whether oncogenic RAS is a main driver of intrinsic Akt activity and to analyze its role for MM cell viability. Oncogenic RAS is a potential upstream activator of the Akt pathway,26-29 and activating mutations in the K-and N-RAS genes are generally found in up to 50% of MM cases.12-17 Our study, based on the correlation of the RAS mutation status of primary MM samples with sensitivity to pharmacologic Akt inhibition and with the phosphorylation status of Akt in corresponding bone marrow biopsies, again denoted the presence of phosphorylated Akt as indicator for Akt dependency. However, it also showed that even if RAS-mutated MM samples tended to express phosphorylated Akt more frequently, no correlation between Akt dependency and the presence of oncogenic RAS existed. Accordingly, knockdown of oncogenic RAS in cell lines MM.1S and JJN-3 failed to attenuate the intrinsic phosphorylation of Akt or of Akt substrates, such as GSK-3β or FOXO1/3a, also indicating that the presence of oncogenic RAS is not required to sustain endogenous Akt activity. This is in contrast to studies in RAS wild-type ANBL-6 MM cells, where stable overexpression of oncogenic RAS entailed increased levels of Akt phosphorylation.26 However, our results are in agreement with loss-of-function analyses in solid tumor entities where oncogenic RAS was not found to be a primary driver for constitutive Akt activation.39,40 Notwithstanding its statistical lack of significance, a larger number of RAS mutant primary MM samples did also show constitutive activation of Akt. We would therefore, not rule out that oncogenic RAS may well support Akt activity in a subset of MM cases. It is nowadays known that neither the preclinical efficacy nor the clinical failure of farnesyltransferase inhibitors was attributable to effects on oncogenic RAS,20-23 and currently no selective pharmacologic RAS inhibitors are available. Whereas extensive knowledge has been acquired on the incidence and prevalence of RAS mutations in MM and their clinical context, functional information on the actual relevance of oncogenic RAS in this disease is less prevalent. Insight into RAS function has mainly been gained through overexpression studies of either K- or N-RAS in ANBL-6 cells,24-26 but depletion of endogenous oncogenic RAS has not yet been performed. Because the high frequency of RAS mutation renders it a convenient genetic marker for classification of potential therapeutic subgroups, we decided to perform an isoform-specific loss-of-function analysis in MM cell lines to investigate whether oncogenic RAS still maintains a role for survival in these late-stage representations of MM. Specific K- or N-RAS knockdown did indeed lead to strong decreases in viability if MM cells harbored the respective oncogenic isoform, whereas little or no effects were observed if only the wild-type isoform was present. These experiments therefore are the first to directly demonstrate that MM cells actually display a particular dependence on oncogenic RAS for their survival.
Our study suggests that in MM oncogenic RAS mutation and Akt dependency are not 2 sides of the same medal: neither does oncogenic RAS necessarily activate Akt nor does constitutive activation of Akt depend on the presence of oncogenic RAS. This indicates that intrinsically active RAS and Akt may delineate distinct oncogenic pathways, which independently contribute to MM cell survival. Such a notion is supported by our observation that depletion of K-RAS, when combined with Akt blockade (via knockdown of Akt1 or via pharmacologic inhibition of Akt), led to enhanced cell death in the K-RAS mutant/Akt-dependent cell line MM.1S but was without effect in RAS wild-type/Akt-independent AMO-1 cells.
Oncogenic RAS is known to sustain activity of the RAS/MAPK pathway,26,41,42 and in our hands, too, knockdown of oncogenic RAS led to a strong decrease in the level of phosphorylated ERK1/2 in MM.1S and JJN-3 cells. Nevertheless, although sustained blockade of this pathway with suitable concentrations of MEK inhibitor PD184352 abrogated ERK1/2 phosphorylation, it had little or no effect on the survival of MM cells regardless of their RAS mutation status. To that end, we have previously shown that knockdown of ERK1&2 did not impair survival of N-RAS mutated INA-6 cells.31 This indicates that the prosurvival effects of oncogenic RAS in MM are unlikely to be exclusively transmitted via the MEK/ERK module. Pharmacologic intervention at the level of MEK is thus not a substitute for direct RAS blockade. Oncogenic K-RAS has recently been shown to be associated with the presence of activated nuclear factor-κB signaling in various human carcinoma cell lines.43 However, comparison of the RAS mutation status of the MM cell lines represented here with their ranking for nuclear factor-κB activity (nuclear factor-κB transcriptional signature index) recently published by Demchenko et al,44 does not suggest a clear pattern. RAS mutant cell lines appear equally well represented among those ranking highest (eg, MM.1 and JJN-3) or lowest (eg INA-6, NCI-H929) in the index.
In conclusion, our study shows, for the first time, that oncogenic RAS isoforms sustain the survival of MM cells and that this effect is unlikely to be mediated via either the Akt or the MAPK pathway. The oncogenic RAS isoforms therefore constitute potential therapeutic targets in their own right. Although RAS is currently not drugable, the development of strategies to block oncogenic RAS function is warranted and highly desirable for novel treatment approaches, which would be of immediate relevance for a well-defined patient subgroup.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB-TR17 and KFO 216) and by the MD/PhD program of the Interdisciplinary Center for Clinical Research of the University of Würzburg (Würzburg, Germany).
Authorship
Contribution: T. Steinbrunn designed, performed, and analyzed experiments and wrote the paper; T. Stühmer designed and analyzed experiments and wrote the paper; S.G., A.R., and A.M. provided biopsies and conducted and analyzed the immunohistochemical experiments; C.U. and H.E. provided patient samples; M.C. provided patient samples and wrote the paper; and R.C.B. initiated and designed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Torsten Steinbrunn, Medizinische Klinik und Poliklinik II, Josef-Schneider-Straße 2, 97080 Würzburg, Germany; e-mail: steinbrunn_t@medizin.uni-wuerzburg.de.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal