Abstract
The LIM only protein 2 (LMO2) is a key regulator of hematopoietic stem cell development whose ectopic expression in T cells leads to the onset of acute lymphoblastic leukemia. Through its LIM domains, LMO2 is thought to function as the scaffold for a DNA-binding transcription regulator complex, including the basic helix-loop-helix proteins SCL/TAL1 and E47, the zinc finger protein GATA-1, and LIM-domain interacting protein LDB1. To understand the role of LMO2 in the formation of this complex and ultimately to dissect its function in normal and aberrant hematopoiesis, we solved the crystal structure of LMO2 in complex with the LID domain of LDB1 at 2.4 Å resolution. We observe a largely unstructured LMO2 kept in register by the LID binding both LIM domains. Comparison of independently determined crystal structures of LMO2 reveals large movements around a conserved hinge between the LIM domains. We demonstrate that such conformational flexibility is necessary for binding of LMO2 to its partner protein SCL/TAL1 in vitro and for the function of this complex in vivo. These results, together with molecular docking and analysis of evolutionarily conserved residues, yield the first structural model of the DNA-binding complex containing LMO2, LDB1, SCL/TAL1, and GATA-1.
Introduction
The LIM-only protein 2 (LMO2) is an essential transcriptional regulator in hematopoiesis, whose inappropriate regulation frequently contributes to the development of leukemia. LMO2 was initially cloned from a T-cell acute lymphoblastic leukemia (T-ALL) patient sample bearing a (t(11;14)(p13;q11)) translocation,1,2 and its aberrant expression can be detected in 9% of all pediatric cases of T-ALL.3 T-ALL is characterized by an accumulation in the bone marrow of immature T cells that are altered in their capacity to terminally differentiate. Ectopic activation of LMO2 is thought to initiate leukemia by promoting self-renewal of preleukemic thymocytes through reactivation of expression of hematopoietic stem cell-specific genes.4 LMO2 has also been shown to be activated in T cells by retroviral insertion in a cohort of young patients undergoing gene therapy treatment for X-linked severe combined immunodeficiency syndrome, thereby deregulating its transcription and triggering clonal proliferation of thymocytes.5
LMO2 belongs to a subset of the large zinc finger protein family, the LMO proteins.6 LMO1, LMO2, LMO3, and LMO4 are characterized by the presence of 2 LIM domains (LIM1 and LIM2), each one of them composed of 2 zinc fingers. Despite the presence of zinc finger motifs, no in vivo interaction of LMO2 with DNA has been shown to date, whereas LMO2 interacts with a plethora of proteins. These include the bHLH protein SCL/Tal1, thought to act synergistically with LMO2 in hematopoietic development and, like LMO2, involved in the pathophysiology of T-ALL,7-12 SCL's heterodimerization partner E2A (E47/E12), the hematopoietic transcription factors GATA-1/2,13 AF6, the fusion partner of chromatin remodeling and leukemogenic protein MLL,14 and the LIM-domain interacting protein LDB1.15,16
LMO proteins are thought to act as scaffolding proteins involved in multiprotein complex formation in a myriad of developmental processes through their interaction with LDB1.9,17,18 LDB1 is a widely expressed nuclear adaptor protein that dimerizes through its N-terminal domain and binds LMO proteins and several other LIM homeodomains through its C-terminal LID domain (LIM-interacting domain).19 Dimerization of LDB1 is thought to give rise to various multiple competitive binding events in which different tetrameric or higher-order complexes with LMO and LIM homeodomains are created in different cell types, specifying distinct cell fates.20 Differences in affinities of LMO proteins for LDB1 are at the basis of the delicate equilibrium of complex formation, needed for correct transcriptional control. Enforced expression of LMO2 by chromosomal translocations or transgenesis may cause T-cell tumorigenesis, for example, by displacement of LMO4 as the normal binding partner of LDB1 in maturing T cells.21
Loss-of-function studies in mouse and zebrafish have revealed critical functions for LMO2 in the establishment of primitive and definitive hematopoiesis during embryonic development.12,22,23 In normal blood development, the LMO2-LDB1 dimer mediates interaction between the heterodimer E2A/SCL and GATA-1 to form a multiprotein complex that bridges bipartite DNA motifs in erythroid cells (respectively, Eboxes and GATA sites with 8-, 9-, or 10-bp separation).18 Several combinations of bipartite motifs have been described in gene promoter regions, including Ebox/Gata,18,24 Ebox/Ebox25 (Figure 1A-B), and Gata/Ebox/Gata.26 It is possible that various LMO2-containing complexes might carry different DNA recognition motifs, which in turn bind to specific genomic targets resulting in transcriptional activation or repression. Variations in composition of the oligomeric complexes and in DNA-binding site recognition can thereby yield a precise control of target gene expression patterns during hematopoietic differentiation. Importantly, ectopic expression of LMO2 in T cells may lead to leukemogenesis through aberrant gene regulation.27
LMO2 is also a key regulator of angiogenesis, where it has a specific role in the remodeling of the existing capillary network into mature vasculature.28 Angiogenic remodeling is essential for tumor oxygenation and expansion, and LMO2 expression is up-regulated in tumor endothelium, such as mouse thymomas and human lung tumors.29 Furthermore, it has been suggested that LMO2 is associated with the progression of prostate cancer, possibly by repressing E-cadherin expression.30 Inhibiting tumor proliferation by controlling LMO2 protein levels and activity might therefore constitute an efficient strategy for cancer treatment.
Given the crucial role of LMO2 scaffolding activity in the control of hematopoiesis and its potential therapeutic use, understanding the atomic basis of its intermolecular interactions is of primary interest. A nuclear magnetic resonance structure of the first LIM domain of LMO2 bound to LDB1-LID has been previously solved31 ; however, both LIM domains are required for LMO2 functions.32 To address this, we have now crystallized and determined the atomic structure of human LMO2 containing both LIM1 and LIM2 domains in complex with the LDB1-LID at 2.4 Å resolution. Analysis of the structure leads to a possible model for the assembly of a multiprotein hematopoietic transcription factor complex, opening the way for the design of small molecules antileukemia drugs targeting protein-protein interfaces.
Methods
Protein production
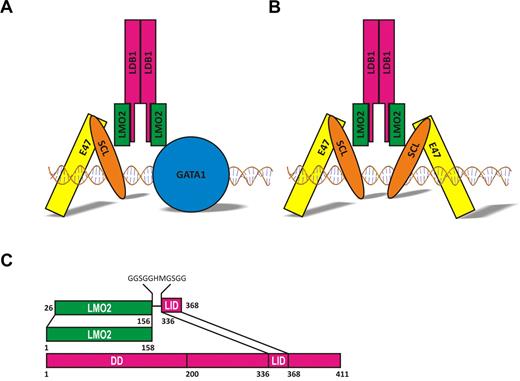
To express a soluble form of LMO2, a fusion to LDB1-LID domain residues 336 to 368 was designed to generate a construct encoding LMO2 (residues 26-156) coupled C-terminally to LID via an 11-residue linker (GGSGGHMGSGG; Figure 1C).33,34 This construct was cloned, expressed, and purified as described elsewhere.35 Briefly, LMO2:LDB1-LID was expressed as a 6xHis-tagged protein and purified by affinity and size exclusion chromatography. The fusion protein behaves as a monomer in solution as shown by multiangle light scattering (data not shown). The fusion protein was concentrated to 20 mg/mL and used for crystallization.
Crystallization, structure determination
LMO2:LDB1-LID crystallized in 2 different conditions. The first condition appeared in a range of acidic pH and high NaCl concentrations, typically 1.6M NaCl, 100mM 2-(N-morpholino)ethanesulfonic acid, pH 5, and 1mM dithiothreitol. Crystals were cryoprotected in 4M sodium malonate, pH 5, and diffracted to 3 Å resolution only after multiple cycles of crystal annealing. Diffraction data, collected on beamline BM14 at the European Synchroton Radiation Facility (Grenoble, France), was indexed, scaled, and merged with HKL2000.36 The space group was determined to be C2 with unit cell dimensions: a = 179.9, b = 51.5, c = 114.7 Å, and β = 90.1°. The asymmetric unit was composed of 5 LMO2:LDB1-LID molecules according to Matthews coefficient calculations corresponding to a VM of 2.4 Å3 Da−1 and a solvent content of 49%. The LMO2:LDB1-LID structure was solved using a 3-wavelength anomalous dispersion experiment at the peak, inflection, and remote wavelengths of the Zn X-ray absorption edge. Details of data collection are described elsewhere.35 PHENIX AutoSol and AutoBuild37 were used to locate the zinc positions, carry out density modification, and build the initial model.
The second crystal form was obtained in 25% PEG 1500 and 100mM succinic acid/phosphate/glycine system) buffer, pH 4.0 (Molecular Dimensions). The crystals were cryoprotected with 20% glycerol for data collection at the European Synchroton Radiation Facility beam line ID23-2. Data were processed with XIA2. The crystals belong to space group P21 with unit cell parameters: a = 25.1, b = 54.3, c = 61.8 Å, and β = 95.5°. The asymmetric unit was composed of one molecule of the LMO2:LDB1-LID, and the structure was solved by molecular replacement using MOLREP38 and the individual LIM domains of the D chain of the previously solved LMO2:LDB1-LID structure as search models.
Model refinement
Manual model building of the structure obtained from the C2 crystal form was done using the program COOT39 and restrained refinement was performed initially with PHENIX-REFINE37 and then AUTOBUSTER,40 taking care of keeping the same Rfree test set in both programs. Noncrystallographic symmetry restrains were imposed on the 5 LMO2:LDB1-LID molecules, except for the domains that showed noticeable differences in the electron density maps. Toward the end of refinement, TLS (Translation/Libration/Screw) vibrational motion refinement was used and water molecules added, giving a final Rwork/Rfree of 22.8/24.4. Data collection and refinement statistics are listed in Table 1.
Data collection and refinement statistics
| . | First crystal form . | Second crystal form . |
|---|---|---|
| Data collection statistics | ||
| Space group | C2 | P21 |
| Cell dimension | a = 179.9 Å; b = 55.5 Å; c = 114.7 Å; β = 90.1° | a = 25.1 Å; b = 54.4 Å; c = 61.8 Å; β = 95.5° |
| Resolution, Å | 50-2.8 (2.85-2.80) | 54.4-2.4 (2.47-2.40) |
| Completeness, % | 96.7 (73.2) | 97.8 (95.8) |
| Redundancy | 7.3 (6.3) | 3.6 (3.6) |
| I/σ(I) | 18.8 (1.8) | 6.7 (1.2) |
| Rmerge, % | 13.9 (84.2) | 12.3 (91.6) |
| Refinement statistics | ||
| Resolution, Å | 48.3-2.8 | 40.7-2.4 |
| No. of reflections | 27 216 | 6395 |
| Rwork/Rfree, % | 22.8/24.4 | 20.1/22.9 |
| RMSD bond, Å | 0.009 | 0.008 |
| RMSD angle, ° | 1.13 | 1.05 |
| Mean B-factors/Wilson plot, Å2 | 80.3/80.8 | 63.2/49.2 |
| Number of molecules in osymmetric unit | 5 | 1 |
| Ramachadran plot, % Favored/allowed/outliers | 95.2/4.5/0.3 | 96.9/3.1/0 |
| . | First crystal form . | Second crystal form . |
|---|---|---|
| Data collection statistics | ||
| Space group | C2 | P21 |
| Cell dimension | a = 179.9 Å; b = 55.5 Å; c = 114.7 Å; β = 90.1° | a = 25.1 Å; b = 54.4 Å; c = 61.8 Å; β = 95.5° |
| Resolution, Å | 50-2.8 (2.85-2.80) | 54.4-2.4 (2.47-2.40) |
| Completeness, % | 96.7 (73.2) | 97.8 (95.8) |
| Redundancy | 7.3 (6.3) | 3.6 (3.6) |
| I/σ(I) | 18.8 (1.8) | 6.7 (1.2) |
| Rmerge, % | 13.9 (84.2) | 12.3 (91.6) |
| Refinement statistics | ||
| Resolution, Å | 48.3-2.8 | 40.7-2.4 |
| No. of reflections | 27 216 | 6395 |
| Rwork/Rfree, % | 22.8/24.4 | 20.1/22.9 |
| RMSD bond, Å | 0.009 | 0.008 |
| RMSD angle, ° | 1.13 | 1.05 |
| Mean B-factors/Wilson plot, Å2 | 80.3/80.8 | 63.2/49.2 |
| Number of molecules in osymmetric unit | 5 | 1 |
| Ramachadran plot, % Favored/allowed/outliers | 95.2/4.5/0.3 | 96.9/3.1/0 |
The structure of the P21 crystal form was built using COOT39 and refined using AUTOBUSTER.40 After TLS refinement and addition of water molecules, the final Rwork/Rfree were, respectively, 20.1/22.9. Data collection and refinement statistics are listed in Table 1. The final refined structure is composed of near full-length LMO2 (residues 26-156) and LDB1-LID (residues 336-368). No electron density was observed for the residues in the linker region, with the exception of the 2 residues (GG) adjacent to LDB1-LID residue 336.
Structure analysis
To identify regions of the molecular surface that may be involved in protein-protein interactions, we examined LMO2 using the WHISCY server,42 which combines surface conservation with structural information. A sequence alignment of human LMO1, LMO2, and LMO4 was used to calculate a conservation score for each surface residue of the 2.4 Å resolution LMO2:LDB1-LID structure; the WHISCY algorithm adjusts the conservation score to account for the surface and interface propensities of each residue. The final score represents the probability of a residue being part of a conserved protein-protein interacting surface. The HINGEprot program43 was used to predict rigid body domains and flexible hinge regions in the LMO2:LDB1-LID complex. Buried surfaces and protein contacts were calculated using the PISA server,44 and structure figures were made in PYMOL.45
Mutagenesis
LMO2 F88A and F88D mutants were generated using the Quickchange II site-mutagenesis kit (Stratagene) using the primers 5′-CTGGTAGGACGGGCCGTTCGGACTCT-3′ and 5′-GTAGGACCGGTAGTTCGGACTCT-3′, respectively.
Mammalian 2-hybrid assays
The CheckMate Mammalian 2-Hybrid System (Promega) was used to conduct mammalian 2-hybrid experiments in HEK293 cells. The cells were transfected with (VP16 activation)-SCL, pM(Gal4 DNA-binding domain)-LMO2 (wild-type or mutants), and the reporter constructs pFR-luc (Gal4-responsive, firefly, Stratagene) and pRL-TK (Renilla, Promega) at a ratio of 1:1:1:0.5 with 10% polyethyleneimine. Twenty-four hours after transfection, both firefly and Renilla luciferase enzyme activities were measured from the same cell lysate sample using the Dual-Luciferase Reporter Assay System, according to the manufacturer's instructions (Promega). Luciferase activities were measured in a Luminoskan Ascent plate reader (Thermo Scientific), and results of firefly luciferase activity were normalized for transfection efficiency with the Renilla luciferase activity. Each transfection was repeated 3 times.
Western analysis
Zebrafish embryo in situ hybridization
Zebrafish embryos were obtained by natural matings, raised, and staged according to Westerfield.48 Murine Lmo2 (wild-type and mutants), and Scl/Tal1 full-length mRNAs were generated as previously described.49 mRNA was transcribed from EcoRI-linearized templates using the T3mMessage-mMachine-kit (Ambion). A total of 100 pg of each mRNA was microinjected into 1-cell stage embryos. Whole-mount in situ hybridizations were carried out on embryos as previously described.50 Riboprobes were labeled with digoxigenin and detected with BM Purple (Roche Diagnostics). Embryos were imaged at 5 times magnification using a Nikon DXM 1200 digital camera attached to a Nikon SMZ 1500. Images were processed with Adobe Photoshop.
Results
Production of the LMO2:LDB1-LID intramolecular complex
As reported for other LMO proteins, it was difficult to produce recombinant full-length LMO2.33 We could express 6xHis-LMO2 in Escherichia coli, but the protein was mostly insoluble. GST-LMO2 could be expressed in soluble form, but it was found to be bound to E coli chaperone protein CPN60 (data not shown). We overcame these problems and were able to express large amounts of soluble protein by tethering LMO2 to the LID domain of LDB1 (Figure 1C), as shown previously.33,34 The artificial fusion protein folds, like the native structure, as demonstrated for LMO4:LDB1-LID and Lhx3:LDB1-LID, where the linker was cleaved and the 15N-HSQC spectra collected before and after cleavage showed no change in conformation.51,52 These observations suggest that, when in isolation, LMO2 is unstable or unable to fold properly and requires a binding partner to obtain and maintain a stable fold. It is known that the interaction of SCL with LMO2 prevents the latter from being degraded by the proteasome.53
Schematic models of LMO2 complexes. (A) model of the pentameric complex bound to DNA through the heterodimer SCL/E47 and GATA1. (B) Complex seen in leukemic T cells isolated from lmo2 transgenic mice. (C) Schematic representation of the LMO2:LDB1-LID fusion protein.
Schematic models of LMO2 complexes. (A) model of the pentameric complex bound to DNA through the heterodimer SCL/E47 and GATA1. (B) Complex seen in leukemic T cells isolated from lmo2 transgenic mice. (C) Schematic representation of the LMO2:LDB1-LID fusion protein.
Overall structure
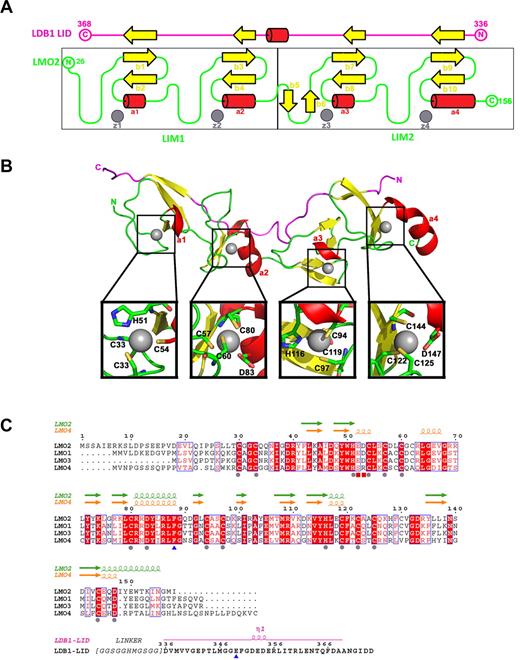
The structure displays a very elongated (80 Å for a protein size of 18 kDa) bar-like shaped LMO2 with a fairly low content of secondary structure: almost 60% of the residues form coils (the average coil content for proteins is ∼ 30%-40%; Figure 2A-B). LMO2 is composed of 2 LIM domains, each of which is composed of 2 zinc fingers. Each LIM domain is composed of 2 small β-sheets and 2 short helices (Figure 2A). The low structural content means that LMO2 heavily relies on the zinc atoms for folding: the first zinc ions of both LIM domains of LMO2 (z1 and z3) are in contact with 3 cysteines and one histidine residue, whereas the second zinc ions (z2 and z4) coordinate 3 cysteines and one aspartate residue (Figure 2B). The essential role of the structural zinc fingers is supported by the fact that the first and second zinc fingers of each LIM domain (z1 and z2, z3 and Z4) are separated by 2 amino acids and that the length of this spacer is conserved and essential for the biologic function of LIM domains.54 Furthermore, sequence alignment of the 4 LMO proteins shows that the residues around the zinc fingers are highly conserved.
Structure of LMO2:LDB1-LID and its sequence alignment with LMO1, LMO3, and LMO4. (A) Schematic representation of the topology of the LMO2:LDB1-LID complex. LMO2 is colored in green, and the LID domain of LDB1 in magenta. β-strands are colored in yellow, helices in red, and zinc atoms in gray. (B) Ribbon diagram showing the LMO2:LDB1-LID complex. Close-up of residues interacting with each zinc finger. (C) Residue conservation between human LMO1, LMO2, LMO3, and LMO4. Conserved amino acids drawn in red boxes; similar amino acids in red type. Residues involved in zinc fingers are marked with a gray sphere, the 2 residues located at the hinge region are marked with a blue triangle, and the residues interacting with LDB1 R360 are marked with a red square.
Structure of LMO2:LDB1-LID and its sequence alignment with LMO1, LMO3, and LMO4. (A) Schematic representation of the topology of the LMO2:LDB1-LID complex. LMO2 is colored in green, and the LID domain of LDB1 in magenta. β-strands are colored in yellow, helices in red, and zinc atoms in gray. (B) Ribbon diagram showing the LMO2:LDB1-LID complex. Close-up of residues interacting with each zinc finger. (C) Residue conservation between human LMO1, LMO2, LMO3, and LMO4. Conserved amino acids drawn in red boxes; similar amino acids in red type. Residues involved in zinc fingers are marked with a gray sphere, the 2 residues located at the hinge region are marked with a blue triangle, and the residues interacting with LDB1 R360 are marked with a red square.
In addition to the zinc fingers, LMO2 uses the LID peptide of LDB1 as a scaffold protein. The LID domain of LDB1 extends across the 2 LMO2 LIM domains (Figure 2A-B) and interacts as a fibronectin-like tandem β-zipper, adding an extra short strand to each β-sheet in the LIM domains of LMO2, as previously seen in LMO4 and Lhx3.51,52 These β-strand interactions are mediated through backbone-backbone contacts, which are generally not affected by amino acid side chains. The LDB1-LID binds to a single face, leaving the zinc finger side of the LMO2 free to interact with other proteins (Figure 2B).
Conformational flexibility of the LMO2:LDB1 complex
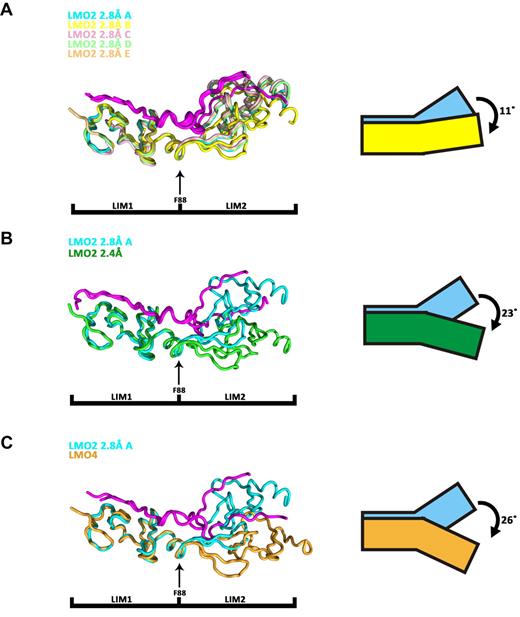
A superimposition of the 5 noncrystallographically related copies of LMO2:LDB1-LID found in the asymmetric unit of the 2.8 Å structure suggests that, despite a very close structural similarity at the level of the individual LIM domains, the relative orientation of the LIM1 and LIM2 can vary. Specifically, the angle between LIM1 and LIM2 varies to a maximum of 11° between copy A and copy B (Figure 3A). This results in an overall root mean square deviation (RMSD) of 1.2 Å between the 2 copies despite an RMSD of only 0.8 and 0.3 Å comparing the individual LIM domains. A more pronounced variation in the relative orientations of the LIM domains is seen in the 2 independently determined structures of LMO2:LDB1-LID: the 2.8 Å solved in space group C2 and the 2.4 Å solved in space group P21. Each LIM domain behaves like a rigid body, and a rotation of 23° is observed comparing the 2 structures, resulting in an RMSD of 2.6 Å (Figure 3B). After this rotation, the rather flat bar-like appearance of the 2.4 Å structure of LMO2 becomes more cusp-like in the 2.8 Å structure.
Structural superimposition of LMO2:LDB1-LID molecules and LMO4:LDB1-LID. Superimpositions are done on the first LIM domain, and the LID domain is colored in magenta. (A) Superimposition of the 5 LMO2:LDB1-LID molecules present in the asymmetric unit of the C2 space group. (B) Superimposition of LMO2:LDB1-LID from the C2 space group (cyan, molecule A) and LMO2:LDB1-LID from the P21 space group (green). (C) Superimposition of LMO2:LDB1-LID (C2 space group, cyan, molecule A) on LMO4:LDB1-LID (yellow).
Structural superimposition of LMO2:LDB1-LID molecules and LMO4:LDB1-LID. Superimpositions are done on the first LIM domain, and the LID domain is colored in magenta. (A) Superimposition of the 5 LMO2:LDB1-LID molecules present in the asymmetric unit of the C2 space group. (B) Superimposition of LMO2:LDB1-LID from the C2 space group (cyan, molecule A) and LMO2:LDB1-LID from the P21 space group (green). (C) Superimposition of LMO2:LDB1-LID (C2 space group, cyan, molecule A) on LMO4:LDB1-LID (yellow).
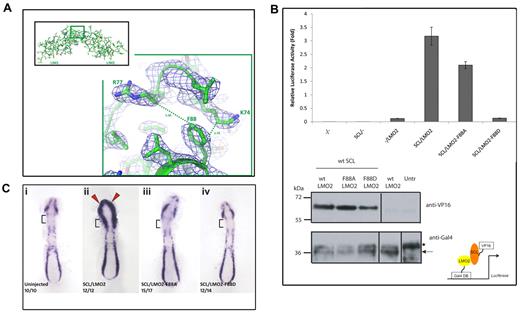
A large motion is also observed when superimposing the structure of LMO2:LDB1-LID 2.8 Å structure to that of LMO4:LDB1-LID,52 with an angle of 26° and an RMSD of 3.1 Å (Figure 3C). From these observations, we conclude that LMO2:LDB1-LID exhibits a hinge-like motion at the region connecting 2 LIM domains. Hinge motions, similar to rotations around an articulated joint, are characterized by large changes in main-chain torsional angles occurring at a localized region, which is called a hinge. Using HingeProt, an algorithm specifically designed to predict protein hinges using elastic network models, it was possible to identify the hinge region of LMO2:LDB1-LID as consisting of 2 residues only: F88 in LMO2 and E349 in LDB1 (Figures 2C, 4A). The HingeProt algorithm predicts hinge regions based on the analysis of individual structures, in this case, the 2.4 Å structure of the LMO2:LDB1-LID. The identified residues F88 in LMO2 and E349 in LDB1-LID are located at the tip of helices at the boundary between the 2 LIM domains exactly at the point where the experimental rotational freedom is observed. Because the same hinge motion is observed in 2 different crystal forms of LMO2:LDB1-LID as well as in the LMO4:LDB1-LID structure, it is unlikely that such movement is caused by crystal-packing artifacts. The F88 and its adjacent residues are conserved among the 4 LMO proteins (Figure 2C), suggesting that the hinge motion of the protein is important for its function. The hinge residue F88 is also in the best determined and most rigid part of the protein, as attested by low B-factors. A similar hinge motion is seen comparing different oligomeric forms of LIM homeobox protein 3 (Lhx3) in complex with LDB1-LID.51 Given the known role of LIM domains as interaction motifs, LMO proteins are thought to act as bridges, bringing together several other proteins to form regulatory complexes. LMO2 could therefore need to undergo hinge motions at the tip of helix a2 (Figure 2A-B) to allow the 2 LIM domains to independently bind different proteins, such as SCL and GATA1. The observed hinge motions could also occur on activation/deactivation of the protein.
The hinge motif F88 in LMO2 is required for functional interaction with SCL/TAL1. (A) Details of the LMO2 structure model around hinge residue F88 (region in green inset) shown in a balls-and-stick representation and the corresponding electron density map (contoured at a σ level of 1). The structurally proximal residues K74 and R77 belonging to the LIM1 domain are also shown. (B) Mammalian 2-hybrid assays in HEK293 cells were carried out to detail the effect of mutating the hinge motif F88. SCL/TAL1 is expressed as a VP16 fusion protein and LMO2 (wild-type and mutants) as Gal4DBD fusion proteins (diagram bottom right). (Top) Interaction between SCL/TAL1 and wild-type LMO2 activates luciferase gene expression. Mutation-dependent effects are observed when the LMO2 mutants F88A and F88D are coexpressed with SCL/TAL1. Error bars represent SD. (Bottom) Western blot analysis of expression of SCL-VP16 (top panel) and LMO2-GAL4DBD (wild-type and mutated forms, bottom panel) in transfected HEK293 cells. Antibodies are indicated on the right. Untr indicates untransfected cells. *Nonspecific band; arrow, the LMO2-Gal4DBD fusion protein. (Ci) Overexpression assay in zebrafish embryos. The hemangioblast marker Fli1 is expressed in the anterior and posterior lateral plate mesoderm in uninjected wild-type embryos. Note that Fli1 expression is absent from the cardiac mesoderm (square brackets). (Cii) Co-overexpression of wild-type Scl/Tal1 and Lmo2 mRNA causes an expansion of Fli1 expression in the head mesoderm (red arrowheads) and cardiac mesoderm (square brackets). This is abolished when Scl/Tal1 is coexpressed with the Lmo2 mutants LMO2-F88A (Ciii) or LMO2-F88D (Civ). Embryos were staged at 10 somites and flat mounted. Anterior to the top. The number of embryos displaying the phenotype is shown in each panel.
The hinge motif F88 in LMO2 is required for functional interaction with SCL/TAL1. (A) Details of the LMO2 structure model around hinge residue F88 (region in green inset) shown in a balls-and-stick representation and the corresponding electron density map (contoured at a σ level of 1). The structurally proximal residues K74 and R77 belonging to the LIM1 domain are also shown. (B) Mammalian 2-hybrid assays in HEK293 cells were carried out to detail the effect of mutating the hinge motif F88. SCL/TAL1 is expressed as a VP16 fusion protein and LMO2 (wild-type and mutants) as Gal4DBD fusion proteins (diagram bottom right). (Top) Interaction between SCL/TAL1 and wild-type LMO2 activates luciferase gene expression. Mutation-dependent effects are observed when the LMO2 mutants F88A and F88D are coexpressed with SCL/TAL1. Error bars represent SD. (Bottom) Western blot analysis of expression of SCL-VP16 (top panel) and LMO2-GAL4DBD (wild-type and mutated forms, bottom panel) in transfected HEK293 cells. Antibodies are indicated on the right. Untr indicates untransfected cells. *Nonspecific band; arrow, the LMO2-Gal4DBD fusion protein. (Ci) Overexpression assay in zebrafish embryos. The hemangioblast marker Fli1 is expressed in the anterior and posterior lateral plate mesoderm in uninjected wild-type embryos. Note that Fli1 expression is absent from the cardiac mesoderm (square brackets). (Cii) Co-overexpression of wild-type Scl/Tal1 and Lmo2 mRNA causes an expansion of Fli1 expression in the head mesoderm (red arrowheads) and cardiac mesoderm (square brackets). This is abolished when Scl/Tal1 is coexpressed with the Lmo2 mutants LMO2-F88A (Ciii) or LMO2-F88D (Civ). Embryos were staged at 10 somites and flat mounted. Anterior to the top. The number of embryos displaying the phenotype is shown in each panel.
Restricting the conformational flexibility of the hinge region of LMO2 hinders interaction with SCL/TAL1 in vitro and in vivo
To address the role of the conformational flexibility of LMO2 around the hinge residue F88, we produced conformationally impaired mutants of LMO2 by mutating the hinge residue phenylalanine 88 in either alanine (A88) or an aspartic acid (D88). The rationale behind this choice of mutants was to decouple the role of the aromatic side chain of the phenylalanine from that of the conformational flexibility. The more conservative alanine mutation would indeed lose the aromatic side-chain character while preserving the hydrophobic character of phenylalanine, as well as its size being advantageous for being in a hinge region.55 The presence of an arginine (R77) at a distance of 5.4 Å and of a lysine (K74) at bonding distance (3.7 Å) to residue 88 (Figure 4A) means that mutation of phenylalanine to aspartic acid would probably induce the creation of one or more salt bridges, effectively locking LIM1 and LIM2 domains in a fixed conformation.
To test the effect of these mutations, we assessed the ability of LMO2 to bind to SCL/TAL1 using 2-hybrid luciferase reporter assays. HEK293 cells were cotransfected with plasmids expressing LMO2 wild-type, LMO2-F88A, or LMO2-F88D fused to the Gal4DBD (DNA-binding domain) and SCL/TAL1 fused to VP16 activation domain (prey) with luciferase reporter plasmids (Figure 4B). The luciferase activation in the presence of SCL/TAL1 prey and LMO2-F88A was reduced by a third compared with SCL/TAL1 prey and LMO2 wild-type, although in the presence of LMO2-F88D, no activation was observed. This indicated that SCL/TAL1-LMO2 interactions are strongly affected when the conformational flexibility of LMO2 is altered (Figure 4B).
Taking advantage of an overexpression strategy in the zebrafish,49 we next established whether the LMO2 hinge residue F88 was important in vivo. Coinjection of wild-type Scl/Tal1 and Lmo2 mRNA causes an expansion of Fli1+ hemangioblasts (blood/endothelial precursors) into the head and cardiogenic mesoderm of zebrafish embryos (Figure 4C), as previously described.49 Overexpression of wild-type SCL/TAL1 with the LMO2 F88A mutant or with the LMO2 F88D mutant abolished this expansion (Figure 4C), presumably as the ability of the LMO2 mutants to interact with SCL/TAL1 is impaired. This is in agreement with the results obtained in the mammalian 2-hybrid assay. Therefore, our mutagenesis studies indicate that, although the aromatic side chain of hinge residue F88 might increase the binding affinity of LMO2 for SCL/TAL1, it is the conformational flexibility that is absolutely required for such interaction.
LMO2:LDB1-LID interactions
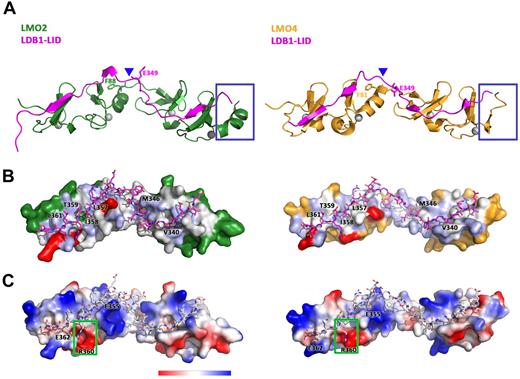
LMO2 and LDB1-LID, as seen in the 2.4 Å structure, interface extensively, burying a surface area of 3505 Å2 and dedicating, respectively, 16% (1608 Å2) and 40% (1898 Å2) of their total surface to this binding. LDB1-LID stretches over LMO2, engaging the LIM domains as separate binding domains with very little hydrogen bonding in the inter-region domain (Figure 5A). Aside from the hydrogen-binding network created by the backbone-backbone β-strand interactions, the LID domain binds LIM1 and LIM2 via extensive regions of hydrophobic contacts and a network of electrostatic interactions (Figure 5B-C). Both LIM domains contribute, in similar extent, to the hydrogen-bond network and hydrophobic contacts; however, the LIM2 addition to the buried surface area is slightly higher than that of LIM1 (LIM1 1561 Å,2 LIM2 1767 Å2). Despite burying less surface area, LIM1 contributes all of the electrostatic interactions between LMO2 and LDB1-LID: R40, E52, and R70 of LIM1 are engaged in salt bridges with E362, R360, and E355 of LDB1-LID (Figure 5C).
Structural comparison of the binding of LDB1-LID to LMO2 and LMO4. (A) Ribbon diagram showing the structural differences between LMO2:LDB1-LID (green/magenta) and LMO4:LDB1-LID (yellow/magenta). (B) Surface hydrophobicity of LMO2 (left) and LMO4 (right) with Ldb1-LID drawn as sticks representation. (C) Surface charge representation of LMO2 (left) and LMO4 (right) with Ldb1-LID drawn as sticks representation.
Structural comparison of the binding of LDB1-LID to LMO2 and LMO4. (A) Ribbon diagram showing the structural differences between LMO2:LDB1-LID (green/magenta) and LMO4:LDB1-LID (yellow/magenta). (B) Surface hydrophobicity of LMO2 (left) and LMO4 (right) with Ldb1-LID drawn as sticks representation. (C) Surface charge representation of LMO2 (left) and LMO4 (right) with Ldb1-LID drawn as sticks representation.
The hinge motion does not heavily impact on the extent of the LMO2:LDB1-LID interface, as even after a rotation of 26° around helix a2, as seen in the LMO2 2.8 Å structure, the total buried surface area remains the same. The relative contributions of LIM1 and LIM2 to the total buried surface area are, however, further shifted, with LIM2 having now an approximately 20% larger interaction surface (LIM1 1557 Å,2 LIM2 1855 Å2). This can be partially explained by an increase in the extent of hydrogen-bonding and electrostatic interactions brought in play by the creation of 2 new salt bridges in the LIM2-interacting domain: K111 and R109 of LIM2 with E349 and D336 of LDB1-LID. The creation of 2 new salt bridges in the LIM2 domain coincides with the disappearance of the first salt bridge in the LIM1 domain of the arched LMO2 structure (between R40 of LIM1 and E362 of LDB1-LID).
Yeast 2-hybrid analysis previously suggested that residues 336 to 366 and/or 350 to 375 of LDB1 are sufficient to bind LMO2, whereas no detectable interaction is seen between residues 336 to 349 and LMO2. Similarly, although LIM1 interacted with LDB1 336 to 375 and 350 to 375, interactions between LIM2 and any LDB1 construct could not be detected. This led to the conclusion that contacts between LIM1 and LDB1-LID are the primary determinants of binding for the LMO2:LDB1-LID interaction.34 These results confirmed previous mutation studies by Terano et al32 showing that deletion of LIM1 in LMO2 abolished the LMO2:LDB1 interaction, whereas deletion of LIM2 did not. Despite its secondary role, several additional pieces of data indicate that LIM2 does make significant contributions to the overall stability of the LMO2:LDB1-LID structure and that its presence increases binding affinity of LMO2 to LDB1-LID by one order of magnitude.34 Our results advocate an equal if not bigger contribution of the LIM2 domain of LMO2 to the binding interface to LDB1-LID. This contribution becomes more prominent as the LMO2:LDB1-LID undergoes the hinge motion around residue F88, suggesting a dynamic modulation of the LDB1-LID affinity for LIM1 and LIM2.
It was suggested that the most essential residue for interaction of LDB1 with LMO2 is isoleucine 358 (I358). I358A is the sole single mutant that prevents binding to LMO2-LIM1 in yeast 2-hydrid experiments, and isothermal calorimetry experiments indicate that the affinity of LDB1-LID for LMO2-LIM1 is reduced by at least 20-fold if I358 is mutated to an alanine.34 From the structures presented here, I358 is buried in a hydrophobic pocket between the 2 zinc fingers of the first LIM domain, with 4 leucine residues from LMO2 (L43, L55, L64, and L71; Figure 5B). Interestingly, these 4 leucines are conserved in LMO1, LMO2, LMO3, and LMO4 (Figure 2C), apart from L71 replaced in LMO4 by a cysteine. Sequence conservation supports the essential role of LDB1 I358 in the specificity of binding LMO proteins. The other aromatic residues around I358 appear to be more permissive for mutation as phage display experiments at LDB1 positions L357, T359, and L361 varied but were always hydrophobic. In our structure of the LMO2:LDB1-LID, as previously observed in the LMO4-LIM1:LDB1-LID, these LDB1 residues lie flat on the surface of LMO2, indicating that aromatic side-chain substitution would be tolerated (Figure 5B). The side chain of residue R360, on the other hand, aside from extensive hydrophobic contacts, seems to insert deeply into a pocket of negative charges formed by LMO2 residues E52, D53, and E66, suggesting an important role for this residue (Figure 5C, green box). This pocket is absent in LMO4 where E52 and D53 are replaced by S45 and R46 (Figure 2C), creating a much smaller negative patch in the equivalent position (Figure 5C, green box). Yeast 2-hybrid screen experiments, however, indicated that mutation of R360 to A360 has little effect on the binding of LMO2 to LDB1, so the role of this residue might be that to confer specificity to the binding rather than strength. Two other clusters of hydrophobic contacts are created between LMO2-LIM2 and LDB1-LID residues V340, which is buried in a hydrophobic pocket between the 2 zinc fingers of LIM2, and M346 (Figure 4B).
Comparison of the LMO2:LDB1-LID and LMO4:LDB1-LID structures shows a conformation-dependent modulation of the LIM domain's affinity for LDB1
LMO2 and LMO4 share extensive structural homology of the individual LIM domains with 128 residues superimposing within an RMSD of 2.7 Å when using the LMO2:LDB1-LID 2.4 Å structure as a reference (Figure 5A). Despite their structural homology, LMO2 and LMO4 differ in the extent of their interactions with LDB1-LID. For comparison, the LMO4:LDB1-LID buries only 3138 Å2 (1375 Å2 from LMO4 and 1763 Å2 from LDB1),52 suggesting that the LID domain of LDB1 shares a more extensive interaction surface with LMO2 (total buried surface 3505 Å2) than with LMO4. This difference in the extent of LDB1 interfacing is small comparing LMO4-LIM1 with LMO2-LIM1, but it becomes noticeable comparing LMO4-LIM2 with LMO2-LIM2 (buried surface area of 1532 Å2 and 1767 Å2, respectively, a decrease of 13%). The difference in the amount of LMO2 interfacing becomes even more substantial (17%) comparing LMO4:LDB1-LID with the 2.8 Å structure of LMO2:LDB1-LID. The overall smaller interaction surface could make a difference in the binding affinity of LMO4 to LDB1-LID compared with LMO2. On the other hand, using homologous competition enzyme-linked immunosorbent assay based on LMO2:LDB1-LID and LMO4:LDB1-LID chimeras, it was previously shown that the binding affinity of LMO2 for LDB1-LID is approximately 2-fold lower than that of LMO4 for the same protein.34 Furthermore, using yeast 2-hybrid analysis, interactions could be readily detected between LMO4-LIM2 and LDB1-LID but not between LMO2-LIM2 and LDB1-LID. In our analysis, the difference in interface area and perhaps in the binding affinity seems to correlate with the relative conformation of the LIM domains.
Despite the high sequence identity of LMO1, LMO2, LMO3, and LMO4 proteins in their core domain composing LMO2 residues 30 to 147, the N- and C-termini are poorly conserved (Figure 2C). The first 26 N-terminal residues are predicted to be disordered and/or flexible in both LMO2 and LMO4 and were truncated from the LMO2:LDB1-LID and LMO4:LDB1-LID constructs. Structural differences are predicted for the C-terminal ends of LMO2 and LMO4 as secondary structure and disorder prediction algorithms predict a short helix and an essentially disordered 16-residue C-terminal tail for LMO4, and a long ordered helix for LMO2. Our results corroborate these predictions as structural differences emerge comparing LMO2 and LMO4 at their C-terminal ends, which are present in both constructs (Figure 5A, blue boxes). LMO4 presents a short 3-amino acid helix, followed by a flexible 8-residue coil, although there is no evidence for the last 19, presumably disordered, residues. On the other hand, all but 2 residues of the C-terminal end of LMO2 are present in our structure, which displays an 11-residue-long rigid (as judged by the low b-factors) helix (Figure 5A). Glutamate 150 (E150) in LMO2 is replaced by a proline in LMO4 (P142) (Figure 2C): prolines are known to be structural disruptors of secondary structures. On the other hand, LMO1 and LMO3 share a glutamate at the equivalent position of LMO2 (Figure 2C) and are predicted to harbor an ordered helix and a short and more ordered C-terminal tail.
Structural analysis reveals binding hot spots on LMO2
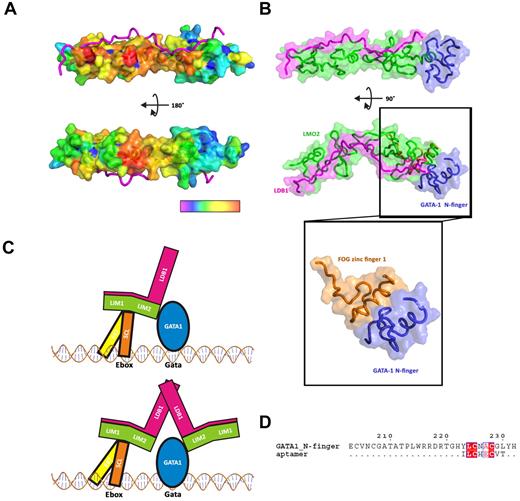
We have analyzed the LMO2 structure to find regions of the molecular surface that may contain hot-spots for protein-protein interactions. The sequence alignment of human LMO1, LMO2, and LMO4 was used as input in the WHISCY server42 to probe conserved residues at the surface of LMO2 for their propensity to be involved in protein interactions. Interestingly, a very clear picture emerges where a whole patch of residues on the side of LMO2 opposite to the LDB1-LID-binding regions and concentrated around the hinge region scores very high, creating a continuous surface that could provide a docking area for LMO-binding partners (Figure 6A). LMO proteins, like many other LIM proteins, can bind multiple partner proteins. SCL/TAL1 is one of the main binding partners of LMO proteins as it can interact in vivo with LMO proteins and these interactions appear to be mediated by sequences within the LIM17 and bHLH domains.56 Furthermore, LMO proteins and SCL/TAL1 are regularly coexpressed in T-ALL and act synergistically, most probably by forming a complex (“Introduction”). It is therefore probable that some, if not all, of the residues exposed by the analysis might provide docking points for SCL/TAL1 and/or SCL/TAL1 complexes. Specifically, the interaction hot-spot residues are concentrated in the hinge and LIM1 region, whereas the LIM2 region is remarkably clear from potential SCL/TAL1 interacting residues (Figure 6A). Intriguingly, our analysis reveals a cluster of potential docking residues in the region around hinge residue F88 (amino acids 84-89; Figure 5A), in agreement with our mammalian 2-hydbrid and in vivo SCL/TAL1 binding studies with the F88 mutants.
Putative interaction of LMO2 with SCL/TAL1-E47 and GATA-1. (A) Surface representation of the residues predicted to be involved in protein-protein interaction, from high probability (red) to low probability (violet). (B) Docking of GATA-1 N-finger (blue) onto the C-terminal zinc finger of LMO2 (green), LDB1 is colored in magenta. The GATA-1:FOG structure (PDB code 1YOJ) was used as a model for the docking. (C) Schematic representation of a possible complex of SCL/TAL1:E47:LMO2:LDB1:GATA-1 on Ebox-GATA motifs. (D) Alignment of an aptamer peptide with GATA_N-finger.
Putative interaction of LMO2 with SCL/TAL1-E47 and GATA-1. (A) Surface representation of the residues predicted to be involved in protein-protein interaction, from high probability (red) to low probability (violet). (B) Docking of GATA-1 N-finger (blue) onto the C-terminal zinc finger of LMO2 (green), LDB1 is colored in magenta. The GATA-1:FOG structure (PDB code 1YOJ) was used as a model for the docking. (C) Schematic representation of a possible complex of SCL/TAL1:E47:LMO2:LDB1:GATA-1 on Ebox-GATA motifs. (D) Alignment of an aptamer peptide with GATA_N-finger.
Deletion of the LIM2 domains, but not of the LIM1 domain in LMO2, has been shown to abolish binding to GATA proteins by immunoprecipitation experiments.32 Furthermore, deletion of the LIM2 domain, or even mutation in its zinc finger domain, inhibited hematopoiesis in mice probably by interfering with the formation of the physiologic complex.32 Thus, it is very probable that GATA proteins bind LMO2 via the zinc finger region of its second LIM domain. The erythroid protein GATA-1 contains 2 zinc fingers: the C-finger, which is sufficient for sequence-specific DNA-binding, and the N-finger, which appears both to modulate DNA-binding and to interact with other transcription factors and partner proteins, including LMO2.57 The structure of the N-finger of GATA-1 bound to zinc finger 1 (F1) of friend of GATA (FOG), a 9-zinc finger-containing protein, revealed how zinc fingers can act as protein recognition motifs (Figure 6B insert).58 Using the FOG F1 structure as a guide, we have docked the structure of the GATA-1 N-finger onto the C-terminal zinc finger of LMO2 (L4; Figure 6B). The structural superimposition between the FOG F1 and the LMO2 L4 is excellent, and the resulting docked model of GATA-1 interacts with the C-terminal helix of LMO2 without clashes. It also appears evident from our LMO2:LDB1-LID structure that, just as for SCL/TAL1, LDB1-LID would not prevent GATA-1 from binding this region (Figure 6B). Furthermore, the predicted binding sites for SCL/TAL1 and GATA-1 are located in different regions of the extended LMO2 protein, making it possible to envisage a situation where both proteins could simultaneously bind (Figure 6C).
The second LIM domain of LMO2 has also been shown to bind an aptamer peptide.59 This peptide disrupts the function of LMO2 in mouse T-cell tumor transplantation assays, supposedly by interfering with the binding of partner proteins. An in silico model of the complex was generated from the structure of LMO4:LDB1-LID and LMO2-LIM1 where the aptamer peptide (sequence ILCHECVT) was predicted to bind L4 of LMO2. According to our docking analysis, this would prevent GATA-1 from binding to LMO2. Interestingly, the aptamer peptide binds specifically to LMO2, whereas no binding to LMO1 and LMO4 is observed. This is in agreement with the lack of observed interactions between LMO proteins other than LMO2 and GATA-1. The peptide aptamer binding modality would therefore be based on mimicking the GATA-1 N-finger. Significant sequence homology between the aptamer peptide and a 6-amino acid stretch of the GATA-1 N-finger (YLCNAC, GATA-1 residues 223-228) can be detected (Figure 6D).
Discussion
Here we report the structure of human LMO2 in complex with the LID domain of LDB1 from 2 crystal forms. Isolated recombinant LMO2 protein is largely insoluble and prone to aggregation in solution. The structure reveals an elongated molecule poor in secondary structure, which relies heavily on zinc ions and an extended interface with the LDB1-LID domain for folding. The LDB1-LID binds to a single face, leaving the zinc fingers side of the LMO2 free to interact with other proteins.
The comparison of the independently determined molecules within one asymmetric unit and between the 2 space groups reveals that LMO2 is very flexible and is able to undergo a hinge rotation of 26° around conserved residue F88, in the region between the 2 LIM domains. This flexibility might reflect the fact that LMO2 is a scaffolding protein that needs to be able to mediate protein-protein interactions. Indeed, LMO2 is able to bind to and mediate binding between numerous protein partners, such as SCL/TAL1, a basic-helix-loop-helix transcription factor, GATA1, a zinc finger transcription factor protein, and LDB1 a nuclear adaptor protein. Hinge motions are also known to occur on activation/deactivation of proteins, again pointing out a regulatory role for the observed flexibility. The hinge motion, revealed from the analysis of our structures, is therefore probably essential for the biologic role of LMO2 as scaffolding protein. Mutagenesis analysis of the hinge motif F88 reveals that the conformational flexibility of LMO2 is crucial for binding to SCL/TAL1 in mammalian 2-hybrid assays and that mutation in this region disrupts the formation of SCL/TAL1-LMO2 complexes in vivo.
Our structure further confirms the crucial role of the LID domain of LDB1 for LMO2: by binding across both LIM domains on the opposite face of the zinc fingers, the LID domain forms a tandem β-zipper, which increases the stability of LMO2 and provides multiple contact points. Along its length, the LID domain is involved in several backbone-backbone hydrogen contacts with LMO2, which do not involve specific side chains and therefore do not require strong sequence conservation. As previously seen for LMO4, this explains why LDB1 can interact with different LMO and LIM homeodomain proteins.60 On the other hand, specific side-chain interactions are also necessary to orient the LIM domains and to drive higher-affinity binding. For example, insertion of the LID residue R360 deeply into a pocket of negative charges formed by LMO2 residues contrasts the much smaller negative patch found in the equivalent position on LMO4. Further, our results advocate an equal, if not bigger, contribution of the LIM2 domain of LMO2 to the binding interface to LDB1-LID. This contribution becomes more prominent as the LMO2:LDB1-LID undergoes the hinge motion around residue F88 and can be explained by the disruption of salt bridges at the LIM1 terminal end and the creation of new salt bridges at the LIM2 terminal end of LMO2. This shift in the tightness of the interaction between the LID domain and the 2 LIM domains, brought in part by the zipping and unzipping of the electrostatic interactions, suggests a dynamic modulation of the LDB1-LID affinity for LMO2, which could be regulated by the binding of the complex to other substrate proteins.
By comparing our structure of LMO2 bound to LDB1-LID with the analogous structure of LMO4,52 we observe, as expected, a high level of structural homology between the 2 proteins. LMO2 and LMO4 differ, however, in the details of their interactions with the LID domain and, more importantly, in the extent to which they interface with the LID domain. LMO2 interfaces more extensively with the LID domain, a difference brought in play by an increase in LIM2 binding compared with LMO4. Again, we postulate that a modulation might exist of the affinity of LMO proteins for the LID domain of LDB1, which emerges from the inherent conformational flexibility around the tandemly arranged LIM interaction motifs and might be driven by the coupling of the scaffolding LMO proteins to their various substrates.
Using a combination of sequence conservation and surface propensity algorithm, together with molecular docking, we have obtained a rough map of hot-spots on the surface of LMO2. A patch, conserved among LMO proteins, which extends mainly across the LIM1 domain and around the hinge region, is a probable anchoring point for SCL/TAL1, the main interacting partner of LMO2 in hematopoiesis. These results are in excellent agreement with our mutagenesis analysis of the hinge motif. Interactions with SCL/TAL1 leave the LIM2 domain essentially free to associate with other binding partners, such as GATA-1. In a region, composed of the fourth zinc finger of LMO2 and the long C-terminal helix, which our analysis suggests being absent in LMO4, we have identified a probable spot for interactions of LMO2 with GATA-1. In this protein-protein interaction, hot-spot map of LMO2 simultaneous binding of the LID domain of LDB1, SCL/TAL1, and GATA-1 would not be precluded. As shown in Figure 6C, the potential of binding SCL/TAL1 and GATA-1 on the same LMO2, together with the dimerization capacity of LDB1, could lead to formation of oligomeric transcription factor complexes possessing recognition domains for 2, 3, or even 4 DNA motifs. This in turn could yield a precise control of target gene expression patterns during differentiation, although misregulation of this fine equilibrium of protein-protein interactions could lead to tumorigenesis.
Our results provide specific insights into the potential interactions of LMO2 with LDB1-LID, SCL/TAL1, and GATA-1. Given its central role as scaffolding protein at the core of the SCL/TAL1, LDB1, GATA-1, and DNA-binding complex, its pivotal function in primitive as well as in definitive hematopoiesis, and its aberrant expression in T-ALL and other cancers, LMO2 has long been considered an attractive therapy target. Structural information that can be translated into chemicals that disrupt specific protein-protein interactions are at the very basis of such quest.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff at beamline BM14 (European Synchroton Radiation Facility, Grenoble) for their support during data collection.
This work was supported by the Leukemia and Lymphoma Research, the Medical Research Council, and Jesus College, Oxford (Major Research Grant). E.J.M. is a Royal Society University Research Fellow. P.V. was supported by the MRC Disease Team Award, the MRC Molecular Hematology Unit, and the Oxford Partnership Comprehensive Biomedical Research Center (Department of Health's National Institute for Health Research, Biomedical Research Centres).
Authorship
Contribution: K.E.O. performed experiments, analyzed data, and wrote the paper; S.J.H., D.K., and K.T. performed experiments; P.V. and R.P. contributed to data analysis; and C.P. and E.J.M. designed and supervised the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Erika J. Mancini, Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Dr, Oxford, OX3 7BN, United Kingdom; e-mail: erika@strubi.ox.ac.uk.