Abstract
The type I interferons (IFNs) normally suppress tumor growth by phosphorylating and activating the signal transducer and activator of transcription 1 (STAT1), but also briefly activate STAT3, which promotes cell growth. In chronic lymphocytic leukemia (CLL) cells, the duration of IFN-mediated STAT3 phosphorylation was found to exhibit significant interpatient variability and was prolonged in cells with high risk features, such as 11q− and 17p− deletions involving ataxia telangiectasia mutated (ATM) and p53. This aberrant signaling pattern was associated with a paradoxical increase in cell size and number in response to IFN and similar alterations in IFN-signaling and responses were seen in cell lines that developed in the absence of p53 or ATM. However, direct inhibition of p53 or ATM failed to cause these changes, and CLL cells with aggressive clinical features were found to also express high levels of reactive oxygen species (ROS), which decrease tyrosine phosphatase activity. Prolonged IFN-mediated STAT3 phosphorylation and lowered phosphatase activity could be reversed by antioxidants. These findings suggest that increased ROS levels may corrupt IFN-signaling processes in aggressive CLL cells, causing IFN to be used as a growth factor rather than a tumor suppressor. Antioxidants or STAT3 kinase inhibitors might improve the outcome of IFN therapy in CLL by restoring normal signaling.
Introduction
Chronic lymphocytic leukemia (CLL) is often benign but aggressive forms of the disease are lethal and generally incurable with conventional therapy.1 Like other cancers, aggressive CLL cells develop adaptive mechanisms that allow them to resist immunotherapies and chemotherapies2 and continue to grow despite such obstacles as hypoxia, poor vasculature, and natural host defenses including tumor-reactive T cells.3 A greater understanding of how CLL cells survive in these conditions is needed to improve treatment outcomes.
The type I interferons (IFNs) are natural tumor suppressor molecules that act by inhibiting cell proliferation and increasing the susceptibility of cancer cells to cytotoxic immune effectors.4,5 Exogenous IFN also has therapeutic activity in CLL, particularly in patients with low risk disease.6 However, IFN is generally inactive in more aggressive disease, and may even accelerate tumor growth,7 suggesting that IFN-signaling may be altered in tumor cells from such patients.
Upon activation of the interferon-α/β receptor (IFNAR), signaling processes are initiated that lead to phosphorylation and activation of signal transducer and activator of transcription 1 (STAT1), via the Janus kinase (JAK), JAK1, and a more transient phosphorylation and activation of STAT3, mainly via tyrosine kinase 2 (TYK2) as shown in Figure 1A.8 STAT1 is a transcription factor with properties of a tumor suppressor, whereas STAT3 is oncogenic and promotes cell growth and resistance to proapoptotic stimuli.5,9 Variations in the magnitude and timing of STAT1 and STAT3 activation9 might then affect the responses of tumor cells to IFN.10 Our studies were designed to investigate IFN-signaling in leukemia cells from patients on different parts of the clinical CLL spectrum.
Methods
Blood samples
Heparinized blood was obtained from healthy volunteers and consenting patients with CLL (diagnosed by a persistent monoclonal expansion of CD19+CD5+IgMlo lymphocytes11 ). Patients were untreated for at least 3 months at the time of analysis. Protocols were approved by the Sunnybrook Ethics Review Board and informed consent was obtained in compliance with the Declaration of Helsinki. Clinical characteristics of the patients are described in Tables 1 and 2.
Clinical properties of patients with CLL
| Patient no.* . | Sex . | Rai stage† . | Age, y . | WBC, × 106/mL . | Disease duration, y . | Treatment‡ . | CD38, % . | Cytogenetics§ . |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 4 | 56 | 122 | 7 | C/P, FCR | 10 | 13q− |
| 2 | F | 2 | 57 | 42 | 11 | C/P | 8 | 13q− |
| 6 | M | 0 | 63 | 27 | 5 | None | 9 | Normal |
| 7 | F | 0 | 44 | 24 | 6 | None | 4 | Normal |
| 9 | M | 0 | 69 | 34 | 9 | None | 5 | Normal |
| 13 | F | 4 | 34 | 55 | 11 | C/P, FCR | 25 | T12 |
| 22 | M | 2 | 58 | 52 | 6 | C/P, FC, BMT | 45 | 11q− |
| 23 | M | 2 | 70 | 125 | 12 | CVP, S,FC (three ×) | 2 | 11q− |
| 25 | F | 3 | 60 | 87 | 11 | S, CP | 1 | 13q− |
| 27 | F | 3 | 80 | 122 | 10 | P | 8 | T12 |
| 28 | M | 4 | 64 | 77 | 8 | S, CP | NA | 13q− |
| 30 | F | 3 | 70 | 142 | 4 | C | 3 | T12 |
| 31 | M | 4 | 59 | 59 | 10 | F, Rev, CHOP | 14 | 13q− |
| 44 | F | 4 | 54 | 65 | 17 | None | 2 | 13q− |
| 55 | M | 0 | 75 | 42 | 6 | C/P, S, FC | 40 | T12 |
| 56 | M | 3 | 55 | 142 | 11 | C/P, Rev | 22 | Normal |
| 57 | M | 3 | 47 | 33 | 6 | C/P, BMT | 8 | T12 |
| 71 | F | 4 | 74 | 120 | 2 | P | NA | 17p− |
| 73 | M | 0 | 85 | 42 | 3 | None | 1 | 17p− |
| 74 | M | 3 | 69 | 93 | 5 | C/P | 4 | 11q− |
| 78 | M | 2 | 71 | 122 | 5 | C/P, S, FCR | 13 | normal |
| 81 | F | 3 | 50 | 22 | 6 | S | 7 | 17p− |
| 82 | M | 4 | 86 | 43 | 5 | CVP, FC, Rads | 71 | 11q− |
| 83 | M | 2 | 49 | 63 | 3 | C/P, FC, BMT | 30 | 11q− |
| 86 | M | 2 | 58 | 65 | 6 | C, FC, Rev | 3 | 13q− |
| 89 | M | 4 | 75 | 72 | 8 | F, R, P, S | NA | 17p− |
| 92 | M | 4 | 56 | 42 | 6 | CHOP, P, R | 81 | 17p− |
| 93 | M | 3 | 62 | 70 | 2 | Rev | NA | 11q− |
| 95 | F | 4 | 60 | 80 | 8 | C/P, FC, FCR | 2 | T12 |
| 96 | M | 2 | 54 | 71 | 6 | C/P | 15 | Normal |
| 105 | F | 3 | 68 | 153 | 9 | Ch, FC | NA | Normal |
| 106 | F | 3 | 69 | 229 | 7 | C/P, FC | 1 | 13q− |
| 109 | F | 3 | 64 | 139 | 7 | C/P, FCR | 51 | Normal |
| 110 | F | 3 | 62 | 98 | 7 | C/P | 20 | T12 |
| 111 | F | 0 | 74 | 54 | 4 | None | 4 | 17p− |
| 114 | M | 4 | 54 | 14 | 11 | C/P, F, CHOP, Rev | 38 | Normal |
| 115 | M | 4 | 57 | 100 | 9 | C/P, FC, S, CHOPR | 54 | 17p− |
| 116 | M | 4 | 55 | 120 | 14 | C, CVP, FC | 1 | Normal |
| 119 | M | 2 | 74 | 42 | 5 | None | 2 | 11q− |
| 120 | M | 4 | 72 | 200 | 6 | C/P, S, CHOP | NA | 17p− |
| 176 | F | 3 | 74 | 17 | 5 | C, CP | 43 | T12 |
| 141 | F | 4 | 62 | 520 | 27 | S | 18 | 11q− |
| 175 | M | 4 | 75 | 183 | 6 | P | 25 | 13q− |
| 134 | M | 3 | 80 | 5 | 7 | C/P, S | NA | 13q− |
| 158 | M | 3 | 56 | 126 | 5 | C/P/R, S | 14 | T12 |
| 132 | M | 2 | 58 | 134 | 3 | None | 1 | T12 |
| 125 | F | 3 | 65 | 76 | 4 | None | NA | T12 |
| Patient no.* . | Sex . | Rai stage† . | Age, y . | WBC, × 106/mL . | Disease duration, y . | Treatment‡ . | CD38, % . | Cytogenetics§ . |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 4 | 56 | 122 | 7 | C/P, FCR | 10 | 13q− |
| 2 | F | 2 | 57 | 42 | 11 | C/P | 8 | 13q− |
| 6 | M | 0 | 63 | 27 | 5 | None | 9 | Normal |
| 7 | F | 0 | 44 | 24 | 6 | None | 4 | Normal |
| 9 | M | 0 | 69 | 34 | 9 | None | 5 | Normal |
| 13 | F | 4 | 34 | 55 | 11 | C/P, FCR | 25 | T12 |
| 22 | M | 2 | 58 | 52 | 6 | C/P, FC, BMT | 45 | 11q− |
| 23 | M | 2 | 70 | 125 | 12 | CVP, S,FC (three ×) | 2 | 11q− |
| 25 | F | 3 | 60 | 87 | 11 | S, CP | 1 | 13q− |
| 27 | F | 3 | 80 | 122 | 10 | P | 8 | T12 |
| 28 | M | 4 | 64 | 77 | 8 | S, CP | NA | 13q− |
| 30 | F | 3 | 70 | 142 | 4 | C | 3 | T12 |
| 31 | M | 4 | 59 | 59 | 10 | F, Rev, CHOP | 14 | 13q− |
| 44 | F | 4 | 54 | 65 | 17 | None | 2 | 13q− |
| 55 | M | 0 | 75 | 42 | 6 | C/P, S, FC | 40 | T12 |
| 56 | M | 3 | 55 | 142 | 11 | C/P, Rev | 22 | Normal |
| 57 | M | 3 | 47 | 33 | 6 | C/P, BMT | 8 | T12 |
| 71 | F | 4 | 74 | 120 | 2 | P | NA | 17p− |
| 73 | M | 0 | 85 | 42 | 3 | None | 1 | 17p− |
| 74 | M | 3 | 69 | 93 | 5 | C/P | 4 | 11q− |
| 78 | M | 2 | 71 | 122 | 5 | C/P, S, FCR | 13 | normal |
| 81 | F | 3 | 50 | 22 | 6 | S | 7 | 17p− |
| 82 | M | 4 | 86 | 43 | 5 | CVP, FC, Rads | 71 | 11q− |
| 83 | M | 2 | 49 | 63 | 3 | C/P, FC, BMT | 30 | 11q− |
| 86 | M | 2 | 58 | 65 | 6 | C, FC, Rev | 3 | 13q− |
| 89 | M | 4 | 75 | 72 | 8 | F, R, P, S | NA | 17p− |
| 92 | M | 4 | 56 | 42 | 6 | CHOP, P, R | 81 | 17p− |
| 93 | M | 3 | 62 | 70 | 2 | Rev | NA | 11q− |
| 95 | F | 4 | 60 | 80 | 8 | C/P, FC, FCR | 2 | T12 |
| 96 | M | 2 | 54 | 71 | 6 | C/P | 15 | Normal |
| 105 | F | 3 | 68 | 153 | 9 | Ch, FC | NA | Normal |
| 106 | F | 3 | 69 | 229 | 7 | C/P, FC | 1 | 13q− |
| 109 | F | 3 | 64 | 139 | 7 | C/P, FCR | 51 | Normal |
| 110 | F | 3 | 62 | 98 | 7 | C/P | 20 | T12 |
| 111 | F | 0 | 74 | 54 | 4 | None | 4 | 17p− |
| 114 | M | 4 | 54 | 14 | 11 | C/P, F, CHOP, Rev | 38 | Normal |
| 115 | M | 4 | 57 | 100 | 9 | C/P, FC, S, CHOPR | 54 | 17p− |
| 116 | M | 4 | 55 | 120 | 14 | C, CVP, FC | 1 | Normal |
| 119 | M | 2 | 74 | 42 | 5 | None | 2 | 11q− |
| 120 | M | 4 | 72 | 200 | 6 | C/P, S, CHOP | NA | 17p− |
| 176 | F | 3 | 74 | 17 | 5 | C, CP | 43 | T12 |
| 141 | F | 4 | 62 | 520 | 27 | S | 18 | 11q− |
| 175 | M | 4 | 75 | 183 | 6 | P | 25 | 13q− |
| 134 | M | 3 | 80 | 5 | 7 | C/P, S | NA | 13q− |
| 158 | M | 3 | 56 | 126 | 5 | C/P/R, S | 14 | T12 |
| 132 | M | 2 | 58 | 134 | 3 | None | 1 | T12 |
| 125 | F | 3 | 65 | 76 | 4 | None | NA | T12 |
NA indicates not available; and WBC, white blood cell.
Corresponding patient numbers are maintained throughout.
Rai stage classification: 0 = lymphocytosis; 1 = with adenopathy; 2 = with hepatosplenomegaly; 3 = with anemia; 4 = with thrombocytopenia.11
Treatment by C indicates chlorambucil; P, prednisone; F, fludarabine; S, splenectomy; Rads, local radiation; R, rituxan; CHOP, cyclophosphamide/vincristine/adriamycin/prednisone; FC, fludarabine/cyclophosphamide; CVP, cyclophosphamide/vincristine/prednisone; BMT, allogeneic bone marrow transplantation; and Rev, revlimid.
In the case of multiple cytogenetic lesions, data for the higher risk lesion were used (eg, 11q− or 17p−)
Clinical properties of patients who had tumor ROS levels measured
| Patient no. . | Sex . | Age, y . | Disease duration, y . | Rai stage . | CD3, % . | WBC, × 106/mL . | Cytogenetics . | LDT, mo . | No. of treatments† . | ROS‡ . |
|---|---|---|---|---|---|---|---|---|---|---|
| 201 | M | 30 | 2 | 0 | 1 | 26 | Normal | 24 | 0 | 730 |
| 202 | M | 72 | 1 | 4 | 1 | 51 | 13q− | 30 | 0 | 833 |
| 203 | F | 73 | 1 | 0 | 1 | 27 | NA | NA | 0 | 972 |
| 204 | M | 57 | 3 | 0 | 35 | 32 | NA | 16 | 0 | 699 |
| 205 | F | 68 | 2 | 3 | 67 | 50 | NA | 11 | 0 | 742 |
| 206 | F | 61 | 4 | 3 | 10 | 82 | 11q | 3 | 0 | 1078 |
| 207 | F | 64 | 2 | 0 | 0 | 27 | 13q | 30 | 0 | 691 |
| 208 | M | 62 | 7 | 0 | 4 | 22 | NA | 60 | 0 | 414 |
| 209 | M | 83 | 8 | 4 | 1 | 111 | Normal | 12 | 1 | 653 |
| 210 | F | 62 | 27 | 4 | 18 | 752 | 11q−,13q− | 15 | 2 | 1780 |
| 211 | M | 61 | 11 | 4 | 191 | 17p− | 2 | 4 | 2008 | |
| 212 | M | 74 | 4 | 4 | 95 | 82 | NA | 12 | 0 | 931 |
| 213 | F | 35 | 5 | 4 | 8 | 135 | 13q− | 12 | 0 | 841 |
| 214 | M | 71 | 13 | 4 | 5 | 138 | 13q− | 7 | 4 | 1204 |
| 215 | M | 57 | 8 | 1 | 40 | NA | 40 | 0 | 639 | |
| 216 | M | 67 | 2 | 4 | 1 | 65 | 13q− | 13 | 0 | 962 |
| 217 | M | 55 | 6 | 4 | 81 | 40 | 13q−,17p− | 6 | 4 | 1896 |
| 218 | M | 64 | 6 | 4 | 20 | 152 | 13q−,17p−, 11q− | 4 | 4 | 2066 |
| 219 | M | 55 | 1 | 4 | 1 | 58 | 13q− | 8 | 0 | 1529 |
| 220 | F | 58 | 2 | 4 | 1 | 74 | NA | 18 | 0 | 667 |
| 221 | M | 57 | 7 | 4 | 21 | 27 | 11q− | 3 | 4 | 1379 |
| 222 | M | 59 | 8 | 2 | 25 | 43 | Normal | 40 | 0 | 696 |
| 223 | M | 64 | 7 | 2 | 15 | 43 | Normal | 17 | 1 | 740 |
| 224 | M | 60 | 15 | 4 | 49 | 66 | 11q− | 5 | 6 | 1712 |
| 225 | F | 79 | 2 | 0 | 73 | 13 | T12 | 45 | 0 | 830 |
| 226 | M | 70 | 8 | 4 | 4 | 20 | Normal | 7 | 1 | 1256 |
| 227 | M | 54 | 6 | 3 | 3 | 37 | 13q− | 36 | 1 | 1016 |
| 228 | F | 79 | 6 | 0 | 16 | 47 | 13q− | 48 | 0 | 695 |
| 229 | M | 75 | 6 | 4 | 31 | 248 | NA | 11 | 1 | 1852 |
| 230 | M | 53 | 1 | 0 | 1 | 13 | NA | NA | 0 | 299 |
| 231 | M | 57 | 1 | 0 | 2 | 27 | 13q− | 30 | 0 | 734 |
| 232 | M | 67 | 6 | 0 | 1 | 24 | NA | 55 | 0 | 422 |
| 233 | F | 62 | 7 | 3 | 20 | 109 | T12 | 9 | 1 | 734 |
| 301 | F | 69 | 8 | 3 | 77 | 97 | Normal | 16 | 0 | NA |
| 302 | M | 49 | 9 | 4 | 12 | 151 | NA | 4 | 0 | NA |
| 303 | M | 61 | 3 | 4 | 2 | 70 | NA | 4 | 1 | NA |
| 304 | F | 83 | 13 | 3 | 8 | 206 | T12 | 10 | 1 | NA |
| 305 | F | 71 | 6 | 4 | 5 | 142 | NA | 8 | 2 | NA |
| 306 | F | 87 | 2 | 4 | 6 | 31 | NA | 24 | 0 | NA |
| 307 | M | 59 | 6 | 4 | 1 | 115 | 13q− | 30 | 1 | NA |
| Patient no. . | Sex . | Age, y . | Disease duration, y . | Rai stage . | CD3, % . | WBC, × 106/mL . | Cytogenetics . | LDT, mo . | No. of treatments† . | ROS‡ . |
|---|---|---|---|---|---|---|---|---|---|---|
| 201 | M | 30 | 2 | 0 | 1 | 26 | Normal | 24 | 0 | 730 |
| 202 | M | 72 | 1 | 4 | 1 | 51 | 13q− | 30 | 0 | 833 |
| 203 | F | 73 | 1 | 0 | 1 | 27 | NA | NA | 0 | 972 |
| 204 | M | 57 | 3 | 0 | 35 | 32 | NA | 16 | 0 | 699 |
| 205 | F | 68 | 2 | 3 | 67 | 50 | NA | 11 | 0 | 742 |
| 206 | F | 61 | 4 | 3 | 10 | 82 | 11q | 3 | 0 | 1078 |
| 207 | F | 64 | 2 | 0 | 0 | 27 | 13q | 30 | 0 | 691 |
| 208 | M | 62 | 7 | 0 | 4 | 22 | NA | 60 | 0 | 414 |
| 209 | M | 83 | 8 | 4 | 1 | 111 | Normal | 12 | 1 | 653 |
| 210 | F | 62 | 27 | 4 | 18 | 752 | 11q−,13q− | 15 | 2 | 1780 |
| 211 | M | 61 | 11 | 4 | 191 | 17p− | 2 | 4 | 2008 | |
| 212 | M | 74 | 4 | 4 | 95 | 82 | NA | 12 | 0 | 931 |
| 213 | F | 35 | 5 | 4 | 8 | 135 | 13q− | 12 | 0 | 841 |
| 214 | M | 71 | 13 | 4 | 5 | 138 | 13q− | 7 | 4 | 1204 |
| 215 | M | 57 | 8 | 1 | 40 | NA | 40 | 0 | 639 | |
| 216 | M | 67 | 2 | 4 | 1 | 65 | 13q− | 13 | 0 | 962 |
| 217 | M | 55 | 6 | 4 | 81 | 40 | 13q−,17p− | 6 | 4 | 1896 |
| 218 | M | 64 | 6 | 4 | 20 | 152 | 13q−,17p−, 11q− | 4 | 4 | 2066 |
| 219 | M | 55 | 1 | 4 | 1 | 58 | 13q− | 8 | 0 | 1529 |
| 220 | F | 58 | 2 | 4 | 1 | 74 | NA | 18 | 0 | 667 |
| 221 | M | 57 | 7 | 4 | 21 | 27 | 11q− | 3 | 4 | 1379 |
| 222 | M | 59 | 8 | 2 | 25 | 43 | Normal | 40 | 0 | 696 |
| 223 | M | 64 | 7 | 2 | 15 | 43 | Normal | 17 | 1 | 740 |
| 224 | M | 60 | 15 | 4 | 49 | 66 | 11q− | 5 | 6 | 1712 |
| 225 | F | 79 | 2 | 0 | 73 | 13 | T12 | 45 | 0 | 830 |
| 226 | M | 70 | 8 | 4 | 4 | 20 | Normal | 7 | 1 | 1256 |
| 227 | M | 54 | 6 | 3 | 3 | 37 | 13q− | 36 | 1 | 1016 |
| 228 | F | 79 | 6 | 0 | 16 | 47 | 13q− | 48 | 0 | 695 |
| 229 | M | 75 | 6 | 4 | 31 | 248 | NA | 11 | 1 | 1852 |
| 230 | M | 53 | 1 | 0 | 1 | 13 | NA | NA | 0 | 299 |
| 231 | M | 57 | 1 | 0 | 2 | 27 | 13q− | 30 | 0 | 734 |
| 232 | M | 67 | 6 | 0 | 1 | 24 | NA | 55 | 0 | 422 |
| 233 | F | 62 | 7 | 3 | 20 | 109 | T12 | 9 | 1 | 734 |
| 301 | F | 69 | 8 | 3 | 77 | 97 | Normal | 16 | 0 | NA |
| 302 | M | 49 | 9 | 4 | 12 | 151 | NA | 4 | 0 | NA |
| 303 | M | 61 | 3 | 4 | 2 | 70 | NA | 4 | 1 | NA |
| 304 | F | 83 | 13 | 3 | 8 | 206 | T12 | 10 | 1 | NA |
| 305 | F | 71 | 6 | 4 | 5 | 142 | NA | 8 | 2 | NA |
| 306 | F | 87 | 2 | 4 | 6 | 31 | NA | 24 | 0 | NA |
| 307 | M | 59 | 6 | 4 | 1 | 115 | 13q− | 30 | 1 | NA |
NA indicates not available; and WBC, white blood cell.
*Normal range is 0.6 to 2.3 mg/mL.
Splenectomy, conventional chemotherapeutic regimens, such as fludarabine/cyclophosphamide (FC), and steroid-based regimens are each considered as a treatment.
Mean fluorescence intensity of DCFH staining.
Peripheral blood mononuclear cells, normal B cells, and CLL cells were isolated as previously described by negative selection with the Rosette Sep Human B cell enrichment cocktail (StemCell Technologies) and density centrifugation with Ficoll-Paque (Amersham Pharmacia Biotech).12 This method of purification yields percentages of CD19+ and CD19+CD5+ cells of > 98% and 96%, respectively.12
Antibodies and reagents
The 7-aminoactinomycin D (7AAD) used was from Pharmingen. Phorbol dibutyrate (PDB) was from Sigma-Aldrich and stock solutions (5 mg/mL) were made in dimethylsulfoxide. Cycloheximide (CHX), N-acetyl-cysteine (NAC), and β-actin antibodies were also from Sigma-Aldrich. Clinical grade IFN-α2b (Schering Canada) was purchased from the hospital pharmacy. Go6976 (classic protein kinase C [PKC] isozyme inhibitor), KU-55933 (ataxia telangiectasia mutated [ATM] inhibitor),13 AG9, and Piceatannol (TYK2 inhibitors) were from Calbiochem, and stock solutions were made in dimethylsulfoxide. Pifithrin-α (p53 inhibitor) and Nutlin-3 (p53 activator) were from Cayman Chemical. Antibodies to p53, p21, STAT1, STAT3, phosphotyrosine 701-specific, STAT1, phosphotyrosine 705-specific, STAT3, JAK1, and TYK2 were from Cell Signaling Technology. TK6 and NH32 cells were from Liber and colleagues14 and BL41tsp53 cells were from Nagy and colleagues.15 Recombinant murine IFN-β was synthesized as described previously.16 The p53−/−, ATM−/−, and control mice on the C57BL/6J background were obtained from The Jackson Laboratory (stock nos. 002101, 008536, and 000664, respectively) and used at 6 weeks of age.
Cell culture and activation
Purified CLL cells (1.5 × 106 cells/mL) were cultured in serum-free AIM-V medium (GibcoBRL) plus 2-mercaptoethanol (2-ME; Sigma-Aldrich) at a final concentration of 5 × 10−5 M in 6- or 24-well plates (BD Labware) for 3 to 4 days at 37°C in 5% CO2. TK6, NH32, and BL41 temperature sensitive p53 cell lines (BL41ts53) were cultured in RPMI 1640 medium (Invitrogen) with 5% fetal bovine serum (FBS; Wisent) at 37°C in 5% CO2. BL41ts53 cells were cultured at 32°C in 5% CO2 to induce p53 activity. Lymphocytes from spleens and lymph nodes of wild-type (wt) p53, wt ATM, p53−/−, and ATM−/− mice were cultured in RPMI 1640 with 5% FBS for the times indicated in the experiment. Human IFN-α2b and murine IFN-β were used at 1000 U/mL. In preliminary experiments this dose of IFN was found to be above the amount that caused maximal phosphorylation of STAT proteins, suggesting that plasma membrane IFNARs had been saturated. PDB was used at 100 ng/mL as before.17 Signaling inhibitors (ie, Go6976, NAC, AG9, and Piceatannol) were added at the concentrations indicated in the experiments.
The ATM+/+ and ATM−/− fibroblast lines, GM16666 and GM16667, were grown at 37°C in 5% CO2 in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% FBS and 100 μg/mL hygromycin (Invitrogen) to maintain transgene expression.
Cell staining
Fibroblasts were visualized with a DiffQuik staining kit (Baxter Diagnostics), which yields a modified Romanowski stain, according to the manufacturer's instructions.
Mixed lymphocyte responses
T cells were isolated from normal donors and adjusted to 5 × 105 cells/mL in AIM-V medium. Activated CLL cells were washed at least 4 times (to remove residual immunomodulators), irradiated (2500 cGy), and suspended at 5 × 105 cells/mL (or lower) concentrations in AIM-V. Responders and stimulators were then mixed in a 1:1 (vol/vol) ratio and cultured in 96-well round bottom plates (BD Labware) without additional cytokines or serum. Proliferation was measured 4 to 6 days later with a colorimetric assay.17
Flow cytometry
Cell were stained with 7AAD and analyzed as described previously.18 DNA content was also measured as before.17 Briefly, CLL cells (∼ 2 × 106/mL) were transferred to conical tubes, pelleted, and resuspended in 50 μL phosphate-buffered saline/Hank balanced salt solution with 2% FBS, and then fixed in 1 mL of ice-cold 80% ethanol. Cells were pelleted again and resuspended in 500 μL of 0.1 mg/mL propidium iodide (Sigma-Aldrich) with 0.6% Nonidet P-60 to which 500 μL of 2 mg/mL RNAse was then added. After incubation for 30 minutes at room temperature, the cells were filtered through nylon mesh and kept on ice in the dark before analysis on a FACSCalibur flow cytometer (BD Biosciences) using CellQuest flow.
Western blot analysis
Proteins were extracted from activated CLL cells, cell lines and mouse lymphocytes using RIPA buffer plus protease, and phosphatase inhibitors. Immunoblotting was performed as described before.17 Blots were stripped for 60 minutes at 37°C in Restore Western blot Stripping buffer (Pierce), washed twice in Tris-buffered saline plus 0.05% Tween-20 at room temperature, blocked with 10% milk for 1 hour, and reprobed, as required. Signals were detected with Supersignal horseradish peroxidase enhanced chemiluminescence reagent (Pierce), and blots were exposed to Kodak Biomax MR film.
Determination of relative pSTAT3 levels
The intensity of phosphorylated STAT3 (pSTAT3) and total STAT3 bands on a blot were quantified with GeneTools Analysis software from Syngene Genius 2 Bioimager (Version 4.00; Syngene). To control for experimental variations that arise when comparing multiple samples on different blots, intensity values were normalized to the results obtained with 50 μg of a reference standard from Patient 92 (Pt. 92), which was always run with each gel. To further control for experimental variation, 2 separate gels were run. The average of the results from the 2 gels was called the “relative pSTAT3 level at 4 hours.”
Para-nitrophenyl phosphate assay for phosphatase activity
CLL cells were lysed in a RIPA buffer containing 50mM Tris-HCl (pH 7.4), 150mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1mM ethylenediaminetetraacetic acid, 1mM phenylmethylsulfonyl fluoride, and aprotinin and leupeptin each at 1 mg/mL. Phosphatase activity in each lysate was assayed in 96-well plates (BD Labware) in a buffer (50mM Tris, 10mM NaCl, and 1mM ethylenediaminetetraacetic acid, and 10mM dithiothreitol [pH 7.2]) using para-nitrophenyl phosphate (10 mg/mL; Sigma-Aldrich), which is a chromogenic substrate for most phosphatases. The reaction yields para-nitrophenol, which becomes an intense yellow soluble product under alkaline conditions measurable at 405 nm on a spectrophotometric plate reader (model 3550; Bio-Rad).
Measurement of intracellular ROS
Formation of reactive oxygen species (ROS) was indicated by 2′7′-dichlorofluorescin diacetate (DCFH2-DA; Molecular Probes). Intracellular esterases cleave the acetyl groups from the molecule to produce nonfluorescent 2′7′-dichlorofluorescein (DCFH2), which is trapped inside the cell. In the presence of ROS, DCFH2 is oxidized to DCF, which emits fluorescence at 530 nm, after excitation at 488 nm. Activated CLL cells were incubated with 10μM DCFH2-DA at 37°C for 30 minutes. Samples were then washed in phosphate-buffered saline and analyzed on a FACSCalibur flow cytometer (BD Biosciences). DCFH2 oxidation was measured as “green” (FL1) fluorescence on a log-scale for 10 000 events.
To measure ROS production in circulating CLL cells, the cells were first purified from the blood and then cultured in serum-free medium for 2 to 3 days. This time was chosen on the basis of preliminary experiments showing that DCFH staining and cell viability decreased in normal B cells after day 5 in these conditions, but was relatively similar immediately after isolation or 1 and 3 days of culture.
Cytokine measurement
Cytokines in culture supernatants were measured in a Luminex-100 System as before.17 A kit for human interleukin-6 (IL-6), interleukin-8 (IL-8), interleukin-10 (IL-10), and tumor necrosis factor-α (TNF-α) was used, according to the manufacturer's instructions (R&D Systems). Individual cytokine concentrations were determined from standard curves using Bio-Plex 2.0 software (Bio-Rad). The assay was linear between 30 and 1000 pg/mL for each cytokine.
Statistical analysis
The Student t test was used to determine P values for differences between sample means.
Results
Variable IFN-mediated STAT3 phosphorylation and functional outcomes in CLL cells
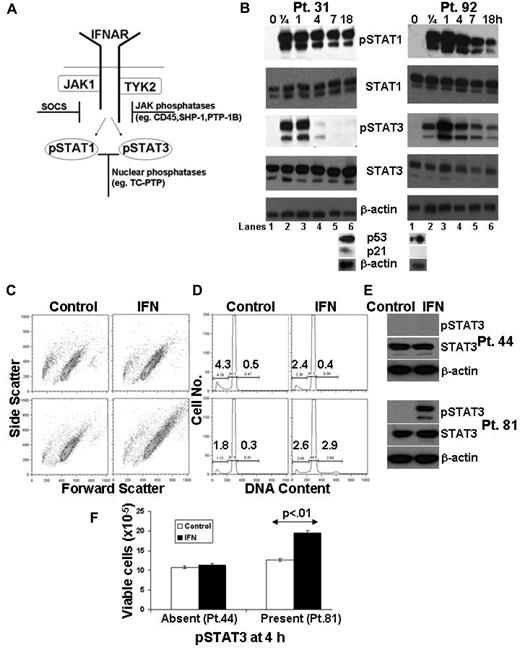
IFN-signaling in purified, circulating CLL cells was assessed by serial measurements of pSTAT1 and pSTAT3 proteins. Both STAT1 and STAT3 became phosphorylated within minutes of stimulation with IFN-α2b and pSTAT1 levels remained high for many hours. However, the decay of IFN-induced STAT3 phosphorylation exhibited significant interpatient variation. For example, pSTAT3 essentially disappeared after 4 hours in CLL cells from Pt. 31, whereas high levels persisted for up to 18 hours in cells from Pt. 92 (Figure 1B).
Variations in IFN-mediated STAT3 phosphorylation and proliferation in CLL cells. (A) Schematic diagram of IFN-signaling. (B) Primary CLL cells were treated with IFN-α2b (1000 U/mL) for the indicated times, and whole cell lysates were immunoblotted with antibodies to phosphotyrosine 701-specific STAT1 or phosphotyrosine 705-specific STAT3, as well as total STAT1, STAT3, and β-actin as loading controls. Cell aliquots were also exposed to ionizing radiation (2.5 Gy) and p53 and p21 levels were determined by immunoblotting 18 hours later (bottom blots). Tumor cells from Pt. 31 and Pt. 92 had intact and absent p53 axes, respectively, according to this assay.19 (C) Tumor cells from 2 other patients (Pt. 81 bottom, and Pt. 44 top) were cultured for 4 days in the presence or absence of IFN. Cell size was determined by the forward scatter parameter of flow cytometry. (D) DNA content was also determined by flow cytometry at this time. The number on the right of the histogram represents the percentage of cells with DNA content greater than 2N and the number on the left represents the percentage with subdiploid DNA content.39 (E) Immunoblots showing measurable pSTAT3 levels at 4 hours in CLL cells from Pt. 81 but not Pt. 44. F. Viable cells were counted in a hemocytometer after 4 days of culture. The results show that IFN induced a proliferative response when STAT3 phosphorylation persisted longer than 4 hours.
Variations in IFN-mediated STAT3 phosphorylation and proliferation in CLL cells. (A) Schematic diagram of IFN-signaling. (B) Primary CLL cells were treated with IFN-α2b (1000 U/mL) for the indicated times, and whole cell lysates were immunoblotted with antibodies to phosphotyrosine 701-specific STAT1 or phosphotyrosine 705-specific STAT3, as well as total STAT1, STAT3, and β-actin as loading controls. Cell aliquots were also exposed to ionizing radiation (2.5 Gy) and p53 and p21 levels were determined by immunoblotting 18 hours later (bottom blots). Tumor cells from Pt. 31 and Pt. 92 had intact and absent p53 axes, respectively, according to this assay.19 (C) Tumor cells from 2 other patients (Pt. 81 bottom, and Pt. 44 top) were cultured for 4 days in the presence or absence of IFN. Cell size was determined by the forward scatter parameter of flow cytometry. (D) DNA content was also determined by flow cytometry at this time. The number on the right of the histogram represents the percentage of cells with DNA content greater than 2N and the number on the left represents the percentage with subdiploid DNA content.39 (E) Immunoblots showing measurable pSTAT3 levels at 4 hours in CLL cells from Pt. 81 but not Pt. 44. F. Viable cells were counted in a hemocytometer after 4 days of culture. The results show that IFN induced a proliferative response when STAT3 phosphorylation persisted longer than 4 hours.
This behavior of pSTAT3 appeared to be linked to defects in the p53 axis, which are associated with clinically aggressive disease.19 In Figure 1B, the sample in which IFN-induced pSTAT3 levels were maintained, had defective p53 function (Pt. 92), in that p21 was not transcribed when the cells were radiated.19 In the sample in which pSTAT3 disappeared within 4 hours of stimulation (Pt. 31), the p53 axis was intact by these criteria. Moreover, IFN induced different behaviors in CLL cells depending on the duration of STAT3 phosphorylation. If IFN-induced STAT3 phosphorylation persisted longer than 4 hours (Figure 1E bottom), the tumor cells became larger (Figure 1C bottom), entered cell cycle (Figure 1D bottom), and increased in number almost 2-fold within 4 days (Figure 1F). In contrast, cells in which IFN-mediated STAT3 phosphorylation disappeared within 4 hours did not enlarge, and there was no increase in DNA content (Figure 1C-E top panels and F).
Increased viable cell numbers could be caused by effects of IFN on proliferation or on survival, or both. However, because these experiments were carried out in serum-free medium, little death occurred in control cells from the majority of patient samples during the culture period. Treatment with IFN did not increase the number of apoptotic cells (indicated by the subdiploid peak of cell-cycle analysis) as shown by Figure 1D and supplemental Figure 3 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) or necrotic cells, which is indicated by staining with the DNA dye, 7AAD (for example, as illustrated in Figure 7E). We conclude that the increase in cell numbers associated with prolonged IFN-mediated STAT3 phosphorylation was mainly from the increase (albeit small) in the number of cycling cells (Figure 1D). Accordingly, the phenotype of CLL cells that exhibited prolonged increases in pSTAT3 levels after treatment with IFN was characterized by an increase in cell size and proliferation, both of which are consistent with the known oncogene-like functions of STAT3.9,10
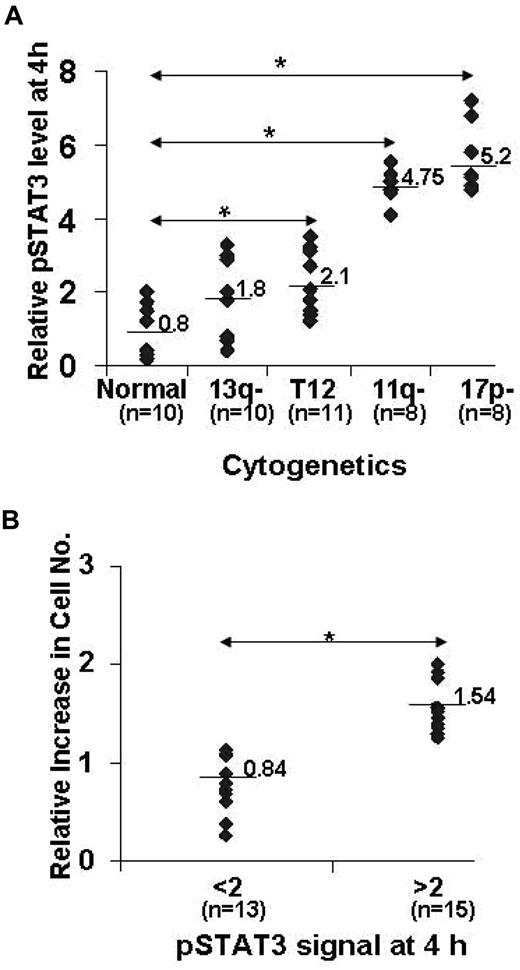
The 5 subtypes of CLL are distinguished by fluorescence in situ hybridization including types with no detectable cytogenetic lesions, trisomy 12, or deletions at 13q14, 11q22-q23 (with loss of the ATM gene) and 17p13 (with loss of p53).20 To more fully explore the relationship of the temporal characteristics of IFN-mediated STAT3 phosphorylation with underlying cytogenetic abnormalities, pSTAT3 and total STAT3 levels were measured in 47 different CLL samples after stimulation with IFN-α2b for 4 hours. This time was based on the initial analysis of the disappearance of IFN-mediated pSTAT3 proteins in individual patient samples. The amount of pSTAT3 relative to STAT3 at this time was then related to the most severe cytogenetic abnormality determined by fluorescence in situ hybridization (eg, patients with both 13q− and 17p− deletions were classed as 17p−). As shown in Figure 2A, IFN-induced pSTAT3 levels were higher in CLL cells with 11q− or 17p− deletions compared with samples with low-risk cytogenetic abnormalities and were not related to altered expression of IFN-signaling pathway components (Figure 1A and data not shown). This larger study confirmed that CLL cells that exhibited prolonged STAT3 phosphorylation (defined as a densitometric ratio of pSTAT3 to STAT3 greater than 2 after 4 hours of stimulation with IFN) increased in number, whereas cells with a shorter duration of STAT3 phosphorylation (pSTAT3 to STAT3 ratio less than 2 after 4 hours) did not show an increase (Figure 2B).
Correlation of IFN-mediated pSTAT3 duration and proliferation with cytogenetic lesions. (A) Circulating CLL cells from 47 consecutive patients presenting to the CLL clinic at Sunnybrook Health Sciences Center, as described in Table 1, were isolated and stimulated with IFN-α2b. After 4 hours, levels of pSTAT3 relative to total STAT3 were determined by immunoblotting and densitometry as described in “Western blot analysis.” This number was then plotted as a function of the highest-risk cytogenetic abnormality.38 The median value for each subgroup is shown.*P < .001 (B) Cell cultures were continued for another 4 days. Viable cells were then counted in a hemocytometer and the ratio of the numbers obtained with and without IFN-α2b was plotted against the pSTAT3 to pSTAT1 ratio after 4 hours. Median values for the 2 groups are indicated. *P < .01).
Correlation of IFN-mediated pSTAT3 duration and proliferation with cytogenetic lesions. (A) Circulating CLL cells from 47 consecutive patients presenting to the CLL clinic at Sunnybrook Health Sciences Center, as described in Table 1, were isolated and stimulated with IFN-α2b. After 4 hours, levels of pSTAT3 relative to total STAT3 were determined by immunoblotting and densitometry as described in “Western blot analysis.” This number was then plotted as a function of the highest-risk cytogenetic abnormality.38 The median value for each subgroup is shown.*P < .001 (B) Cell cultures were continued for another 4 days. Viable cells were then counted in a hemocytometer and the ratio of the numbers obtained with and without IFN-α2b was plotted against the pSTAT3 to pSTAT1 ratio after 4 hours. Median values for the 2 groups are indicated. *P < .01).
Effect of p53 loss on IFN-mediated pSTAT3 levels in B-lymphoma cells
Results with primary CLL cells (Figures 1–2) suggested that adverse oncogenic lesions were associated with prolonged IFN-mediated STAT3 phosphorylation, leading to paradoxical use of IFN-α2b as a growth factor rather than a cytostatic molecule. To study this finding in more detail, we focused on p53 because 17p− deletions are associated with the most aggressive forms of CLL.21 As a model, IFN responses were studied in an Epstein-Barr virus –transformed lymphoblastoid B-cell line with wt p53 (TK6) and a double p53 knockout cell line derived from TK6 using a promoterless gene targeting approach.14 Because Epstein-Barr virus proteins activate similar signaling pathways as inflammatory mediators encountered by CLL cells22 in proliferation centers,1 we reasoned that these 2 cell lines might provide insights into how signaling and responsiveness of malignant B cells to IFN are affected by loss of p53.
The status of p53 in the cell lines was confirmed by the increased expression of p53 and p21 in irradiated TK6 cells and their absence in irradiated NH32 cells (Figure 3A, top).19 IFN-α2b induced phosphorylation of STAT1 and STAT3 within 1 hour in both TK6 and NH32 cells. However, consistent with the findings in primary CLL cells, pSTAT3 levels remained high for more than 7 hours after stimulation with IFN-α2b in NH32 cells, which lacked functional p53, whereas these levels decayed much more rapidly in TK6 cells (Figure 3A, middle and bottom). Similarly, treatment with IFN-α2b produced a higher number of NH32 cells after 4 days of culture, whereas TK6 numbers were significantly lower (Figure 3B).
Increased IFN-mediated STAT3 phosphorylation and proliferation in p53−/− B-cell lines. (A) The p53 axes of TK6 (left) and NH32 cells (right) were assessed by measuring p53 and p21 levels 18 hours after irradiation. Other cells were treated with IFN-α2b (1000 U/mL) for the indicated times, and whole cell lysates were then immunoblotted with phosphotyrosine 701-specific STAT1 or phosphotyrosine 705-specific STAT3 antibodies. The pSTAT3 level relative to pSTAT1 was quantified by densitometry and shown below the blot. (B) TK6 and NH32 (5 × 104 cells/mL) were cultured with or without IFN-α (1000 U/mL) for 48 hours and then counted manually in a hemocytometer. The average (+ SE) of 3 separate measurements is shown. (C) Supernatants were also collected after 48 hours and the concentrations of IL-6, IL-10, IL-8, and TNF-α were determined as described in “Cytokine measurement.”
Increased IFN-mediated STAT3 phosphorylation and proliferation in p53−/− B-cell lines. (A) The p53 axes of TK6 (left) and NH32 cells (right) were assessed by measuring p53 and p21 levels 18 hours after irradiation. Other cells were treated with IFN-α2b (1000 U/mL) for the indicated times, and whole cell lysates were then immunoblotted with phosphotyrosine 701-specific STAT1 or phosphotyrosine 705-specific STAT3 antibodies. The pSTAT3 level relative to pSTAT1 was quantified by densitometry and shown below the blot. (B) TK6 and NH32 (5 × 104 cells/mL) were cultured with or without IFN-α (1000 U/mL) for 48 hours and then counted manually in a hemocytometer. The average (+ SE) of 3 separate measurements is shown. (C) Supernatants were also collected after 48 hours and the concentrations of IL-6, IL-10, IL-8, and TNF-α were determined as described in “Cytokine measurement.”
Production of various cytokines and chemokines, including IL-6 and IL-10, which are regulated by JAK-STAT signaling modules,23 was measured to determine whether the altered kinetics of pSTAT3 reflected enhanced STAT3 activity. NH32 cells constitutively produced IL-10, which was greatly enhanced by IFN-α2b. Low level production of IL-6 was also increased somewhat by IFN-α2b in NH32 cells, whereas TNF-α and IL-8 production were not affected. In contrast, IL-10 and IL-6 production by TK6 cells were not changed by IFN-α2b, whereas IL-8 was only modestly increased (Figure 2C).
NH32 is derived from TK6 but the 2 cell lines have been subjected to the stresses of tissue culture24 for many generations and may have evolved different genetic and epigenetic alterations. Accordingly, an aberrant pathway other than p53 could account for prolonged IFN-mediated STAT3 phosphorylation in NH32 cells. Consistent with this possibility, IFN-signaling patterns were not different in spleen cells from p53+/+ and p53−/− mice (supplemental Figure 1B) or at permissive and nonpermissive temperatures for functional p53 activity in human B cells expressing a temperature-sensitive p53 mutant15 (supplemental Figure 1A). Similarly, IFN-signaling in CLL cells was not altered by the p53 inhibitor, pifithrin (supplemental Figure 2).25
Effect of ATM loss on IFN-mediated pSTAT3 levels
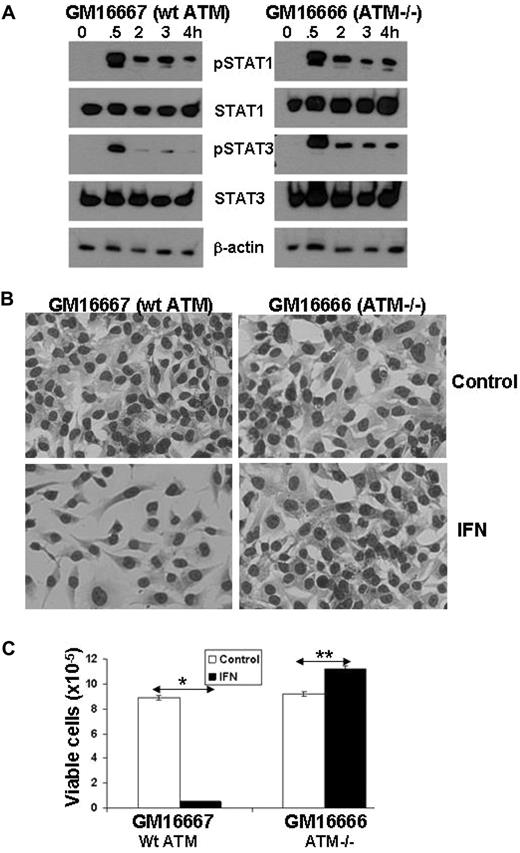
The results shown in Figure 2 suggested that 11q− deletions and loss of ATM were also associated with prolonged IFN-mediated phosphorylation of STAT3. Accordingly, IFN-signaling was studied in the GM16666 and GM16667 cell lines derived from fibroblasts of a patient with ataxia telangiectasia (caused by a mutant ATM gene) transfected with either an ATM expression construct (GM16667) or an empty vector (GM16666).26 Treatment of GM16667 cells (that express wt ATM) with IFN-α2b caused only a brief phosphorylation of STAT3 (Figure 4A) and strongly inhibited cell growth (Figure 4B-C). Remarkably, STAT3 phosphorylation was prolonged in ATM-deficient GM16666 cells (Figure 4A), which grew in response to IFN (Figure 4B-C).
Increased IFN-mediated STAT3 phosphorylation and proliferation in ATM−/− cell lines. (A) GM16667 (wt ATM) and GM16666 (absent ATM) cells in exponential growth phase were treated with IFN-α2b (1000 U/mL) for the indicated times, and whole cell lysates were then immunoblotted with antibodies to phosphotyrosine 701-specific STAT1, phosphotyrosine 705-specific STAT3, total STAT1 and STAT3, or β-actin. (B) After 3 days of culture, the cells were stained and images were taken at a magnification of ×10. Pictures were taken with a Canon PowerShot G11 Digital Camera equipped with a Carl Zeiss 426126 lens. The microscope was an Axiovert 40 C equipped with an A-plan 10×/0.25 objective, both from Zeiss. (C) The cells were also counted manually in a hemocytometer. The average (+ SE) of 3 separate measurements is shown and the experiment was performed twice with similar results. *P < .01; **P < .05
Increased IFN-mediated STAT3 phosphorylation and proliferation in ATM−/− cell lines. (A) GM16667 (wt ATM) and GM16666 (absent ATM) cells in exponential growth phase were treated with IFN-α2b (1000 U/mL) for the indicated times, and whole cell lysates were then immunoblotted with antibodies to phosphotyrosine 701-specific STAT1, phosphotyrosine 705-specific STAT3, total STAT1 and STAT3, or β-actin. (B) After 3 days of culture, the cells were stained and images were taken at a magnification of ×10. Pictures were taken with a Canon PowerShot G11 Digital Camera equipped with a Carl Zeiss 426126 lens. The microscope was an Axiovert 40 C equipped with an A-plan 10×/0.25 objective, both from Zeiss. (C) The cells were also counted manually in a hemocytometer. The average (+ SE) of 3 separate measurements is shown and the experiment was performed twice with similar results. *P < .01; **P < .05
However, analogous to the situation with p53, IFN-signaling was similar in the spleen and thymus of ATM+/+ and ATM−/− mice (supplemental Figure 1B right) and treatment of CLL cells with the ATM inhibitor, KU-55933, did not alter IFN-signaling responses (supplemental Figure 2). Taken together, these results suggested that loss of p53 and ATM were necessary but not sufficient to corrupt IFN-signaling patterns in CLL cells.
Effect of phorbol esters on IFN-signaling in CLL cells
The association of aberrant IFN-signaling with high-risk cytogenetic lesions that could not be reproduced by inhibitors of p53 or ATM raised the possibility that other aspects of the cellular phenotype of aggressive tumors are associated with prolonged IFN-mediated STAT3 phosphorylation. Increased ROS levels are associated with CLL cells that are sufficiently aggressive to have warranted chemotherapy.27 Other features of high-risk CLL cells include activation of PKC28,29 and enhanced signaling through immunoreceptors.30 Because phorbol esters activate PKC, increase ROS and CD38 expression,31 and promote immunoreceptor signaling,30 exposing CLL cells to these tumor promoters may model some of the behavioral aspects of aggressive tumor cells.
Similar to CLL cells with high-risk cytogenetic lesions, IFN-mediated STAT3 phosphorylation was prolonged by concomitant activation with phorbol esters (Figure 5A). This result was not an indirect effect of cytokines induced by IFN because it was still observed in the presence of the protein translation inhibitor CHX (Figure 5B). Phorbol esters increase ROS levels partly through classic PKC isozymes,32 and both the antioxidant, NAC, and the PKC inhibitor, Go6976, shortened the duration of IFN-mediated STAT3 phosphorylation but did not otherwise affect pSTAT1 levels (Figure 5C-D).
Effect of phorbol esters on IFN-signaling in CLL cells. (A). CLL cells were cultured for 24 hours with or without PDB (100 ng/mL) and/or IFN-α2b (1000 U/mL). pSTAT1, pSTAT3, and total STAT1 and STAT3 levels were then determined by immunoblotting. Immunoblots were stripped and reprobed with β-actin antibodies to assess loading. The results were similar for 21 additional samples. (B) CLL cells were cultured alone or with PDB for 48 hours (to allow abatement of cytokine production caused by PDB alone), washed, and recultured with IFN in the presence and absence of CHX (10 μg/mL) for 6 hours before determining pSTAT1 and pSTAT3 levels. The pSTAT3 levels were higher in cells treated with PDB and IFN than in cells treated with IFN alone and were not affected by the presence of CHX to block autocrine cytokine production. Similar results were obtained with 4 additional patient samples. (C-D) CLL cells were cultured alone or with PDB, IFN, or PDB and IFN with or without the classic PKC isozyme inhibitor, Go6976 (1μM) or the antioxidant (C), NAC (30μM) added 1 hour before stimulation (D). Cell lysates were collected 4 hours later and pSTAT1 and pSTAT3 levels determined by immunoblotting. Go6976 and NAC shortened STAT3 phosphorylation in CLL cells treated with PDB and IFN. This experiment was repeated with 8 different patient samples with similar results. (E) After 24 hours of culture, global phosphatase levels were determined using para-nitrophenyl phosphate as described in “Para-nitrophenyl phosphate assay for phosphatase activity.” This experiment was repeated with 8 different CLL patient samples and showed that global phosphatase activity was decreased in PDB-treated CLL cells, which could be reversed by blocking PKC and ROS. *P < .05.
Effect of phorbol esters on IFN-signaling in CLL cells. (A). CLL cells were cultured for 24 hours with or without PDB (100 ng/mL) and/or IFN-α2b (1000 U/mL). pSTAT1, pSTAT3, and total STAT1 and STAT3 levels were then determined by immunoblotting. Immunoblots were stripped and reprobed with β-actin antibodies to assess loading. The results were similar for 21 additional samples. (B) CLL cells were cultured alone or with PDB for 48 hours (to allow abatement of cytokine production caused by PDB alone), washed, and recultured with IFN in the presence and absence of CHX (10 μg/mL) for 6 hours before determining pSTAT1 and pSTAT3 levels. The pSTAT3 levels were higher in cells treated with PDB and IFN than in cells treated with IFN alone and were not affected by the presence of CHX to block autocrine cytokine production. Similar results were obtained with 4 additional patient samples. (C-D) CLL cells were cultured alone or with PDB, IFN, or PDB and IFN with or without the classic PKC isozyme inhibitor, Go6976 (1μM) or the antioxidant (C), NAC (30μM) added 1 hour before stimulation (D). Cell lysates were collected 4 hours later and pSTAT1 and pSTAT3 levels determined by immunoblotting. Go6976 and NAC shortened STAT3 phosphorylation in CLL cells treated with PDB and IFN. This experiment was repeated with 8 different patient samples with similar results. (E) After 24 hours of culture, global phosphatase levels were determined using para-nitrophenyl phosphate as described in “Para-nitrophenyl phosphate assay for phosphatase activity.” This experiment was repeated with 8 different CLL patient samples and showed that global phosphatase activity was decreased in PDB-treated CLL cells, which could be reversed by blocking PKC and ROS. *P < .05.
Tyrosine phosphatases are inhibited by oxidation,33 which could account for prolonged IFN-mediated STAT3 phosphorylation and its normalization by antioxidants. Consistent with this effect, total phosphatase activity was diminished in phorbol ester–activated CLL cells and restored in the presence of NAC or Go6976 (Figure 5E). However, we were unable to implicate inactivation of specific tyrosine phosphatases (such as CD45 and PTP1b; Figure 1A) that are known to regulate aspects of IFN receptor signaling.23
ROS levels in CLL cells
The association of prolonged IFN-mediated STAT3 phosphorylation with ROS in phorbol ester–activated CLL cells (Figure 5) suggested that higher ROS levels might be found in aggressive CLL cells that exhibit similar changes in IFN-signaling (Figure 2). Accordingly, circulating CLL cells from an additional 32 patients (Table 2) were purified and stained with DCFH after a brief period of culture in serum-free conditions. ROS levels in CLL cells were found to be much higher than in normal B cells (Figure 6A) and exhibited significant interpatient variability. Remarkably, the pattern of DCFH staining mirrored the relative 4-hour pSTAT3 levels observed in the different cytogenetic subtypes of CLL (Figures 2A,6A). ROS levels were significantly higher in CLL cells with 11q− or 17p− deletions, compared with the other subtypes. Consistent with an association of increased ROS levels with more aggressive disease, DCFH staining was also significantly higher in tumor cells with higher CD38 expression (Figure 6C). Lymphocyte doubling times (LDTs) were significantly faster (< 12 months) in patients whose tumor cells had high ROS levels (Figure 6B). These patients also had a higher Rai Stage classification (Figure 6D) and were much more likely to require treatment for their disease (Figure 6E).
Correlation of intracellular ROS levels with clinical parameters. The mean fluorescence intensity (mfi) of DCFH staining (indicative of ROS levels) in short-term cultured CLL cells was measured as described in “Measurement of intracellular ROS” for 33 patients whose clinical features are shown in Table 2. Cytogenetic abnormalities (A), LDTs (B), percentage of CD38+ cells (C), clinical Rai Stage classification (D), and number of treatments for symptomatic disease (E) were determined from patients medical records. The average values and standard errors for the different groups are shown. Tumor cells from patients with more aggressive disease had significantly higher ROS levels, as indicated by the P values in the respective panels.*P < .001; **P < .02. The open circles in panel A represent ROS levels in B cells from 4 normal donors, handled in the same manner as CLL cells.
Correlation of intracellular ROS levels with clinical parameters. The mean fluorescence intensity (mfi) of DCFH staining (indicative of ROS levels) in short-term cultured CLL cells was measured as described in “Measurement of intracellular ROS” for 33 patients whose clinical features are shown in Table 2. Cytogenetic abnormalities (A), LDTs (B), percentage of CD38+ cells (C), clinical Rai Stage classification (D), and number of treatments for symptomatic disease (E) were determined from patients medical records. The average values and standard errors for the different groups are shown. Tumor cells from patients with more aggressive disease had significantly higher ROS levels, as indicated by the P values in the respective panels.*P < .001; **P < .02. The open circles in panel A represent ROS levels in B cells from 4 normal donors, handled in the same manner as CLL cells.
Normalization of IFN-signaling and responses by TYK2 inhibition
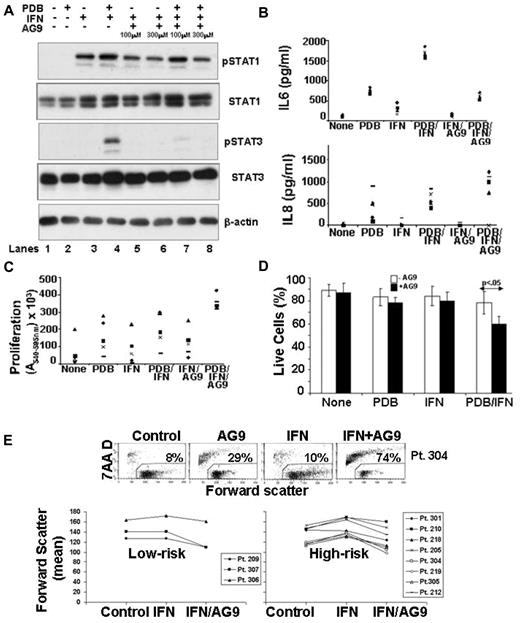
Because prolonged IFN-mediated STAT3 phosphorylation was associated with cell growth (Figures 1D,F and 2B) and production of immunosuppressive cytokines (Figure 3C), a clinically relevant method to inhibit IFN-mediated phosphorylation of STAT3, while preserving STAT1 phosphorylation, might improve the therapeutic effects of IFN-α2b in high-risk patients with CLL. Current models of IFNAR signaling suggest that STAT1 is phosphorylated by JAK1, whereas STAT3 is phosphorylated by TYK2 (Figure 1A).8 Accordingly the effect on IFN responses of AG9, a tyrphostin that inhibits IL-2–stimulated TYK2 phosphorylation in T cells,34 was studied in CLL cells treated concomitantly with phorbol esters to mimic features of high-risk disease (Figure 5).
The levels of pSTAT3 normally seen after 5 hours of treatment with PDB and IFN were abrogated by AG9 in a dose-dependent manner, with maximal inhibition achieved at 300μM (Figure 7A compare lanes 4, 7, and 8). Consistent with direct inhibition of TYK2, early (ie, within 10 minutes) IFN-mediated STAT3 phosphorylation was also prevented by AG9 (data not shown).
Effects of a TYK2 inhibitor on IFN-signaling and responses. (A) CLL cells were cultured for 1 hour alone, or with different concentrations of AG9, before treatment with PDB (100 ng/mL), IFN-α2b (1000 U/mL), or PDB and IFN-α. After 4 hours, pSTAT1 and total STAT1 and STAT3 levels were determined by immunoblotting. STAT3 phosphorylation was abrogated by AG9, whereas STAT1 activation remained intact. Similar results were obtained with 5 different patient samples. (B) Concentrations of IL-6 (top) and IL-8 (bottom) were measured in culture supernatants after 48 hours. IL-6 production was increased by PDB and inhibited by AG9 without significantly affecting IL-8. (C) CLL cells cultured for 3 to 4 days with or without PDB and/or IFN in the presence or absence of AG9 were obtained from 5 patients and used to stimulate T cells from healthy donors to proliferate as measured 5 to 6 days later by a colorimetric assay. Averages (+ SE) from triplicate mixed lymphocyte reactions cultures (after subtracting the averages from control wells containing T cells and stimulators, alone) are shown. The different symbols represent the results obtained using individual patient stimulator cells. Prevention of IFN-mediated STAT3 phosphorylation by AG9 made CLL cells, treated concomitantly with PDB and IFN, better able to stimulate T cell proliferation. (D) The percentage of viable cells that excluded 7-AAD was determined by flow cytometry after 48 hours. The average (+ SE) of the results from 5 different patient samples are shown. The arrow indicates a statistically significant value of P < .05. (E) CLL cells from Pt. 304 were cultured with or without IFN and/or AG9 (top) and cell viability after 3 days was indicated by measuring exclusion of 7-AAD by flow cytometry. The number in each dot blot is the percentage of 7-AAD+ dead cells. Mean values of the forward scatter parameter for 10 other patient samples are summarized (bottom graphs). “High-risk” cells are defined on the basis of 11q− or 17p− cytogenetic lesions, over 20% CD38+ cells, or LDTs < 12 months. AG9 counters the increased cell size caused by IFN in aggressive CLL cells.
Effects of a TYK2 inhibitor on IFN-signaling and responses. (A) CLL cells were cultured for 1 hour alone, or with different concentrations of AG9, before treatment with PDB (100 ng/mL), IFN-α2b (1000 U/mL), or PDB and IFN-α. After 4 hours, pSTAT1 and total STAT1 and STAT3 levels were determined by immunoblotting. STAT3 phosphorylation was abrogated by AG9, whereas STAT1 activation remained intact. Similar results were obtained with 5 different patient samples. (B) Concentrations of IL-6 (top) and IL-8 (bottom) were measured in culture supernatants after 48 hours. IL-6 production was increased by PDB and inhibited by AG9 without significantly affecting IL-8. (C) CLL cells cultured for 3 to 4 days with or without PDB and/or IFN in the presence or absence of AG9 were obtained from 5 patients and used to stimulate T cells from healthy donors to proliferate as measured 5 to 6 days later by a colorimetric assay. Averages (+ SE) from triplicate mixed lymphocyte reactions cultures (after subtracting the averages from control wells containing T cells and stimulators, alone) are shown. The different symbols represent the results obtained using individual patient stimulator cells. Prevention of IFN-mediated STAT3 phosphorylation by AG9 made CLL cells, treated concomitantly with PDB and IFN, better able to stimulate T cell proliferation. (D) The percentage of viable cells that excluded 7-AAD was determined by flow cytometry after 48 hours. The average (+ SE) of the results from 5 different patient samples are shown. The arrow indicates a statistically significant value of P < .05. (E) CLL cells from Pt. 304 were cultured with or without IFN and/or AG9 (top) and cell viability after 3 days was indicated by measuring exclusion of 7-AAD by flow cytometry. The number in each dot blot is the percentage of 7-AAD+ dead cells. Mean values of the forward scatter parameter for 10 other patient samples are summarized (bottom graphs). “High-risk” cells are defined on the basis of 11q− or 17p− cytogenetic lesions, over 20% CD38+ cells, or LDTs < 12 months. AG9 counters the increased cell size caused by IFN in aggressive CLL cells.
Treatment of CLL cells with AG9 decreased production of IL-6 in response to PDB and IFN, as well to IFN alone (Figure 7B top), whereas IL-8 levels remained the same or were increased (Figure 7B bottom). Changes in IL-10 production tended to follow the pattern of IL-6 (data not shown).
The type I IFNs exert their normal tumor-suppressor functions in part by altering the expression of cytokines and costimulatory molecules to increase the immunogenicity of tumor cells, allowing them to effectively stimulate and be killed by tumor-reactive T cells.2,4 The capacity of CLL cells to stimulate T-cell proliferation, as measured in allogeneic mixed lymphocyte reactions, is poor but can be improved by phorbol esters.35 IFN did not change significantly the stimulatory ability of PDB-activated CLL cells (Figure 7C). However, blockade of STAT3 activation by AG9 caused CLL cells treated with PDB and IFN to acquire strong stimulatory ability (Figure 7C). Similar results were obtained with another putative TYK 2 inhibitor, Piceatannol8 (data not shown).
Changing the balance of IFN-mediated gene transcription to favor STAT1 over STAT3 responses might be expected to cause cell death.9,10 Consistent with this possibility, significantly more cells died after 24 hours of treatment with PDB and IFN in the presence of AG9 (Figure 7D). Because phorbol ester–activated CLL cells exhibit properties associated with aggressive CLL cells (ie, increased ROS, prolonged IFN-mediated STAT3 phosphorylation), these findings suggested that AG9 might also allow IFN to kill aggressive CLL cells directly. Accordingly, CLL cells from 3 low- and 8 high-risk patients (defined by 11q− or 17p− deletions, high CD38 expression, or short LDTs) were stimulated with IFN, with or without AG9, for 4 days. Cell death was then measured by uptake of the DNA dye 7-AAD, and size was measured by the forward scatter parameter of flow cytometry. The increase in size associated with prolonged IFN-mediated STAT3-phosphorylation in high-risk CLL cells (Figure 1C) was prevented by AG9 (Figure 7E bottom right panel). As expected (Figure 1C), this IFN-mediated size increase was not seen in samples from low-risk patients (Figure 7E, bottom left panel). Unlike phorbol ester-activated cells, in only 1 case (Pt. 304) did IFN and AG9 act in a synergistic manner to kill cells in vitro (Figure 7E top). These findings suggest that phorbol esters have additional effects on tumor cells that affect responses to IFN. However, the results also suggest that TYK2 inhibitors can counter some of the aberrant effects of IFN on high-risk CLL cells.
Discussion
This study shows that IFN-signaling is altered in leukemia cells from CLL patients with aggressive clinical disease (Figure 1). The normally brief period of activation of STAT3 induced by IFN-α in normal cells becomes sustained, and the outcome switches from a tumor-suppressor phenotype involving growth arrest and enhanced immunogenicity to a phenotype involving proliferation and production of immunosuppressive cytokines, such as IL-10, consistent with the oncogenic effects of STAT3-mediated gene expression (Figures 1,Figure 2,Figure 3,Figure 4–5).
“Corrupted” IFN-signaling appears to be a phenotype associated with aggressive disease, intertwined with the processes of tumor progression but not linked solely to a single oncogene (Figures 3,4). A variety of tumorigenic pathways appear to yield this phenotype. Increased ROS levels associated with more aggressive disease (Figure 6) may lower the activity of many phosphatases (Figure 5) and enhance the survival (and activity) of phosphorylated proteins in general, and pSTAT3 in particular. Although p53 and ATM do not appear to directly affect IFN-signaling (supplemental Figures 1-2), growth of tumor cells, unrestrained by these tumor suppressors, appears to result in gene expression patterns and elevated ROS levels (Figure 6) that impart aberrant IFN-signaling (Figures 1–2).
Where do the elevated ROS levels in aggressive CLL cells originate? One possibility is receptors on the B-cell surface that transduce signals that stimulate mitochondria to provide energy for functional cellular responses. Activated mitochondria also make ROS, which can be measured by flow cytometry with DCFH.36 A hallmark of aggressive CLL cells is their enhanced responsiveness to environmental signals30,37 and increased ROS levels in these cells may reflect these signaling processes.
It seems unlikely that corruption of IFN-signaling in aggressive tumor cells is unique to CLL.10 Why might altered IFN-signaling be involved in the development of a cancer? Potentially, intact IFN-signaling leading to STAT1-dominated gene expression allows the immune system to initially control an evolving tumor.4 Rewiring of IFN-signaling pathways may negate the normal tumor-suppressor function of IFN, causing it to become a growth and immunosuppressive factor that promotes cancer development. Assessment of the duration of pSTAT3 persistence after stimulation with IFN-α2b in vitro might then be an independent method to assess prognosis in patients with CLL (Figure 1).38 Larger, prospective studies are needed to address this possibility.
Our identification of an association of altered IFN-signaling pathways with high-risk disease characteristics may help explain the clinical observation that IFN-α2b is most effective in patients with relatively low-risk CLL.6 Our results also suggest that inhibiting the activation of STAT3 with small molecule TYK2 inhibitors (Figure 7) or possibly antioxidants (Figure 5) might be a strategy to improve the therapeutic efficacy of IFN-α2b in patients with more aggressive disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Howard Liber (Colorado State University) for TK6 and NH3 cells, Noemi Nagy (Karolinska Institutet) for BL41tsp53 cells, Greg Downey and Vera Cherepanov (University of Toronto) for help with phosphatase assays, Stephen Meyn and Paul Bradshaw (Hospital for Sick Children and University of Toronto) for GM16666 and GM16667 cell lines, Jayne Danska (Hospital for Sick Children) for ATM−/− mice, and Gisele Knowles (Sunnybrook Research Institute) for help with cell-cycle analysis.
This work was supported by the Ontario Institute of Cancer Research grant no. 07Nov-61, the Canadian Institutes of Health Research grant no. 190633, the Leukemia and Lymphoma Society of Canada (D.E.S.), and the Harold E. Johns Studentship award of the National Cancer Institute of Canada (J.T.).
National Institutes of Health
Authorship
Contribution: J.T. and D.E.S. designed and performed research, analyzed data, and wrote the paper; and B.L. made murine IFN-β and helped design research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David E. Spaner, Division of Molecular and Cellular Biology, Research Institute, S-116A, Research Bldg, Sunnybrook Health Sciences Center, 2075 Bayview Ave, Toronto, ON, Canada M4N 3M5; e-mail: spanerd@sri.utoronto.ca.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal