Abstract
Several drugs used for diffuse large B-cell lymphoma (DLBCL) treatment rely on DNA damage for tumor cell killing. We verified the prognostic impact of the host DNA repair genotype in 2 independent cohorts of DLBCL treated with R-CHOP21 (training cohort, 163 cases; validation cohort, 145 cases). Among 35 single nucleotide polymorphisms analyzed in the training series, MLH1 rs1799977 was the sole predicting overall survival. DLBCL carrying the MLH1 AG/GG genotype displayed an increased death risk (hazard ratio [HR] = 3.23; P < .001; q =0 .009) compared with patients carrying the AA genotype. Multivariate analysis adjusted for International Prognostic Index identified MLH1 AG/GG as an independent OS predictor (P < .001). The poor prognosis of MLH1 AG/GG was the result of an increased risk of failing both R-CHOP21 (HR = 2.02; P = .007) and platinum-based second-line (HR = 2.26; P = .044) treatment. Survival analysis in the validation series confirmed all outcomes predicted by MLH1 rs1799977. The effect on OS of MLH1, a component of the DNA mismatch repair system, is consistent with its role in regulating the genotoxic effects of doxorubicin and platinum compounds, which are a mainstay of DLBCL first- and second-line treatment.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is characterized by a heterogeneous clinical course, which may be predicted to some extent by the International Prognostic Index (IPI).1,2 In addition to clinical prognosticators, several biologic markers have been proposed as tools for refining outcome stratification in DLBCL.3-6 Most of these markers rely on features of the tumor clone.3-6
The genetic background of the host may also be relevant for DLBCL outcome. At present, evidence pointing to a role of the genetic background of the host in driving DLBCL prognosis is restricted to a limited number of studies that have focused on single nucleotide polymorphisms (SNPs) of immune system genes and of genes involved in drug metabolism.7-11
DNA damage is one of the mainstays of cancer treatment. Several drugs used in DLBCL treatment rely on DNA damage as part of the mechanisms of tumor cell killing.12-17 These include drugs of the R-CHOP regimen (rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone)18-23 as well as drugs of second-line regimens containing platinum compounds.24,25
Tumor cells respond to DNA damage induced by chemotherapy by activating 5 major DNA repair pathways: (1) mismatch repair (MMR),12-14 (2) base excision repair,12-14 (3) nucleotide excision repair,12-14 (4) double strand break repair,15 and (5) direct reversal.16
MMR, of which MLH1 is a major component, recognizes and removes mismatched or unmatched DNA base pairs or insertion-deletion loops.12-14 MMR is tightly involved in mediating cytotoxicity of doxorubicin and platinum compounds.13,17 DNA adducts produced by doxorubicin and platinum compounds exert a double genotoxic effect because they: (1) induce nucleotide mismatches and (2) interfere with normal MMR activity, thus preventing the repair of nucleotide mismatches from being completed.13,17 In cells characterized by MMR competence, the excess of unrepaired DNA triggers apoptosis. In contrast, when MMR is deficient, cells become chemorefractory and proliferate despite DNA damage caused by doxorubicin and platinum compounds.13,17 Chemorefractoriness resulting from alterations of DNA repair mechanisms may also involve base excision repair in the case of doxorubicin and alkylating agents,12-14 nucleotide excision repair in the case of platinum compounds and alkylating agents,12-14 double strand break repair in the case of doxorubicin and etoposide,15 and direct reversal in the case of alkylating agents.16
On these grounds, DNA repair mechanisms and DNA repair capacity represent attractive candidates for explaining interindividual heterogeneity in treatment response of DLBCL. This study aimed at verifying whether SNPs of genes involved in DNA repair pathways may contribute to the prognostic stratification of DLBCL patients treated with R-CHOP.
Methods
Patients and study design
The design of the study was based on a training-validation approach. Accordingly, the patient population consisted of 2 cohorts of DLBCL treated with R-CHOP21 from December 2001 through November 2008. Patients were Italian residents and self-reported to be of white ancestry. Diagnosis of DLBCL was based on the World Health Organization classification of Hematopoietic Tumors.1 A prior history of lymphoproliferative disorders was ruled out in all cases.
The first cohort (n = 163) was used as learning series because this cohort: (1) was a consecutive series from a single institution (Amedeo Avogadro University of Eastern Piedmont); (2) was provided with a large and homogeneous dataset of prospectively collected variables; (3) was homogeneously treated with the same chemotherapeutic regimen both at DLBCL diagnosis (R-CHOP21) and at relapse/progression (R-DHAP, rituximab, desamethasone, high-dose cytarabine, cisplatin); and (4) was genotyped on DNA from peripheral blood granulocytes in all cases. The following clinical variables were recorded at presentation: date of diagnosis, age, sex, Eastern Cooperative Oncology Group (ECOG) performance status (PS), Ann Arbor stage,26 number of extranodal sites, B symptoms, bulky disease more than 10 cm, liver, and bone marrow involvement; lactate dehydrogenase (LDH, normal range, 200-450 U/L), absolute neutrophil count, hemoglobin and platelet count, bilirubin (normal range, 0.6-1.2 mg/dL), alkaline phosphatase (normal range, 90-360 U/L), albumin (normal range, 34-48 g/L), glomerular filtration rate; cardiac ejection fraction, abnormalities on echocardiography, abnormalities on electrocardiogram; hepatitis C virus, hepatitis B virus, and HIV serology; and number and type of comorbidities according to the Comorbidity Index and Score of Charlson.27 The following variables were recorded during treatment: total number of courses per patient, date and type of response, and date of progression. In addition, the following variables were also recorded for each course of R-CHOP21: date of administration, doses of rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone, delay, toxicity, and dose reduction. The following variables were recorded during follow-up: date of progression, date and type of second-line treatment, date of stem cell transplantation, date of last follow-up visit, date of death, and cause of death.
The second cohort was used as validation series (n = 145). The validation series of DLBCL was composed of a retrospective multi-institutional series from centers based in Rome and participating in the same national lymphoma group (La Sapienza University, Rome, Italy; and Catholic University of the Sacred Heart, Rome, Italy). Inclusion criteria for the validation series were as follows: (1) diagnosis of DLBCL and (2) first-line treatment with R-CHOP21. SNP genotyping was performed on DNA extracted from formalin-fixed, paraffin-embedded tissue biopsies, or peripheral blood granulocytes when available. Second-line regimens used at relapse/progression included platinum compounds in 36 of 39 patients. The following clinical variables were recorded in the validation cohort: date of diagnosis, age, sex, ECOG PS, Ann Arbor stage, number of extranodal sites, bulky disease more than 10 cm, LDH, date of progression, date and type of second-line treatment, date of stem cell transplantation, date of last follow-up visit, and date of death.
Cases with a diagnosis of primary mediastinal large B-cell lymphoma, cases presenting with central nervous system involvement, and cases harboring HIV infection were excluded from both the training and the validation series.
Involved field radiotherapy was part of the initial treatment plan as consolidation on sites of primary bulky disease for both the training and the validation series. Involved field radiotherapy was administered to 44 of 163 (27.0%) patients in the training series and to 37 of 145 (25.5%) patients in the validation series.
In both training and validation series, response was evaluated at the end of the fourth course and at the end of R-CHOP21 treatment according to the 1999 National Cancer Institute-Sponsored International Working Group guidelines.28 Follow-up visits were scheduled every 3 months for the first 2 years and every 6 months thereafter. In case of nonresponse or progression after R-CHOP21, patients younger than 65 years were offered BEAM (carmustine, etoposide, cytarabine, melphalan) conditioned autologous stem cell transplantation as consolidation after salvage treatment.
Patients provided informed consent in accordance with the Declaration of Helsinki and with approval from the Amedeo Avogadro University of Eastern Piedmont Institutional Review Board.
SNP genotyping
Nonsynonymous SNPs, promoter SNPs, splicing site SNPs, and miRNA binding site SNPs were selected using an educated guess approach based on the following criteria: (1) minor allele frequency more than 5% in whites according to NCBI (http://www.ncbi.nlm.nih.gov/snp; accessed August 29, 2009); (2) previously reported functional consequence in vitro/in vivo; and/or (3) functional consequences predicted in silico according to PupaSuite (http://pupasuite.bioinfo.cipf.es; accessed August 29, 2009), PolyPhen (http://genetics.bwh.harvard.edu/pph; accessed August 29, 2009), or SIFT (http://blocks.fhcrc.org/sift/SIFT.html; accessed August 29, 2009); and/or (4) clinical relevance in settings other than DLBCL.
Sample size calculation was performed according to the assumption of number of patients with the genotype at risk more than or equal to 10 (corresponding to a minor allele frequency of ≥ 6%). Based on this assumption, the sample size of the training series (n = 163) would allow detection of at least 30% difference in 3-year overall survival (OS) with a power of 81% (alpha = 0.05).
A total of 35 SNPs from 18 genes were analyzed (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These included SNPs affecting: (1) MMR genes (MLH1 rs1799977 and MLH1 rs1800734); (2) base excision repair genes (XRCC1 rs1799782, XRCC1 rs25487, and OGG1 rs1052133); (3) nucleotide excision repair genes (ERCC1 rs3212986, ERCC2 rs1052555, ERCC2 rs13181, ERCC2 rs1799793, ERCC2 rs238406, ERCC4 rs1800067, ERCC4 rs3136038, ERCC5 rs17655, ERCC6 rs2228528, ERCC6 rs2228529, ERCC6 rs3793784, XPA rs1800975, XPC rs2227999, XPC rs2228000, XPC rs2607775, and XPC rs2228001); (4) double strand break repair genes (BRCA1 rs4986850, BRCA1 rs1799950, BRCA1 rs799917, BRCA2 rs144848, LIG4 rs1805388, XRCC2 rs3218536, XRCC3 rs1799794, XRCC3 rs861539, XRCC4 rs1805377, XRCC6 rs5751129, and XRCC6 rs132788); and (5) direct reversal genes (MGMT rs16906252, MGMT rs2308321, and MGMT rs12917).
SNP genotyping was performed by SNP minisequencing (ABI Prism SNaPshot Multiplex kit, Applied Biosystems), after validation of this approach by DNA direct sequencing of each SNP in a pilot panel of cases (n = 15; data not shown). Quality control of genotyping was performed by replicate sample analysis. The concordance of SNP genotyping performed on granulocytes and on tumor biopsy from the same patients (n = 100) was 100%.
Deviation of SNP genotype distribution from Hardy-Weinberg equilibrium was tested by χ2 test or Fisher exact test if appropriate. Five SNPs were not in Hardy-Weinberg equilibrium (supplemental Table 1) and therefore were excluded from subsequent analysis. Linkage disequilibrium among SNPs was calculated as r2 values with the use of HAPLOVIEW (http://www.broad.mit.edu/haploview/haploview; accessed August 29, 2009). None of the SNPs was in strong linkage disequilibrium (r2 > 0.8).
Immunohistochemistry
Immunohistochemistry for MLH1 was performed on formalin-fixed, paraffin-embedded DLBCL tissue sections from tissue microarrays (n = 35) and single sections (n = 23) using the mouse monoclonal antibody G168-15 (BD Biosciences PharMingen) against hMLH1. A score of 0 to 3 for stain intensity was assigned: no staining = 0; weakly positive = 1; moderately positive = 2; and strongly positive = 3. Percentage staining was assessed according to the following scoring system: 0% to 5% = 0; 6% to 20% = 1; 21% to 80% = 2; and 81% to 100% = 3. A combined immunohistochemical score was achieved by multiplying the percentage by the intensity score.29 Both immunostaining and scoring were blinded to MLH1 rs1799977 genotype. For the tissue microarray construction, hematoxylin and eosin-stained sections from each block were used to define diagnostic areas. Three representative 0.6-mm cores were obtained from each case and inserted in a grid pattern into a recipient paraffin block using a tissue arrayer (Advanced Tissue Arrayer ATA-100; Chemicon International-Millipore).30 Cell of origin was assigned as previously reported.30 Immunohistochemical studies for CD10, BCL-6, and IRF4 were performed on paraffin-embedded tissue sections as previously described.30,31
Statistical analysis
The impact of DNA repair SNPs on R-CHOP efficacy was evaluated by OS. For the purpose of this study, which focused on DNA repair pathways associated with both R-CHOP and second-line regimen drugs, OS was considered as the primary endpoint because it may be influenced by both R-CHOP and second-line treatments. OS was measured from date of DLBCL diagnosis to date of death as a result of any cause (event), or last follow-up (censoring).28 Progression-free survival (PFS) was measured from date of DLBCL diagnosis to date of disease progression or death as a result of any cause (event), or last follow-up (censoring).28 OS after second-line treatment was measured from date of second-line treatment to date of death as a result of any cause (event), transplantation (censoring), or last follow-up (censoring). PFS after second-line treatment was measured from date of second-line treatment to date of progression (event), death as a result of any cause (event), transplantation (censoring) or last follow-up (censoring). Response was assessed according to published guidelines.28 The association between SNPs and efficacy endpoints was evaluated considering the minor allele of each SNP as acting both in a recessive and in a dominant fashion. For SNPs where 10 or fewer minor allele homozygotes were observed, only the combination of minor allele homozygotes with heterozygotes was analyzed. If this combined frequency was still less than 10, then the SNP was removed from the analysis. Survival analysis was performed using the Kaplan-Meier method.32 Univariate and multivariate analyses were performed by Cox regression.33 IPI score entered Cox analysis as an ordinal variable with 4 levels (low, low-intermediate, high-intermediate, and high).2 False discovery rate was used to control for multiple statistical testing for each SNP as acting both in a recessive and in a dominant fashion.34 Categorical variables were compared by χ2 test. Continuous variables were compared by Mann-Whitney test. All statistical tests were 2-sided. Statistical significance was defined as P value less than .05. The analysis was performed with the Statistical Package for the Social Sciences software Version 17.0, R statistical package 2.8.1 (http://www.r-project.org; accessed October 11, 2010).
Results
Clinical characteristics at diagnosis and treatment of the DLBCL training series
All 35 SNPs from 18 genes belonging to 5 different DNA repair pathways were genotyped in the training series of 163 newly diagnosed DLBCL homogeneously treated with R-CHOP21 (supplemental Table 1).
The clinical characteristics at DLBCL diagnosis of the training series are described in Table 1. Overall, 974 courses of R-CHOP21 were administered. The median number of courses per patient was 6 (25th-75th percentile, 5-8) and 120 of 163 (73.6%) patients received at least 6 R-CHOP21 courses. Median relative dose intensity (RDI) of R-CHOP21 drugs was 100% for rituximab, 95.4% for cyclophosphamide, 94.0% for doxorubicin, 100% for vincristine, and 100% for prednisone. Complete remission (CR)/unconfirmed CR after R-CHOP21 was achieved in 127 of 163 (77.9%) patients. PFS at 4 years was 57.9%. OS at 4 years was 72.9%. Forty-six patients failed or progressed after R-CHOP21 and required R-DHAP salvage treatment. Autologous stem cell transplantation consolidation after salvage treatment was performed in 15 patients younger than 65 years.
Clinical characteristics of the DLBCL cohorts at diagnosis
| Clinical characteristic . | Training series (n = 163) . | Validation series (n = 145) . | P . |
|---|---|---|---|
| Age > 60 y | 104/163 (63.8%) | 96/145 (66.2%) | .659 |
| Male:female | 92:71 | 66:79 | .056 |
| ECOG PS > 1 | 21/163 (12.9%) | 23/145 (15.9%) | .456 |
| Ann Arbor stage | .301 | ||
| I | 34/163 (20.9%) | 25/145 (17.2%) | — |
| II | 44/163 (27.0%) | 51/145 (35.2%) | — |
| III | 29/163 (17.8%) | 18/145 (12.4%) | — |
| IV | 56/163 (34.4%) | 51/145 (35.2%) | — |
| B symptoms | 38/163 (23.3%) | ||
| Bulky | 46/163 (28.2%) | 37/145 (25.5%) | .593 |
| Extranodal sites > 1 | 41/163 (25.2%) | 28/145 (19.3%) | .220 |
| LDH > ULN | 54/163 (33.1%) | 66/145 (45.5%) | .931 |
| IPI | .526 | ||
| Low | 80/163 (49.1%) | 65/145 (44.8%) | — |
| Low-intermediate | 34/163 (20.9%) | 33/145 (22.8%) | — |
| High-intermediate | 25/163 (15.3%) | 29/145 (20.0%) | — |
| High | 24/163 (14.7%) | 18/145 (12.4%) | — |
| Non-GC phenotype | 53/127 (41.7%) | ||
| Bone marrow function | |||
| ANC, ×109/L | 4.5 (3.6-5.7) | — | — |
| Hb, g/dL | 12.9 (11.8-14.1) | — | — |
| Platelets, ×109/L | 256 (206-320) | — | — |
| Liver, renal, and cardiac function | |||
| Albumin, g/L | 4.1 (3.7-4.4) | — | — |
| ALP, U/L | 174 (147-233) | — | — |
| Bilirubin, mg/dL | 0.7 (0.5-0.9) | — | — |
| Glomerular filtration rate, mL/min | 61 (48-77) | — | — |
| Cardiac ejection fraction, % | 59 (55-64) | — | — |
| Comorbidities | |||
| Charlson Comorbidity Index | 1 (0-2) | — | — |
| Clinical characteristic . | Training series (n = 163) . | Validation series (n = 145) . | P . |
|---|---|---|---|
| Age > 60 y | 104/163 (63.8%) | 96/145 (66.2%) | .659 |
| Male:female | 92:71 | 66:79 | .056 |
| ECOG PS > 1 | 21/163 (12.9%) | 23/145 (15.9%) | .456 |
| Ann Arbor stage | .301 | ||
| I | 34/163 (20.9%) | 25/145 (17.2%) | — |
| II | 44/163 (27.0%) | 51/145 (35.2%) | — |
| III | 29/163 (17.8%) | 18/145 (12.4%) | — |
| IV | 56/163 (34.4%) | 51/145 (35.2%) | — |
| B symptoms | 38/163 (23.3%) | ||
| Bulky | 46/163 (28.2%) | 37/145 (25.5%) | .593 |
| Extranodal sites > 1 | 41/163 (25.2%) | 28/145 (19.3%) | .220 |
| LDH > ULN | 54/163 (33.1%) | 66/145 (45.5%) | .931 |
| IPI | .526 | ||
| Low | 80/163 (49.1%) | 65/145 (44.8%) | — |
| Low-intermediate | 34/163 (20.9%) | 33/145 (22.8%) | — |
| High-intermediate | 25/163 (15.3%) | 29/145 (20.0%) | — |
| High | 24/163 (14.7%) | 18/145 (12.4%) | — |
| Non-GC phenotype | 53/127 (41.7%) | ||
| Bone marrow function | |||
| ANC, ×109/L | 4.5 (3.6-5.7) | — | — |
| Hb, g/dL | 12.9 (11.8-14.1) | — | — |
| Platelets, ×109/L | 256 (206-320) | — | — |
| Liver, renal, and cardiac function | |||
| Albumin, g/L | 4.1 (3.7-4.4) | — | — |
| ALP, U/L | 174 (147-233) | — | — |
| Bilirubin, mg/dL | 0.7 (0.5-0.9) | — | — |
| Glomerular filtration rate, mL/min | 61 (48-77) | — | — |
| Cardiac ejection fraction, % | 59 (55-64) | — | — |
| Comorbidities | |||
| Charlson Comorbidity Index | 1 (0-2) | — | — |
The 25th to 75th percentiles are reported in parentheses for continuous variables.
CG indicates germinal center; ANC, absolute neutrophil count; Hb, hemoglobin; ALP, alkaline phosphatase; and —, not applicable.
The MLH1 rs1799977 genotype is a predictor of OS in DLBCL
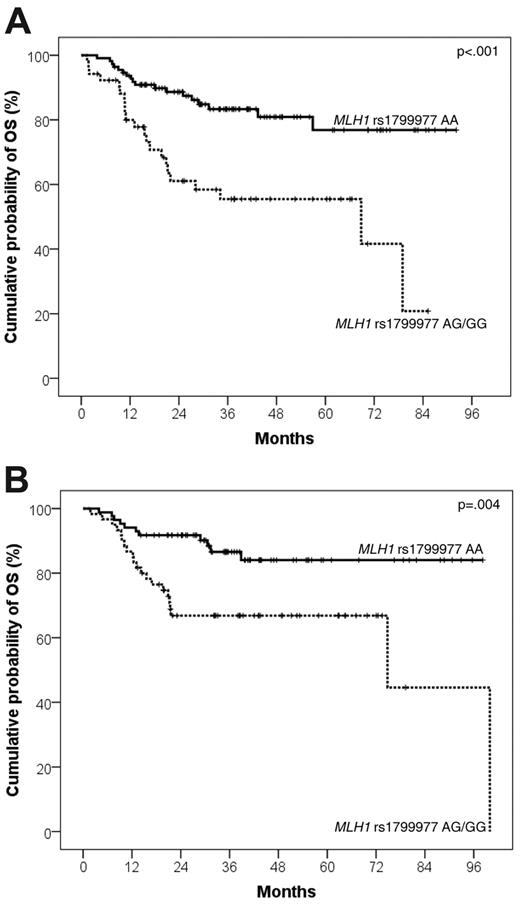
Univariate Cox analysis controlled for multiple comparisons by false discovery rate testing identified MLH1 rs1799977 as the sole SNP predicting OS in the DLBCL training series (supplemental Table 1). DLBCL patients who carried the MLH1 rs1799977 AG/GG genotype displayed an increased risk of death (hazard ratio [HR] = 3.23; events/N = 22/52; 4-year OS, 55.5%: 95% confidence interval [CI], 40.7%-70.3%) compared with patients who carried the MLH1 rs1799977 AA genotype (events/N = 18/111; 4-year OS, 80.9%; 95% CI, 72.1%-89.7%; P < .001; q = .009; Table 2; Figure 1A).
Univariate analysis for OS and PFS after R-CHOP21 in the training and validation DLBCL series
| . | Training series . | Validation series . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | LCI . | UCI . | P . | HR . | LCI . | UCI . | P . | |
| OS | ||||||||
| MLH1 rs1799977 AG/GG | 3.23 | 1.73 | 6.03 | < .001* | 2.97 | 1.42 | 6.21 | .004* |
| Age > 60 y | 2.00 | 0.95 | 4.20 | .068 | 3.71 | 1.29 | 10.62 | .014* |
| Male | 1.59 | 0.83 | 3.04 | .162 | 1.69 | 0.83 | 3.46 | .147 |
| ECOG PS > 1 | 2.79 | 1.32 | 5.92 | .007* | 1.05 | 0.41 | 2.80 | .883 |
| Ann Arbor stage III or IV | 1.23 | 0.66 | 2.30 | .515 | 1.87 | 0.91 | 3.87 | .088 |
| Bulky | 2.92 | 1.55 | 5.48 | < .001* | 1.81 | 0.86 | 3.79 | .114 |
| Extranodal sites > 1 | 2.06 | 1.08 | 3.92 | .027* | 2.56 | 1.22 | 5.35 | .012* |
| LDH > ULN | 2.52 | 1.35 | 4.69 | .003* | 2.77 | 1.30 | 5.89 | .008* |
| IPI score | 1.54 | 1.18 | 2.00 | .001* | 1.88 | 1.37 | 2.58 | < .001* |
| Non-GC phenotype | 1.68 | 0.59 | 4.36 | .352 | NA | NA | NA | NA |
| RDI cyclophosphamide < 90% | 1.69 | 0.91 | 3.14 | .099 | NA | NA | NA | NA |
| RDI doxorubicin < 90% | 2.02 | 1.08 | 3.78 | .028* | NA | NA | NA | NA |
| Radiotherapy | 1.11 | 0.57 | 2.16 | .749 | 1.08 | 0.50 | 2.37 | .830 |
| PFS | ||||||||
| MLH1 rs1799977 AG/GG | 2.02 | 1.20 | 3.38 | .007* | 1.92 | 1.09 | 3.39 | .024* |
| Age > 60 y | 1.74 | 0.97 | 3.12 | .064 | 2.75 | 1.28 | 5.89 | .009* |
| Male | 1.35 | 0.79 | 2.31 | .364 | 1.34 | 0.76 | 2.36 | .311 |
| ECOG PS > 1 | 1.84 | 0.93 | 3.64 | .081 | 1.06 | 0.50 | 2.85 | .863 |
| Ann Arbor stage III or IV | 1.91 | 1.12 | 3.26 | .017* | 2.87 | 1.56 | 5.30 | .001* |
| Bulky | 2.33 | 1.39 | 3.91 | .001* | 1.26 | 0.67 | 2.35 | .460 |
| Extranodal sites > 1 | 1.83 | 1.07 | 3.15 | .028* | 2.98 | 1.64 | 5.39 | < .001* |
| LDH > ULN | 1.91 | 1.14 | 3.22 | .014* | 2.52 | 1.39 | 4.55 | .002* |
| IPI score | 1.50 | 1.21 | 1.86 | < .001* | 1.85 | 1.44 | 2.38 | < .001* |
| Non-GC phenotype | 1.05 | 0.50 | 2.18 | .897 | NA | NA | NA | NA |
| RDI cyclophosphamide < 90% | 1.42 | 0.85 | 2.37 | .183 | NA | NA | NA | NA |
| RDI doxorubicin < 90% | 1.47 | 0.88 | 2.46 | .136 | NA | NA | NA | NA |
| Radiotherapy | .096 | 0.56 | 1.72 | .963 | 1.39 | 0.63 | 3.08 | .406 |
| . | Training series . | Validation series . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | LCI . | UCI . | P . | HR . | LCI . | UCI . | P . | |
| OS | ||||||||
| MLH1 rs1799977 AG/GG | 3.23 | 1.73 | 6.03 | < .001* | 2.97 | 1.42 | 6.21 | .004* |
| Age > 60 y | 2.00 | 0.95 | 4.20 | .068 | 3.71 | 1.29 | 10.62 | .014* |
| Male | 1.59 | 0.83 | 3.04 | .162 | 1.69 | 0.83 | 3.46 | .147 |
| ECOG PS > 1 | 2.79 | 1.32 | 5.92 | .007* | 1.05 | 0.41 | 2.80 | .883 |
| Ann Arbor stage III or IV | 1.23 | 0.66 | 2.30 | .515 | 1.87 | 0.91 | 3.87 | .088 |
| Bulky | 2.92 | 1.55 | 5.48 | < .001* | 1.81 | 0.86 | 3.79 | .114 |
| Extranodal sites > 1 | 2.06 | 1.08 | 3.92 | .027* | 2.56 | 1.22 | 5.35 | .012* |
| LDH > ULN | 2.52 | 1.35 | 4.69 | .003* | 2.77 | 1.30 | 5.89 | .008* |
| IPI score | 1.54 | 1.18 | 2.00 | .001* | 1.88 | 1.37 | 2.58 | < .001* |
| Non-GC phenotype | 1.68 | 0.59 | 4.36 | .352 | NA | NA | NA | NA |
| RDI cyclophosphamide < 90% | 1.69 | 0.91 | 3.14 | .099 | NA | NA | NA | NA |
| RDI doxorubicin < 90% | 2.02 | 1.08 | 3.78 | .028* | NA | NA | NA | NA |
| Radiotherapy | 1.11 | 0.57 | 2.16 | .749 | 1.08 | 0.50 | 2.37 | .830 |
| PFS | ||||||||
| MLH1 rs1799977 AG/GG | 2.02 | 1.20 | 3.38 | .007* | 1.92 | 1.09 | 3.39 | .024* |
| Age > 60 y | 1.74 | 0.97 | 3.12 | .064 | 2.75 | 1.28 | 5.89 | .009* |
| Male | 1.35 | 0.79 | 2.31 | .364 | 1.34 | 0.76 | 2.36 | .311 |
| ECOG PS > 1 | 1.84 | 0.93 | 3.64 | .081 | 1.06 | 0.50 | 2.85 | .863 |
| Ann Arbor stage III or IV | 1.91 | 1.12 | 3.26 | .017* | 2.87 | 1.56 | 5.30 | .001* |
| Bulky | 2.33 | 1.39 | 3.91 | .001* | 1.26 | 0.67 | 2.35 | .460 |
| Extranodal sites > 1 | 1.83 | 1.07 | 3.15 | .028* | 2.98 | 1.64 | 5.39 | < .001* |
| LDH > ULN | 1.91 | 1.14 | 3.22 | .014* | 2.52 | 1.39 | 4.55 | .002* |
| IPI score | 1.50 | 1.21 | 1.86 | < .001* | 1.85 | 1.44 | 2.38 | < .001* |
| Non-GC phenotype | 1.05 | 0.50 | 2.18 | .897 | NA | NA | NA | NA |
| RDI cyclophosphamide < 90% | 1.42 | 0.85 | 2.37 | .183 | NA | NA | NA | NA |
| RDI doxorubicin < 90% | 1.47 | 0.88 | 2.46 | .136 | NA | NA | NA | NA |
| Radiotherapy | .096 | 0.56 | 1.72 | .963 | 1.39 | 0.63 | 3.08 | .406 |
LCI indicates 95% lower confidence interval; UCI, 95% upper confidence interval; NA, not applicable; and GC, germinal center.
Significant value.
Kaplan-Meier curves for DLBCL survival after R-CHOP21 according to MLH1 rs1799977 genotype. OS after R-CHOP21 according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
Kaplan-Meier curves for DLBCL survival after R-CHOP21 according to MLH1 rs1799977 genotype. OS after R-CHOP21 according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
Clinical variables associated with poor OS in the DLBCL training series were ECOG PS > 1 (HR = 2.79; P = .007), bulky disease (HR = 2.92; P < .001), extranodal sites more than 1 (HR = 2.06; P = .027), LDH more than upper limit of normal (ULN; HR = 2.52; P = .003), IPI score (HR = 1.54; P = .001), and RDI of doxorubicin less than 90% (HR = 2.02; P = .028; Table 2; supplemental Figure 1). Multivariate analysis identified the MLH1 rs1799977 AG/GG genotype (HR = 3.14; P < .001) as an independent predictor of OS in DLBCL treated with R-CHOP21, along with IPI score (HR = 1.38; P = .037) and bulky disease (HR = 2.56; P = .004; Table 3).
Multivariate analysis for OS and PFS after R-CHOP21 in the training and validation DLBCL series
| . | Training series . | Validation series . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | LCI . | UCI . | P . | HR . | LCI . | UCI . | P . | |
| OS | ||||||||
| MLH1 rs1799977 AG/GG | 3.14 | 1.67 | 5.91 | < .001* | 3.04 | 1.45 | 6.37 | .003* |
| Bulky | 2.56 | 1.34 | 4.89 | .004* | 1.57 | 0.74 | 3.33 | .233 |
| IPI | 1.38 | 1.02 | 1.88 | .037* | 1.85 | 1.35 | 2.53 | < .001* |
| RDI doxorubicin < 90% | 1.19 | 0.58 | 2.43 | .642 | NA | NA | NA | NA |
| PFS | ||||||||
| MLH1 rs1799977 AG/GG | 1.96 | 1.17 | 3.28 | .010* | 1.98 | 1.12 | 3.50 | .019* |
| Bulky | 1.96 | 1.15 | 3.34 | .012* | 1.01 | 0.54 | 1.91 | .954 |
| IPI | 1.41 | 1.12 | 1.77 | .002* | 1.86 | 1.45 | 2.38 | < .001* |
| . | Training series . | Validation series . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | LCI . | UCI . | P . | HR . | LCI . | UCI . | P . | |
| OS | ||||||||
| MLH1 rs1799977 AG/GG | 3.14 | 1.67 | 5.91 | < .001* | 3.04 | 1.45 | 6.37 | .003* |
| Bulky | 2.56 | 1.34 | 4.89 | .004* | 1.57 | 0.74 | 3.33 | .233 |
| IPI | 1.38 | 1.02 | 1.88 | .037* | 1.85 | 1.35 | 2.53 | < .001* |
| RDI doxorubicin < 90% | 1.19 | 0.58 | 2.43 | .642 | NA | NA | NA | NA |
| PFS | ||||||||
| MLH1 rs1799977 AG/GG | 1.96 | 1.17 | 3.28 | .010* | 1.98 | 1.12 | 3.50 | .019* |
| Bulky | 1.96 | 1.15 | 3.34 | .012* | 1.01 | 0.54 | 1.91 | .954 |
| IPI | 1.41 | 1.12 | 1.77 | .002* | 1.86 | 1.45 | 2.38 | < .001* |
LCI indicates 95% lower confidence interval; UCI, 95% upper confidence interval; and NA, not applicable.
Significant value.
The MLH1 rs1799977 genotype is a predictor of failure after R-CHOP21
In the DLBCL training series, the prognostic role of MLH1 rs1799977 on OS after R-CHOP21 may not be explained by differences in clinical features at presentation or differences in R-CHOP21 feasibility. Indeed, DLBCL patients who carried the MLH1 rs1799977 AG/GG genotype did not differ from DLBCL patients who carried the MLH1 rs1799977 AA genotype in terms of age more than 60 years, ECOG PS more than 1, Charlson Comorbidity Index, Ann Arbor stage III or IV, bulky disease, B symptoms, extranodal sites more than 1, LDH more than ULN, IPI 3 to 5, or cell of origin (P > .05 in all instances; Table 4). In addition, DLBCL patients who carried the MLH1 rs1799977 AG/GG genotype did not differ from DLBCL patients who carried the MLH1 rs1799977 AA genotype in terms of RDI of doxorubicin, RDI of cyclophosphamide, number of R-CHOP21 courses, or R-CHOP21-related mortality (P > .05 in all instances; Table 4). Finally, the CR rate after R-CHOP21 (AG/GG 39/of 52, 75.0% vs AA 88 of 111, 79.3%, P = .539) and after second-line treatment (AG/GG 3 of 18, 16.7% vs AA 14 of 28, 50.0%, P = .072) was not significantly affected by the MLH1 rs1799977 genotype.
Clinical characteristics of the DLBCL training cohort at diagnosis according to the MLH1 rs1799977 genotype
| Clinical characteristic . | MLH1 rs1799977 . | P . | |
|---|---|---|---|
| AA . | AG/GG . | ||
| Age > 60 y | 71/111 (64.0%) | 33/52 (63.5%) | .950 |
| Male | 59/111 (53.2%) | 33/52 (63.5%) | .216 |
| ECOG PS > 1 | 13/111 (11.7%) | 8/52 (15.4%) | .514 |
| Charlson Comorbidity Index | 1 (0-2) | 1 (0-3) | .115 |
| Ann Arbor stage III or IV | 56/111 (50.5%) | 29/52 (55.8%) | .526 |
| B symptoms | 21/111 (18.9%) | 17/52 (32.7%) | .052 |
| Bulky | 30/111 (27.0%) | 16/52 (30.8%) | .620 |
| Extranodal sites > 1 | 28/111 (25.2%) | 13/52 (25.0%) | .975 |
| LDH > ULN | 33/111 (29.7%) | 21/52 (40.4%) | .177 |
| IPI 3-5 | 32/111 (28.8%) | 17/52 (32.7%) | .616 |
| Non-GC phenotype | 35/79 (44.3%) | 18/48 (37.5%) | .450 |
| RDI of doxorubicin, % | 94.8 (81.8-100) | 88.2 (76.1-99.8) | .123 |
| RDI of cyclophosphamide, % | 95.8 (87.5-100) | 92.1 (80.7-100) | .236 |
| No. of R-CHOP courses | 6 (6-8) | 6 (4-8) | .372 |
| 1-year treatment-related mortality | 1.8% | 6.0% | .286 |
| Clinical characteristic . | MLH1 rs1799977 . | P . | |
|---|---|---|---|
| AA . | AG/GG . | ||
| Age > 60 y | 71/111 (64.0%) | 33/52 (63.5%) | .950 |
| Male | 59/111 (53.2%) | 33/52 (63.5%) | .216 |
| ECOG PS > 1 | 13/111 (11.7%) | 8/52 (15.4%) | .514 |
| Charlson Comorbidity Index | 1 (0-2) | 1 (0-3) | .115 |
| Ann Arbor stage III or IV | 56/111 (50.5%) | 29/52 (55.8%) | .526 |
| B symptoms | 21/111 (18.9%) | 17/52 (32.7%) | .052 |
| Bulky | 30/111 (27.0%) | 16/52 (30.8%) | .620 |
| Extranodal sites > 1 | 28/111 (25.2%) | 13/52 (25.0%) | .975 |
| LDH > ULN | 33/111 (29.7%) | 21/52 (40.4%) | .177 |
| IPI 3-5 | 32/111 (28.8%) | 17/52 (32.7%) | .616 |
| Non-GC phenotype | 35/79 (44.3%) | 18/48 (37.5%) | .450 |
| RDI of doxorubicin, % | 94.8 (81.8-100) | 88.2 (76.1-99.8) | .123 |
| RDI of cyclophosphamide, % | 95.8 (87.5-100) | 92.1 (80.7-100) | .236 |
| No. of R-CHOP courses | 6 (6-8) | 6 (4-8) | .372 |
| 1-year treatment-related mortality | 1.8% | 6.0% | .286 |
The 25th to 75th percentiles are reported in parentheses for continuous variables.
GC indicates germinal center.
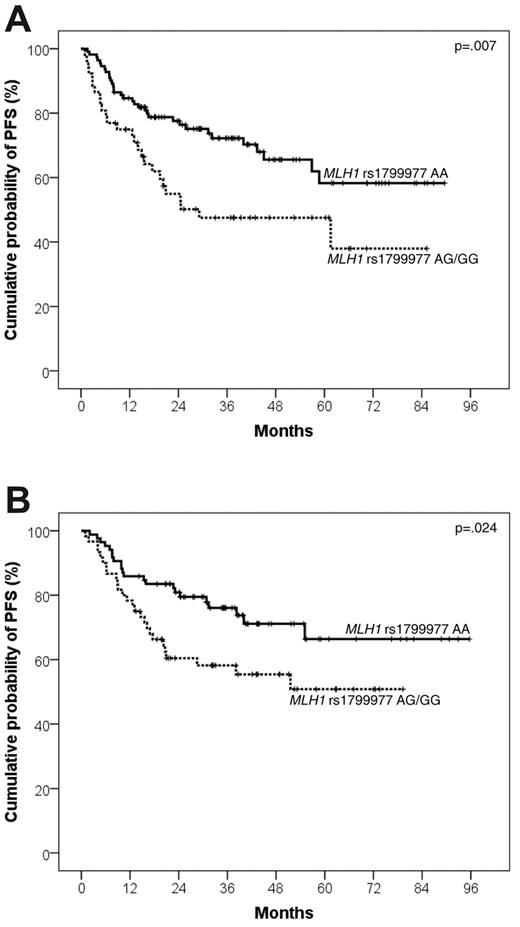
The poor prognosis heralded by MLH1 rs1799977 AG/GG genotype in DLBCL of the training series was the result of an increased risk of progressing after both first- and second-line treatment. Indeed, DLBCL patients carrying the MLH1 rs1799977 AG/GG genotype displayed an increased risk of progression after R-CHOP21 (HR = 2.02; events/N = 26/52; 4-year PFS, 47.5%; 95% CI, 33.0%-62.0%) compared with patients carrying the MLH1 rs1799977 AA genotype (events/N = 33/111; 4-year PFS, 65.6%; 95% CI, 54.7%-76.5%; P = .007; Table 2; Figure 2A).
Kaplan-Meier curves for DLBCL progression after R-CHOP21 according to MLH1 rs1799977 genotype. PFS after R-CHOP21 according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
Kaplan-Meier curves for DLBCL progression after R-CHOP21 according to MLH1 rs1799977 genotype. PFS after R-CHOP21 according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
Clinical variables associated with poor PFS after R-CHOP21 in the DLBCL training series were Ann Arbor stage III or IV (HR = 1.91; P = .017), bulky disease (HR = 2.33; P = .001), extranodal sites more than 1 (HR = 1.83; P = .028), LDH more than ULN (HR = 1.91; P = .014), and IPI score (HR = 1.50; P < .001; Table 2; supplemental Figure 1).
Multivariate analysis identified the MLH1 rs1799977 AG/GG genotype (HR = 1.96; P = .010) as an independent predictor of PFS after R-CHOP21 in the DLBCL training series, along with IPI score (HR = 1.41; P = .002) and bulky disease (HR = 1.96; P = .012; Table 3).
The MLH1 rs1799977 genotype is a predictor of failure after platinum-based second-line regimens
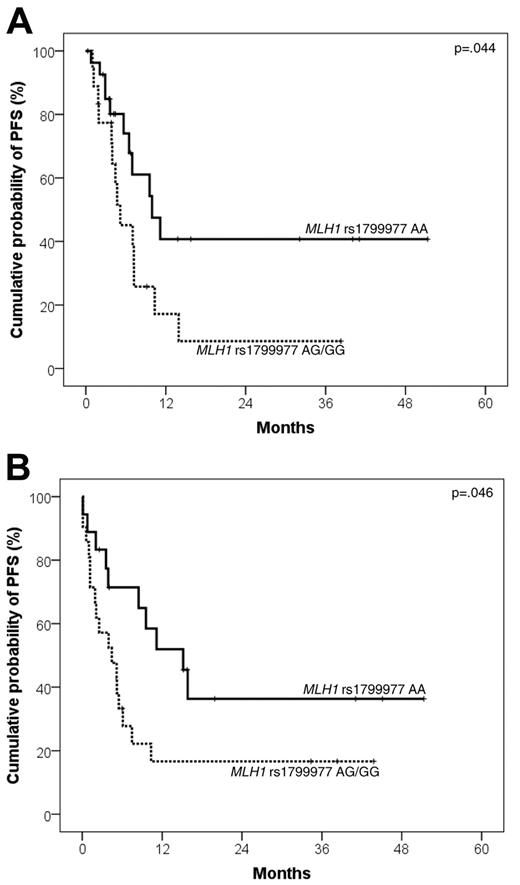
MLH1 rs1799977 AG/GG genotype predicted poor PFS and OS from salvage in patients who failed R-CHOP21 and received R-DHAP as second-line treatment (n = 46). Indeed, DLBCL patients carrying the MLH1 rs1799977 AG/GG genotype displayed an increased risk of progression after R-DHAP (HR = 2.26; 95% CI, 1.02-5.01; events/N = 14/18; 2-year PFS from salvage: 8.6%; 95% CI, 0%-24.0%) compared with patients carrying the MLH1 rs1799977 AA genotype (events/N = 11/28; 2-year PFS from salvage: 40.7%; 95% CI, 16.6%-64.8%; P = .044; Figure 3A).
Kaplan-Meier curves for DLBCL PFS from salvage treatment according to MLH1 rs1799977 genotype. PFS from salvage treatment according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
Kaplan-Meier curves for DLBCL PFS from salvage treatment according to MLH1 rs1799977 genotype. PFS from salvage treatment according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
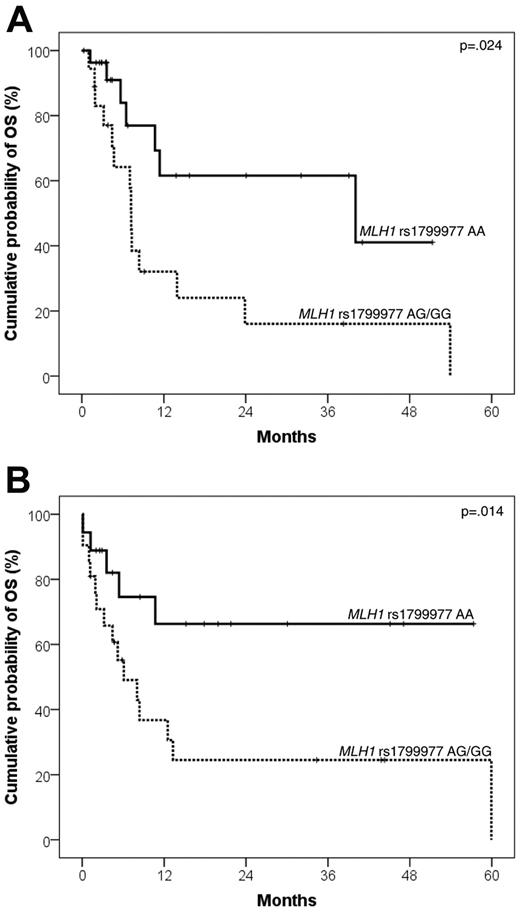
In addition, DLBCL patients who carried the MLH1 rs1799977 AG/GG genotype displayed an increased risk of death after R-DHAP (HR = 2.93; 95% CI, 1.14-7.33; events/N = 14/18; 2-year OS from salvage: 16.0%; 95% CI, 0%-35.4%) compared with patients who carried the MLH1 rs1799977 AA genotype (events/N = 7/28; 2-year OS from salvage: 61.6%; 95% CI, 36.4%-86.8%; P = .024; Figure 4A).
Kaplan-Meier curves for DLBCL survival from salvage treatment according to MLH1 rs1799977 genotype. OS from salvage treatment according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
Kaplan-Meier curves for DLBCL survival from salvage treatment according to MLH1 rs1799977 genotype. OS from salvage treatment according to MLH1 rs1799977 genotype in the training series (A) and in the validation series (B).
Validation of MLH1 rs1799977 genotype as a predictor of outcome in an independent DLBCL series treated with R-CHOP21
The prognostic value of MLH1 rs1799977 as a predictor of outcome in DLBCL patients treated with R-CHOP21 was validated in an independent series of DLBCL (n = 145). The validation series did not differ from the training series in terms of: (1) clinical features at presentation (Table 1); (2) median follow-up (49 vs 48 months, respectively; P = .312); (3) 4-year OS after R-CHOP21 (77.4% vs 72.9%, respectively; P = .370); (4) 4-year PFS after R-CHOP21 (64.8% vs 61.1%, respectively; P = .404); (5) 2-year OS from salvage (46.0% vs 38.2%, respectively; P = .920); (6) 2-year PFS from salvage (25.8% vs 25.3%; P = .610); and (7) prevalence of MLH1 rs1799977 genotypes (AA: 85/145, 58.6%; AG: 51/145, 35.2%; GG: 9/145, 6.2% vs AA: 111/163, 68.1%; AG: 43/163, 26.4%; GG: 9/163, 5.5%, respectively; P = .213).
Survival analysis in the validation series confirmed that MLH1 rs1799977 is a prognostic factor in DLBCL patients treated with R-CHOP21. Indeed, by univariate Cox analysis, the MLH1 rs1799977 AG/GG genotype was associated with poor OS (HR = 2.97; events/N = 21/60; 4-year OS, 66.8%; 95% CI, 54.5%-79.1%) compared with the MLH1 rs1799977 AA genotype (events/N = 11/85; 4-year OS, 84.0%; 95% CI, 74.8%-93.2%; P = .004; Table 2; Figure 1B).
Other variables associated with poor OS in the validation series were age more than 60 years (HR = 3.71; P = .014), extranodal sites more than 1 (HR = 2.56; P = .012), LDH more than ULN (HR = 2.77; P = .008), and IPI score (HR = 1.88; P < .001; Table 2; supplemental Figure 1).
Multivariate analysis selected the MLH1 rs1799977 AG/GG genotype as an independent predictor of OS in the validation series (HR = 3.04; P = .003), after adjusting for IPI score (HR = 1.85; P < .001) and bulky disease (HR = 1.57; P = .233; Table 3).
Analogous to the training series, the MLH1 rs1799977 genotype was associated with poor PFS also in the validation series. Indeed, DLBCL patients carrying the MLH1 rs1799977 AG/GG genotype displayed an increased risk of progression after R-CHOP21 (HR = 1.92; events/N = 26/60; 4-year PFS, 55.4%; 95% CI, 42.1%-68.7%) compared with patients carrying the MLH1 rs1799977 AA genotype (events/N = 22/85; 4-year PFS, 71.1%; 95% CI, 60.0%-82.2%; P = .024; Table 2; Figure 2B).
Other variables associated with poor PFS after R-CHOP21 in the validation series were age more than 60 years (HR = 2.75; P = .009), Ann Arbor stage III or IV (HR = 2.87; P = .001), extranodal sites more than 1 (HR = 2.98; P < .001), LDH more than ULN (HR = 2.52; P = .002), and IPI score (HR = 1.85; P < .001; Table 2; supplemental Figure 1).
Multivariate analysis selected the MLH1 rs1799977 AG/GG genotype as an independent predictor of PFS after R-CHOP21 in the validation series (HR = 1.98; P = .019), after adjusting for IPI score (HR = 1.86; P < .001) and bulky disease (HR = 1.01; P = .954; Table 3).
Finally, the MLH1 rs1799977 AG/GG genotype predicted poor PFS and OS from salvage with second-line regimens also in patients belonging to the validation series who failed R-CHOP21 (n = 39). Indeed, patients carrying the AG/GG genotype had a worse PFS from salvage (HR = 2.25; 95% CI, 1.01-4.99; events/N = 17/21; 2-year PFS from salvage: 16.7%; 95% CI, 0.1%-33.3%) compared with patients carrying the AA genotype (events/N = 10/18; 2-year PFS from salvage: 36.4%; 95% CI, 11.2%-61.6%; P = .046; Figure 3B). Patients carrying the AG/GG genotype had a worse OS from salvage (HR = 3.01; 95% CI, 1.08-8.39; events/N = 15/21; 2-year OS from salvage: 25.4%; 95% CI, 5.1%-45.7%) compared with patients carrying the AA genotype (events/N = 5/18; 2-year OS from salvage: 66.3%; 95% CI, 41.7%-90.9%; P = .014; Figure 4B).
Similarly to the training series, also in the validation series the CR rate after R-CHOP21 (AG/GG 44/60, 73.3% vs AA 71/85, 83.5%, P = .135) and after second-line treatment (AG/GG 5/21, 23.8% vs AA 7/18, 38.9%, P = .377) was not significantly affected by the MLH1 rs1799977 genotype.
The MLH1 rs1799977 genotype correlates with MLH1 protein expression
Immunohistochemical analysis revealed that MLH1 was expressed by all DLBCL patients analyzed (n = 58). Percentage of positive tumor cells and intensity of staining for MLH1 were heterogeneous in DLBCL samples (Figure 5). Indeed, MLH1 was expressed by 6% to 20% of tumor cells in 2 of 58 (3.4%) DLBCLs, by 21% to 80% tumor cells in 25 of 58 (43.1%), and by 81% to 100% tumor cells in 31 of 58 (53.4%). In addition, DLBCL showed a weak expression of MLH1 in 5 of 58 (8.6%) cases, a moderate expression in 14 of 58 (24.1%), and a strong expression in 39 of 58 (67.2%). Heterogeneity of MLH1 expression correlated with the MLH1 rs1799977 genotype. Indeed, the immunohistochemical score for MLH1 expression29 was significantly lower among DLBCL patients harboring the AG/GG genotype (median, 5) compared with cases harboring the AA genotype (median, 9; P = .034).
MLH1 expression inDLBCLs. Immunohistochemical analysis of MLH1 protein expression in paraffin sections from representative DLBCL primary biopsies. (A) A representative DLBCL sample harboring the MLH1 rs1799977 AA genotype and characterized by a strong nuclear positivity for MLH1. (B) A representative DLBCL sample harboring the MLH1 rs1799977 GG genotype and characterized by a weak nuclear positivity for MLH1. Tissue microarray, immunoperoxidase, hematoxylin counterstain. Images were taken using a Nikon Eclipse 80i microscope (Nikon) with a pan fluor 20×/0.13 objective and Nikon digital sight DS-Fi1 camera equipped with control unit-DS-L2 (Nikon). Images were assembled using Adobe Photoshop 6 (Adobe Systems).
MLH1 expression inDLBCLs. Immunohistochemical analysis of MLH1 protein expression in paraffin sections from representative DLBCL primary biopsies. (A) A representative DLBCL sample harboring the MLH1 rs1799977 AA genotype and characterized by a strong nuclear positivity for MLH1. (B) A representative DLBCL sample harboring the MLH1 rs1799977 GG genotype and characterized by a weak nuclear positivity for MLH1. Tissue microarray, immunoperoxidase, hematoxylin counterstain. Images were taken using a Nikon Eclipse 80i microscope (Nikon) with a pan fluor 20×/0.13 objective and Nikon digital sight DS-Fi1 camera equipped with control unit-DS-L2 (Nikon). Images were assembled using Adobe Photoshop 6 (Adobe Systems).
Discussion
The current study on 308 de novo DLBCL documents that the MLH1 rs1799977 SNP: (1) predicts OS in newly diagnosed DLBCL patients treated with R-CHOP21, (2) predicts failure after both R-CHOP21 and platinum-based second-line regimens, (3) exerts a prognostic role independent of IPI, and (4) predicts all DLBCL outcomes in series of patients treated at different institutions, as documented by the training-validation approach chosen for study design.
The association between MLH1 rs1799977 AG/GG genotype and poor DLBCL outcome appears to be biologically plausible based on several lines of evidence. First, the G variant allele of MLH1 rs1799977 associates with a reduction of MLH1 protein expression, as documented in DLBCL (this study) and other types of cancer.35,36 Consistent with the role of MLH1 in promoting apoptosis triggered by chemotherapy, reduced MLH1 protein expression in cancer cells confers in vitro resistance to doxorubicin and platinum compounds.37-39 Second, at variance with other DNA repair genes investigated in this study, MLH1 not only affects DNA repair mechanisms but is also involved in DNA damage signaling, a process that induces cell cycle arrest and can lead to apoptosis in case of major DNA damages by doxorubicin and platinum compounds.12,13,17 Third, the MLH1 rs1799977 AG/GG genotype harbors prognostic relevance also in disease models other than lymphoma, including acute lymphoblastic leukemia, solid tumors, and inflammatory bowel disease.40-47
The cross-validation approach of the current study documents that the MLH1 rs1799977 genotype is an independent prognostic factor retaining its value in 2 DLBCL series treated at different institutions using similar therapeutic strategies. This observation suggests that the prognostic value of MLH1, although detected retrospectively, appears to be independent of a potential bias because of patient referral or patient management at a single center. Confirmation within the frame of prospective controlled studies will be helpful to fully assess the prognostic role of MLH1 in DLBCL.
In both the training and validation series, cases of DLBCL with an unfavorable IPI appear to be underrepresented because of the concomitance of the DLCL-04 national trial run by the Italian Lymphoma Intergroup testing the role of autologous stem cell transplantation at diagnosis in young patients with unfavorable IPI (www.clinicaltrials.gov #NCT00499018; accessed July 13, 2010). Despite the underrepresentation of high-risk DLBCL, the impact of MLH1 rs1799977 genotype is observed also in this DLBCL subgroup (data not shown).
Consistent with the characteristics of our patient series, bulky disease emerged as an independent prognostic factor in the validation series. The prognostic role of bulky disease is not unexpected in DLBCL cohorts with a preponderance of favorable IPI patients, as clearly documented by the MInT trial.48
Although the role of radiotherapy within a combined-modality approach including R-CHOP has not been defined,49 in our study, patients presenting with bulky disease were planned to receive involved field radiotherapy to the sites of initial bulk. In both the training and the validation series, involved field radiotherapy administration to sites of initial bulk did not compromise the prognostic effect of MLH1 rs1799977. Indeed, compared with the MLH1 rs1799977 AA genotype, the MLH1 rs1799977 AG/GG genotype was associated with a significantly shorter PFS independent of having or having not received radiotherapy in both the training series (radiotherapy, P = .037; no radiotherapy, P = .022, respectively) and in the validation series (radiotherapy, P = .049; no radiotherapy, P = .035).
The impact of MLH1 rs1799977 genotype on DLBCL outcome may be related to the chance of obtaining CR with induction therapy and/or to the degree of clearance of residual lymphoma cells, which may be responsible of relapse during follow-up. MLH1 rs1799977 predicted PFS and OS in both the training and validation series. Conversely, the CR rate after R-CHOP21 was not significantly affected by MLH1 rs1799977. Notably, also TP53 disruption, a well-known biomarker of tumor cell chemorefractoriness in lymphoid neoplasia, associates with poor survival but not with CR rate in CHOP-treated DLBCL.50 These data would suggest that the impact of the MLH1 rs1799977 genotype on PFS, rather than on CR rate, is the result of a better clearance of residual tumor cells in patients harboring the MLH1 rs1799977 AA genotype, thus reducing the chance of disease recurrence. It should be noted, however, that the high activity of R-CHOP and the high frequency of CR (127 of 163, 77.9% in the training series; and 115 of 145, 79.3% in the validation series), as well as the sample size of the 2 series might have prevented the detection of a statistical difference in CR rate between MLH1 rs1799977 genotypes, and that larger studies might be required to fully address this issue.
The results of this study point to MLH1 rs1799977 as a potential novel prognosticator for predicting treatment failure and survival of DLBCL. With very few exceptions,7-11 most new prognostic markers of DLBCL have been prompted by investigations of the genetics and of the biology of the lymphoma clone. The example of the predictive value of the MLH1 genotype points to the need of focusing also on the genetic background of the host for a comprehensive assessment of DLBCL prognosis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Fondo per gli Investimenti della Ricerca di Base-Programma Futuro in Ricerca 2008, and Programma di Ricerca di Interesse Nazionale 2008, Ministero della Università e della Ricerca, Rome, Italy; Associazione Italiana per la Ricerca sul Cancro, Special Program Molecular Clinical Oncology, 5 × 1000, No. 10007, Milan, Italy; Progetto Giovani Ricercatori 2008, Ricerca Sanitaria Finalizzata, and Progetto Integrato Oncologia-Advanced Molecular Diagnostics project (RFPS-2006-2-339694.1; RFPS-2006-2-339723.2), Ministero della Salute, Rome, Italy; Ricerca Sanitaria Finalizzata, Regione Piemonte, Torino, Italy; Istituto Toscano Tumori 2008 Grant, Florence, Italy; and Novara-Associazione Italiana contro le Leucemie, Linfomi e Mielomi Onlus, Novara.
Authorship
Contribution: D.R. and G.G. designed the study, analyzed and interpreted data, performed statistical analysis, and drafted the manuscript; S.R., A.B., and D.C. performed experimental research and analyzed data; S.F, L.D.P., M.F., and R.B. collected clinical data of the training series; F.F., A.F., A.D.R., A. Chiappella, C.L.B., M.G., and M.C.T. collected clinical data of the validation series; A.G. performed immunohistochemistry and interpreted data; and E.M.P., F.L., M.L., S.H., M.M., U.V., A. Carbone, and R.F. contributed to data analysis and interpretation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Davide Rossi, Division of Hematology, Department of Clinical and Experimental Medicine & CRIFF, Amedeo Avogadro University of Eastern Piedmont, Via Solaroli 17, 28100 Novara, Italy; e-mail: rossidav@med.unipmn.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal