Abstract
Cell-cycle quiescence in hematopoietic stem cells (HSCs) is essential for maintaining stemness by protecting cells from differentiation or senescence. F-box and WD-40 domain protein 7 (Fbxw7) maintains HSCs and suppresses leukemogenesis by mediating ubiquitin-dependent degradation of cell-cycle activators and oncoproteins. Fbxw7α was shown to be the preferentially expressed Fbxw7 isoform in primitive HSCs. Forced Fbxw7α expression in lineage marker Sca-1+c-Kit+ cells led to cell-cycle dormancy by reducing the protein levels of the Fbxw7 substrates c-Myc, Notch1, and phosphorylated S6 (a key downstream element of mTOR). Hypoxia, an essential factor for HSC quiescence, suppressed c-Myc in an Fbxw7α-dependent manner. Fbxw7α-overexpressing lineage marker Sca-1+c-Kit+ cells sustained high reconstitution capacities during in vitro culture. These data suggest that Fbxw7α sustains HSC dormancy through c-Myc, Notch1, and the mTOR pathways. The modulation of Fbxw7α expression or activity represents a promising new tool for ex vivo HSC maintenance.
Introduction
Quiescence maintains the “stemness” of hematopoietic stem cells (HSCs) by protecting them from differentiation or senescence.1 Various strategies have been used to maintain and amplify HSCs,2-4 such as HoxB4 up-regulation, Bmi1 overexpression, Angptl addition, and Wnt3a supplementation. However, a novel strategy for the maintenance/amplification of HSCs is needed because effective expansion methodologies have not been established.
F-box and WD-40 domain protein 7 (Fbxw7) is an F-box protein component of an stem cell factor–type ubiquitin ligase that contributes to ubiquitin-dependent degradation of cell-cycle activators and oncoproteins.5 Known substrates of Fbxw7 include cyclin E,5,6 c-Myc,5,6 Notch1,5,6 and mTOR,7 which are reportedly important for G0 state maintenance in HSCs.8-10 Deletion of Fbxw7 in adult HSCs results in loss of quiescence in HSCs, defective bone marrow transplantation (BMT) capacity, and the emergence of T-cell acute lymphoblastic leukemia.
In this study, Fbxw7α was identified as the major Fbxw7 isoform in HSCs. Fbxw7α overexpression in HSCs maintained cell-cycle quiescence and supported ex vivo HSC transplantation capacity. These results identify Fbxw7α as a key factor for HSC maintenance.
Methods
Retrovirus transduction
Fbxw7α, β, and γ cDNAs were amplified and subcloned into the pMY-IRES-EGFP retroviral vector.11 Production and transduction of retrovirus were performed as previously described.12 At 48 hours after transduction, enhanced green fluorescence protein-positive (EGFP+) cells were sorted for assays. All experiments were approved by the Animal Care and Use Committee in Keio University School of Medicine.
Cell-cycle analysis
For cell-cycle analysis by bromodeoxyuridine (BrdU) assay, EGFP or Fbxw7α virus-transduced lineage marker (Lin)− Sca-1+ c-Kit+ (LSK) cells were cultured on fibronectin-coated plates in serum-free medium (SF-O3) containing 1.0% bovine serum albumin, 100 ng/mL murine stem cell factor, and 100 ng/mL human thrombopoietin. Cells cultured for 42 hours were labeled for 3 hours with 10μM BrdU. EGFP+ cells were isolated and stained with anti-BrdU antibody followed by secondary antibody and TOTO-3, and the percentage of BrdU+ cells was calculated.
Statistical analysis
P values were calculated using 2-tailed unpaired Student t tests (for normal distribution), Wilcoxon tests (for non-normal distribution) for 2-group experiments, or Tukey multiple comparison tests (for multiple-group experiments).
Additional procedures can be found in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results and discussion
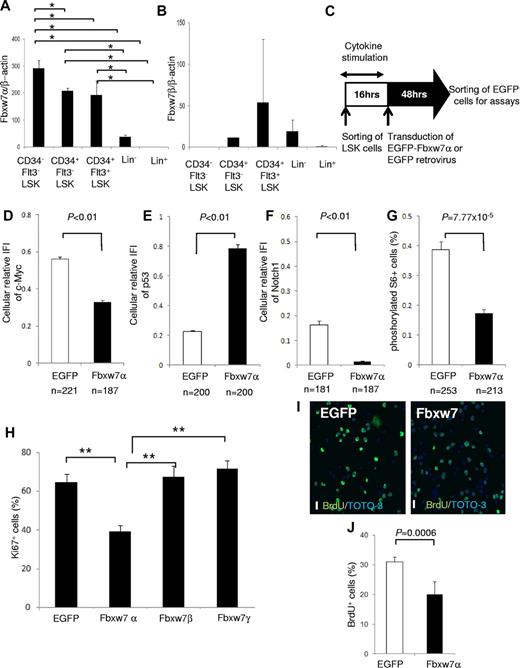
The Fbxw7 locus encodes 3 mRNA isoforms (Fbxw7α, β, and γ), each of which has a unique 5′ exon and 10 shared exons. Fbxw7α localizes to the nucleus, Fbxw7β displays a cytoplasmic distribution, and Fbxw7γ is predominantly nucleolar. To determine the importance of each isoform, the distribution of Fbxw7 mRNAs was investigated in hematopoietic cell fractions of mouse BM by quantitative reverse-transcribed polymerase chain reaction (RT-PCR) with isoform-specific primer sets.13 Fbxw7α mRNA was highly expressed in the more primitive fractions, such as CD34− Flt3− LSK cells, CD34+ Flt3− LSK cells, and CD34+ Flt3+ LSK cells (Figure 1A). By contrast, basal Fbxw7β mRNA was low in each fraction (Figure 1B), whereas Fbxw7γ mRNA was barely detectable in any fraction (data not shown).
Fbxw7α overexpression suppresses Fbxw7-target accumulation and HSC cell-cycle progression. (A-B) Quantitative PCR analysis of Fbxw7α (A) or Fbxw7β (B) transcripts in BM CD34− Flt3− LSK, CD34+ Flt3− LSK, CD34+ Flt3+ LSK, Lin−, or Lin+ fractions from 12-week-old mice. Each value was normalized to β-actin expression and is expressed as the fold induction compared with Lin+ samples (mean ± SD, n = 4). *P < .01. (C) Study design of Fbxw7α overexpression in LSK cells. LSK cells were transduced with pMY-IRES-EGFP (EGFP virus; control) or pMY-Fbxw7α-IRES-EGFP retrovirus (Fbxw7α virus). After 48 hours, EGFP+ cells were sorted and used for in vitro and in vivo assays. (D-F) EGFP+ cells were isolated from EGFP or Fbxw7α virus-transduced LSK cells. Isolated cells were stained with 4,6-diamidino-2-phenylindole (DAPI) and anti-c-Myc, p53, or Notch1 antibody. Expression levels of c-Myc, p53, and Notch1 were quantified by the integrated fluorescence intensity (IFI) of the c-Myc (D), p53 (E), or Notch1 (F) signal normalized to the IFI of the DAPI signal in independent cells (mean ± SEM). (G) EGFP or Fbxw7α virus-transduced EGFP+ cells were stained with DAPI and antiphosphorylated S6 antibody. Phosphorylated S6+ cells were counted (mean ± SEM). (H) EGFP or Fbxw7α, β, or γ virus-transduced cells were harvested, and CD41−CD48− LSK cells were sorted and stained with DAPI and anti-Ki67 antibody. At least 450 cells/sample were counted (mean ± SD, n = 5). **P < .00007. (I-J) Short-term BrdU-labeling experiment with EGFP or Fbxw7α virus-transduced EGFP+ cells. Transduced cells were sorted and cultured on fibronectin-coated plates. After 42 hours of culture, cells were labeled for 3 hours with 10μM BrdU and stained with anti-BrdU antibody (green) and TOTO-3 (blue; I). Images were obtained and analyzed using a confocal laser-scanning microscope (FV1000; Olympus). UPIanApp 20×/0.70 objective lens (Olympus) and FV10-ASW2.0 viewer (Olympus). Scale bars represent 20 μm. Quantification of BrdU-labeled cells (J). At least 1500 cells/sample were counted (mean ± SD, n = 5).
Fbxw7α overexpression suppresses Fbxw7-target accumulation and HSC cell-cycle progression. (A-B) Quantitative PCR analysis of Fbxw7α (A) or Fbxw7β (B) transcripts in BM CD34− Flt3− LSK, CD34+ Flt3− LSK, CD34+ Flt3+ LSK, Lin−, or Lin+ fractions from 12-week-old mice. Each value was normalized to β-actin expression and is expressed as the fold induction compared with Lin+ samples (mean ± SD, n = 4). *P < .01. (C) Study design of Fbxw7α overexpression in LSK cells. LSK cells were transduced with pMY-IRES-EGFP (EGFP virus; control) or pMY-Fbxw7α-IRES-EGFP retrovirus (Fbxw7α virus). After 48 hours, EGFP+ cells were sorted and used for in vitro and in vivo assays. (D-F) EGFP+ cells were isolated from EGFP or Fbxw7α virus-transduced LSK cells. Isolated cells were stained with 4,6-diamidino-2-phenylindole (DAPI) and anti-c-Myc, p53, or Notch1 antibody. Expression levels of c-Myc, p53, and Notch1 were quantified by the integrated fluorescence intensity (IFI) of the c-Myc (D), p53 (E), or Notch1 (F) signal normalized to the IFI of the DAPI signal in independent cells (mean ± SEM). (G) EGFP or Fbxw7α virus-transduced EGFP+ cells were stained with DAPI and antiphosphorylated S6 antibody. Phosphorylated S6+ cells were counted (mean ± SEM). (H) EGFP or Fbxw7α, β, or γ virus-transduced cells were harvested, and CD41−CD48− LSK cells were sorted and stained with DAPI and anti-Ki67 antibody. At least 450 cells/sample were counted (mean ± SD, n = 5). **P < .00007. (I-J) Short-term BrdU-labeling experiment with EGFP or Fbxw7α virus-transduced EGFP+ cells. Transduced cells were sorted and cultured on fibronectin-coated plates. After 42 hours of culture, cells were labeled for 3 hours with 10μM BrdU and stained with anti-BrdU antibody (green) and TOTO-3 (blue; I). Images were obtained and analyzed using a confocal laser-scanning microscope (FV1000; Olympus). UPIanApp 20×/0.70 objective lens (Olympus) and FV10-ASW2.0 viewer (Olympus). Scale bars represent 20 μm. Quantification of BrdU-labeled cells (J). At least 1500 cells/sample were counted (mean ± SD, n = 5).
To analyze the impact of Fbxw7α in HSCs, Fbxw7α was overexpressed in murine LSK cells using a retroviral vector (Figure 1C). Sorted mouse LSK cells were transduced by control EGFP or Fbxw7α virus. After transduction, EGFP+ cells were sorted and used for the following assays. Immunocytochemistry and quantitative RT-PCR revealed that Fbxw7α transcript and protein levels were elevated after transduction (supplemental Figure 1A-C). Overexpressed Fbxw7α in primitive hematopoietic cells predominantly localized in the nucleoplasm and slightly in the cytoplasm but not in the nucleolus (supplemental Figure 2). Overexpressed Fbxw7β was distributed predominantly in the cytoplasm and slightly in the nucleoplasm (supplemental Figure 2). By contrast, overexpressed Fbxw7γ, which was originally identified as a nucleolar isoform of Fbxw7, localized mainly in the nucleoplasm, but not in the nucleolus (supplemental Figure 2). Therefore, the nucleoplasmic localization Fbxw7α might be essential for cellular quiescence in HSCs.
Among the Fbxw7 targets, expression of c-Myc, Notch1 (Figure 1D,F; supplemental Figure 3), and phosphorylated S6 (a downstream element of mTOR; Figure 1G; supplemental Figure 3) was reduced in Fbxw7α virus-transduced HSCs, whereas p53 expression was up-regulated (Figure 1E; supplemental Figure 3). C-Myc, Notch1, and mTOR contribute to cell growth and proliferation in HSCs,7,8,10,14 whereas p53 regulates HSC quiescence.15 Next, the functional effects of Fbxw7α overexpression on HSC cell cycle were investigated. Fbxw7α virus-transduced HSCs contained fewer Ki67+ cells and a reduced short-term BrdU uptake compared with EGFP virus-transduced HSCs (Figure 1H-J). On the other hand, the percentage of Ki67+ cells in Fbxw7β and γ virus-transduced HSCs was equivalent to control (Figure 1H). These data suggest that forced Fbxw7α expression represses HSC cell-cycle progression by promoting c-Myc, Notch1, and mTOR protein degradation and up-regulation of p53.
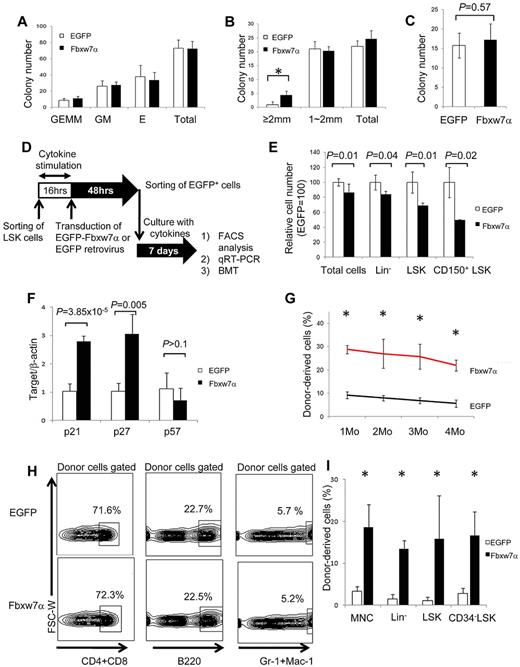
Hematopoietic progenitor assays (CFU-C and CFU-S12) revealed no significant differences between EGFP and Fbxw7α virus-transduced HSCs (Figure 2A,C). By contrast, 4 times more “large” HPP-CFC-derived colonies (> 2 mm in diameter) representing the more primitive hematopoietic cell population were generated from Fbxw7α virus-transduced LSK cells (Figure 2B). Therefore, forced Fbxw7α expression in LSK cells does not affect relatively differentiated hematopoietic progenitors but might affect more primitive hematopoietic progenitors.
Repopulation activity of Fbxw7α-transduced HSCs after ex vivo culture. (A) In vitro colony formation assays of EGFP or Fbxw7α virus-transduced EGFP+ cells. Colonies were counted and classified as granulocyte, erythroid, monocyte, and megakaryocyte (GEMM), granulocyte and monocyte (GM), or erythroid (E) on day 9 of culture (mean ± SD, n = 3). (B) Number of highly proliferative colony-forming EGFP or Fbxw7α virus-transduced EGFP+ cells on day 14 of culture (mean ± SD, n = 3). *P < .005. (C) CFU-S12 assay. EGFP or Fbxw7α virus-transduced EGFP+ cells were transplanted into 8.5-Gy irradiated recipient mice. Spleens were harvested after 12 days, fixed with Bounin solution, and CFU-S12 colonies were counted (mean ± SD, n = 5). (D) Study design for transplantation experiment. EGFP virus or Fbxw7α virus was transduced into LSK cells (CD45.1+). After 48 hours, EGFP+ cells were sorted and cultured for 1 week (18 000 LSK cells/well). Cultured cells were transplanted into lethally irradiated recipient mice (CD45.2+) with 4 × 105 CD45.2+ competitors. PB chimerism was analyzed monthly, and BM chimerism was analyzed at 4 months after transplantation. Donor-derived LSK cells were used for serial transplantation after 4 months. (E) Relative numbers of total, Lin−, LSK, and CD150+ LSK EGFP or Fbxw7α virus-transduced EGFP+ cells after 1 week of culture. Cell number was analyzed by flow cytometry (mean ± SD, n = 6 for EGFP, n = 2 for Fbxw7). (F) Quantitative PCR analysis of various cell cycle–related genes in EGFP or Fbxw7α virus-transduced LSK cells after 1 week of culture (n = 4). (G) PB chimerism in primary BMT recipients of EGFP or Fbxw7α virus-transduced cells after 1 week of culture at the indicated times after BMT (mean ± SEM, n = 4 or 5). *P < .05. (H) Differentiation status (CD3+ or CD4/8+ T cells, B220+ B cells, or Mac-1/Gr-1+ myeloid cells) of PB in primary BMT recipients of cultured EGFP or Fbxw7α virus-transduced cells at 4 months after BMT. (I) Donor-derived (Ly5.1+) chimerism in BM mononuclear cells, Lin−, LSK, or CD34− LSK fractions in primary BMT recipients of cultured EGFP or Fbxw7α virus-transduced cells at 4 months after BMT (mean ± SD, n = 4 or 5). *P < .05.
Repopulation activity of Fbxw7α-transduced HSCs after ex vivo culture. (A) In vitro colony formation assays of EGFP or Fbxw7α virus-transduced EGFP+ cells. Colonies were counted and classified as granulocyte, erythroid, monocyte, and megakaryocyte (GEMM), granulocyte and monocyte (GM), or erythroid (E) on day 9 of culture (mean ± SD, n = 3). (B) Number of highly proliferative colony-forming EGFP or Fbxw7α virus-transduced EGFP+ cells on day 14 of culture (mean ± SD, n = 3). *P < .005. (C) CFU-S12 assay. EGFP or Fbxw7α virus-transduced EGFP+ cells were transplanted into 8.5-Gy irradiated recipient mice. Spleens were harvested after 12 days, fixed with Bounin solution, and CFU-S12 colonies were counted (mean ± SD, n = 5). (D) Study design for transplantation experiment. EGFP virus or Fbxw7α virus was transduced into LSK cells (CD45.1+). After 48 hours, EGFP+ cells were sorted and cultured for 1 week (18 000 LSK cells/well). Cultured cells were transplanted into lethally irradiated recipient mice (CD45.2+) with 4 × 105 CD45.2+ competitors. PB chimerism was analyzed monthly, and BM chimerism was analyzed at 4 months after transplantation. Donor-derived LSK cells were used for serial transplantation after 4 months. (E) Relative numbers of total, Lin−, LSK, and CD150+ LSK EGFP or Fbxw7α virus-transduced EGFP+ cells after 1 week of culture. Cell number was analyzed by flow cytometry (mean ± SD, n = 6 for EGFP, n = 2 for Fbxw7). (F) Quantitative PCR analysis of various cell cycle–related genes in EGFP or Fbxw7α virus-transduced LSK cells after 1 week of culture (n = 4). (G) PB chimerism in primary BMT recipients of EGFP or Fbxw7α virus-transduced cells after 1 week of culture at the indicated times after BMT (mean ± SEM, n = 4 or 5). *P < .05. (H) Differentiation status (CD3+ or CD4/8+ T cells, B220+ B cells, or Mac-1/Gr-1+ myeloid cells) of PB in primary BMT recipients of cultured EGFP or Fbxw7α virus-transduced cells at 4 months after BMT. (I) Donor-derived (Ly5.1+) chimerism in BM mononuclear cells, Lin−, LSK, or CD34− LSK fractions in primary BMT recipients of cultured EGFP or Fbxw7α virus-transduced cells at 4 months after BMT (mean ± SD, n = 4 or 5). *P < .05.
To test this possibility, flow cytometry, quantitative RT-PCR, and competitive BMT were used to evaluate the stem cell capacity of Fbxw7α virus-transduced LSK cells (Figure 2D). The absolute cell numbers in several fractions were compared between EGFP and Fbxw7α virus-transduced HSCs after 1 week of ex vivo culture. The cell numbers of Fbxw7α virus-transduced LSK cells were reduced in all fractions (total cells, Lin−, LSK, and CD150+ LSK; Figure 2E). Quantitative RT-PCR analysis revealed increased expression of Cdk inhibitor, p21cip1and p27Kip1, but not p57kip2, in Fbxw7α virus-transduced HSCs (Figure 2F). Fbxw7α overexpression appeared to inhibit HSC cell cycle progression that attenuates stem cell capacity via p21cip1 and p27Kip1.
The reconstitution capacity of EGFP or Fbxw7α virus-transduced LSK cells was also tested. In primary recipients, Fbxw7α virus-transduced donor cells achieved greater peripheral blood (PB) chimerism up to 4 months after BMT (Figure 2G). Because Fbxw7 deficiency affects differentiation of T cells, the PB differentiation status of Fbxw7α-overexpressing cells was investigated after BMT. Fbxw7α donor-derived PB differentiation showed no abnormalities at 4 months after BMT (Figure 2H; supplemental Figure 4). Fbxw7α donor-derived BM chimerism at this time was significantly greater than control in each fraction (eg, BM mononuclear cells, Lin−, LSK, or CD34− LSK; Figure 2I). Although EGFP virus-transduced donor-derived cells did not show any reconstitution in secondary recipients, significant numbers of Fbxw7α-transduced donor cells were detected in the PB (supplemental Table 1). Collectively, these results indicate that Fbxw7α overexpression represses cell-cycle progression and maintains high stem cell capacity during ex vivo culture. Importantly, Fbxw7α overexpression promoted HSC maintenance without impairing the progenitor capacity or multidifferentiation capability of Fbxw7α-overexpressing LSK cells.
Hypoxia maintains HSCs ex vivo by suppressing the cell cycle and inducing quiescence.12,16 Hypoxia suppresses the activity of c-Myc17 (supplemental Figure 5A,D) and mTOR signaling18 and induces Cdk inhibitors.19 Interestingly, hypoxia up-regulated Fbxw7 protein expression in HSCs (supplemental Figure 5B-C), particularly in the nucleus (supplemental Figure 5C), suggesting that Fbxw7α was the up-regulated isoform. In support of this, whereas the protein levels of neither Notch1 nor phosphorylated S6 were suppressed under hypoxic conditions (supplemental Figure 5F-I), c-Myc was suppressed (supplemental Figure 5D), but not in Fbxw7-deficient LSK cells (supplemental Figure 5E). Therefore, the mechanism of c-Myc suppression during hypoxia is regulated in an Fbxw7α-dependent manner. The results of the present study indicate that the hypoxia-Fbxw7α pathway is one mechanism by which low oxygen tension plays a crucial role in maintaining normal HSC function.20 In addition to hypoxic culture, modulation of Fbxw7α expression or activity may be a promising tool for the ex vivo maintenance of HSCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank T. Kitamura for Plat E packaging cells and pMY-IRES-EGFP vector, M. Suematsu for FACS resources, T. Muraki and T. Hirose for essential support, and N. Tago for FACS operation.
H.I. is a research fellow of JSPS. K.T. is supported by a Grant-in-Aid for Young Scientists (B) and a Grant-in-Aid for Scientific Research on Priority Areas. T.S. was supported by a Grant-in-Aid for Scientific Research on Innovative Areas and a Grant-in-Aid for Scientific Research (S).
Authorship
Contribution: H.I. performed experiments, analyzed data, and wrote the paper; K.T. designed research, performed experiments, analyzed data, and wrote the paper; S.M., I.O., and K.I.N. prepared materials; Y.N. organized research; and T.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Keiyo Takubo, Department of Cell Differentiation, Sakaguchi Laboratory of Developmental Biology, Keio University School of Medicine, 35 Shinano-machi, Shinjuku-ku, Tokyo 160-8582, Japan; e-mail: keiyot@gmail.com; and Toshio Suda, Department of Cell Differentiation, Sakaguchi Laboratory of Developmental Biology, Keio University School of Medicine, 35 Shinano-machi, Shinjuku-ku, Tokyo 160-8582, Japan; e-mail: sudato@sc.itc.keio.ac.jp.
References
Author notes
H.I. and K.T. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal