Abstract
The prognosis of germinal center–derived B-cell (GCB) lymphomas, including follicular lymphoma and diffuse large-B-cell lymphoma (DLBCL), strongly depends on age. Children have a more favorable outcome than adults. It is not known whether this is because of differences in host characteristics, treatment protocols, or tumor biology, including the presence of chromosomal alterations. By screening for novel IGH translocation partners in pediatric and adult lymphomas, we identified chromosomal translocations juxtaposing the IRF4 oncogene next to one of the immunoglobulin (IG) loci as a novel recurrent aberration in mature B-cell lymphoma. FISH revealed 20 of 427 lymphomas to carry an IG/IRF4-fusion. Those were predominantly GCB-type DLBCL or follicular lymphoma grade 3, shared strong expression of IRF4/MUM1 and BCL6, and lacked PRDM1/BLIMP1 expression and t(14;18)/BCL2 breaks. BCL6 aberrations were common. The gene expression profile of IG/IRF4-positive lymphomas differed from other subtypes of DLBCL. A classifier for IG/IRF4 positivity containing 27 genes allowed accurate prediction. IG/IRF4 positivity was associated with young age and a favorable outcome. Our results suggest IRF4 translocations to be primary alterations in a molecularly defined subset of GCB-derived lymphomas. The probability for this subtype of lymphoma significantly decreases with age, suggesting that diversity in tumor biology might contribute to the age-dependent differences in prognosis of lymphoma.
Introduction
The 2 most common lymphoma subtypes in Western countries are diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL), accounting for up to 36% and 32% of all lymphomas in adults, respectively.1 Both subtypes are thought to originate from germinal center (GC)–derived B cells.
Approximately 85% of FL carry the chromosomal translocation t(14;18)(q32;q21) as a primary oncogenic event. This translocation juxtaposes the BCL2 oncogene from 18q21 next to the immunoglobulin heavy chain (IGH) locus in 14q32.2 The t(14;18) has a lower incidence in FL grade 3 than in FL grade 1 or 2 and, remarkably, is almost completely absent in FL below the age of 18.3,4 In contrast to adult cases, pediatric FLs are also more frequently grade 3 or composite FL/DLBCL and have a significantly better 5-year event-free survival.4,5 Therefore, “pediatric FL” has been considered as distinct variant of FL by the updated World Health Organization classification.2
DLBCL may derive from transformation of low-grade lymphoma, such as FL, or occur as a de novo malignancy. By gene expression profiling applying “cell-of-origin” signatures, DLBCLs have been divided in biologic subgroups, with the GC B-cell-like (GCB) and the activated B-cell-like (ABC) subtypes being the most prominent.6 Compared with ABC-DLBCL, the GCB subtype is characterized by a better prognosis and a different pattern of genetic aberrations, including the presence of the t(14;18) in approximately 20% to 30% of cases.7,8 Whereas in adults GCB- and ABC-DLBCLs account for 48% and 30% of all de novo DLBCLs,7 respectively, pediatric DLBCLs are predominantly GCB type.9 Despite this predominance, DLBCLs in children again almost completely lack t(14;18). They have a better prognosis than adult DLBCLs.10,11
There is an ongoing debate whether age-related differences of tumor biology, host characteristics, or treatment-associated factors, alone or in combination, contribute to the better outcome of pediatric compared with adult B-cell lymphomas. Age is a continuous variable with well-established prognostic impact within the adult lymphoma population. Nevertheless, few studies have investigated biologic variables across all lymphoma age groups.8,12
Translocations involving the immunoglobulin (IG) loci are the hallmarks of several subtypes of B-cell lymphoma.13 To determine whether hitherto unidentified recurrent IG translocations occur in B-cell lymphomas, we have initiated systematic FISH screening for IGH translocations in the lymphomas characterized in the network project Molecular Mechanisms in Malignant Lymphomas (MMML). This has allowed the identification of chromosomal translocation juxtaposing the IRF4/MUM1 oncogene next to the IGH locus, which is cytogenetically cryptic and thus probably has been missed in conventional cytogenetic studies. We have shown that IG/IRF4 fusions are recurrent genetic changes in GC-derived lymphomas and identify a previously unrecognized subset of B-cell lymphomas with characteristic clinical, morphologic, immunophenotypic, and gene expression profiles. These lymphomas are significantly associated with disease onset in childhood and young adulthood and have a favorable prognosis.
Methods
Lymphoma samples
A total of 720 lymphomas were studied herein. First, a core group of 427 cases was screened by FISH for IRF4 breaks. This series contained 183 cases from network project MMML,8,14 74 cases from the Berlin-Frankfurt-Münster-NHL trials (www.uniklinikum-giessen.de/nhlbfm),4,9 161 cases from the NHL-B trials of the German High-Grade NHL Study Group (www.lymphome.de/Gruppen/DSHNHL),15 and 9 cases with IG break and unknown partner from the routine cytogenetic diagnostics. A complete description of the population screened by FISH is available in supplemental Table 1 and supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Array-based data of 438 lymphomas were mined, including 145 cases from the core group studied by FISH. For 167 of those 438 lymphomas, gene expression and molecular cytogenetic data have been released previously (GEO accession nos. GSE4475 and GSE10172),8,14 whereas 271 lymphomas (GEO accession no. GSE22470) were newly characterized using the published protocols.8,14
The protocols of the clinical trials and the MMML have been approved by central and local review boards. Part of the study has been registered at www.Clinicaltrials.gov as NCT00324779.
Immunohistochemistry and FISH
Immunohistochemistry and FISH analyses were performed using standard protocols.9 Lymphomas were scored positive for CD5, CD10, MUM1, BCL6, BCL2, and BLIMP1 if > 25% of the tumor cells stained positive as described in previous publications.4,9 Staining for Ki-67 was assessed in percentage of positive tumor cells. The algorithm published by Hans et al was applied to classify B-cell lymphomas into GCB and non-GCB subtypes.16 Complete methods and probes used for the detection of breakpoints or gene fusions affecting the IGH, IGL, IGK, BCL2, BCL6, MYC, and IRF4 loci are described in supplemental Table 2.
Long-distance inverse PCR
Long-distance inverse PCR for the IGH switch regions was performed as described previously17 with modifications. Experimental procedures are described in supplemental Table 3.
Mutation analyses
Sequencing of IRF4 binding sites of BCL6 gene18 and PRDM1/BLIMP1 gene and detection of EZH2 Tyr641 mutation were performed (supplemental Tables 4-5). Genotypes of the SNP rs872071 in the IRF4 locus on genomic and cDNA level were determined by high-resolution melting analysis (supplemental Methods). IGHV mutational status was determined by multiplex polymerase chain reaction using the BIOMED2 protocol followed by direct sequencing or after subcloning and comparison with published germline sequences (www.imgt.org).
Bioinformatics and statistical analyses
Gene expression and copy number profiling data from 143 MMML cases analyzed by FISH for IRF4 aberrations (for sample selection, see supplemental Methods) were investigated.14
Differential gene expression was assessed using the LIMMA software Version 3.2.1 in the context of a linear model that included the ABC/GCB status as a confounding factor.19 A linear classifier of IG/IRF4-positive cases was trained on 143 samples using the shrunken centroid method and subsequently applied to 295 independent MMML cases.20
Association between age of onset and incidence of IG/IRF4 translocations was analyzed by logistic regression. Survival curves were estimated by the Kaplan-Meier method. Survival differences were analyzed with the log-rank test. In addition, Cox regression analyses were performed adjusting for age > 60 years. P values ≤ .05 were considered to indicate statistical significance (supplemental Methods).
Results
Identification and molecular characterization of IGH/IRF4 translocations
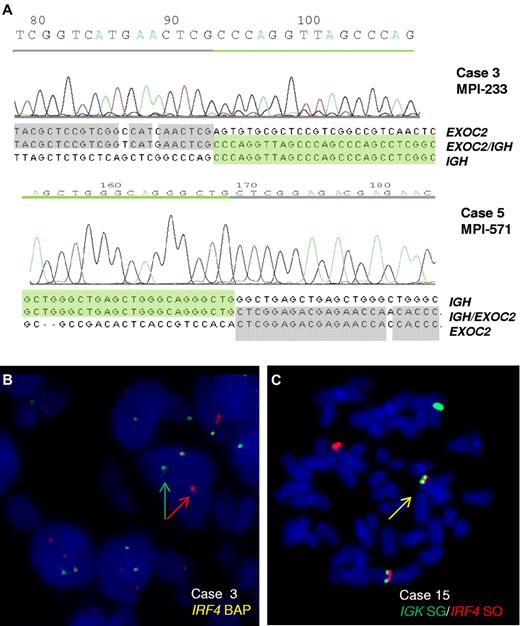
B-cell lymphomas entering the MMML study were screened by FISH for IGH translocations with a hitherto unknown partner. From the cases with FISH pattern indicating an IGH break but lacking known common partners, 28 cases were subjected to long-distance inverse PCR for cloning the IGH partner. In 2 lymphomas, Sμ-long-distance inverse PCR detected a switch μ-associated translocation t(6;14)(p25;q32) (Figure 1A). Both translocations disrupt the coding region of EXOC2, which encodes a component (Sec5) of the exocyst complex, a multiple protein complex essential for targeting exocytic vesicles to specific docking sites on the plasma membrane.21 Sec5 has been linked to cancer as it is required to mediate RalB-dependent survival signals in transformed cells.22 Although we cannot at present exclude a role of EXOC2 in lymphomagenesis, its disruption through the translocation along with the typical mechanism of IG translocations (ie, activation of intact oncogenes) made us search for further candidates. Remarkably, immediately telomeric of EXOC2 maps the IRF4 gene, which through the translocation is juxtaposed to the IGH locus on the der(14)t(6;14) in the same transcriptional direction. Because deregulation of IRF4 expression by a t(6;14)(p25;q32) has already been described as a recurrent event in multiple myeloma23 and because of the well-known function of IRF4 as transcription factor in B cells associated with oncogenic addiction,24 we considered IRF4 to be the probable target of the translocation.
Cloning of the t(6;14)(p25;q32) translocation. (A) Chromatograms and sequences of the different chromosomal junctions of the IGHSμ/EXOC2 translocations in cases 3 and 5. The sequence of IGHSμ is shown according to GenBank accession no. NG_001019.3 and of EXOC2 according to GenBank accession no. NT_007592.15. The der(6) genomic breakpoints were located downstream of exon 22 (case 3) and exon 23 (case 5) of EXOC2 gene at 482 370 and 464 205 bp from 6pter (UCSC, NCBI Build 36.1/hg18). (B) FISH analysis shows IRF4 breaks in case 3. (C) FISH image on metaphase shows an IGK/IRF4 fusion in case 15.
Cloning of the t(6;14)(p25;q32) translocation. (A) Chromatograms and sequences of the different chromosomal junctions of the IGHSμ/EXOC2 translocations in cases 3 and 5. The sequence of IGHSμ is shown according to GenBank accession no. NG_001019.3 and of EXOC2 according to GenBank accession no. NT_007592.15. The der(6) genomic breakpoints were located downstream of exon 22 (case 3) and exon 23 (case 5) of EXOC2 gene at 482 370 and 464 205 bp from 6pter (UCSC, NCBI Build 36.1/hg18). (B) FISH analysis shows IRF4 breaks in case 3. (C) FISH image on metaphase shows an IGK/IRF4 fusion in case 15.
FISH screening for IRF4 translocations
FISH screening of 183 lymphomas of the MMML cohort using probes to IRF4 on chromosome 6 identified 9 cases with signal patterns indicating IRF4 breakpoints, including both index cases (Figure 1B). Six cases displayed an IGH/IRF4 (including both index cases, supplemental Figure 2) and 1 an IGL/IRF4 fusion (Table 1; supplemental Figure 3B). Two cases showed an IRF4 break-apart pattern but lacked the typical IG/IRF4 fusion constellation.
Diagnosis, FISH, and immunohistochemical data of IG/IRF4-positive cases
| Case no. . | Study . | Diagnosis . | FISH . | Immunohistochemistry . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, y/sex . | IRF4 translocation . | BCL6 break . | MYC break . | t(14;18)/BCL2 break . | CD5 . | CD10 . | MUM1 . | BCL6 . | BCL2 . | BLIMP . | ki67 . | Hans* classifier16 . | GEP . | |||
| 1-MPI093 | MMML | DLBCL, NOS | 12/female | IGL/IRF4 | − | − | − | - | NA | ++ | ++ | + | NA | + | NA | GCB |
| 2-MPI124 | MMML | DLBCL, cb | 62/female | IGH/IRF4 | Split | − | − | ++ | - | ++ | ++ | ++ | - | ++ | Non-GC | Unclassified |
| 3-MPI233 | MMML | DLBCL, cb | 8/female | IGH/IRF4† | − | − | − | ++ | - | ++ | ++ | ++ | NA | ++ | Non-GC | GCB |
| 4-MPI276 | MMML | DLBCL, cb | 79/female | IGH/IRF4 | Split | − | − | - | - | ++ | + | - | NA | ++ | Non-GC | GCB |
| 5-MPI571 | MMML | DLBCL, cb | 7/female | IGH/IRF4† | − | − | − | - | ++ | ++ | + | ++ | NA | + | GC | GCB |
| 6-MPI581 | MMML | DLBCL, NOS | 6/female | IGH/IRF4 | − | − | − | - | ++ | ++ | ++ | ++ | - | + | GC | GCB |
| 7-MPI584 | MMML | DLBCL, cb | 15/male | IGH/IRF4 | − | − | − | - | ++ | ++ | ++ | - | NA | ++ | GC | GCB |
| 8 | BFM | FL3B | 7/male | IGH/IRF4 | − | − | − | - | + | + | ++ | ++ | NA | + | GC | NA |
| 9 | BFM | DLBCL, cb | 15/male | IGH/IRF4 | Split | − | − | - | + | ++ | ++ | - | NA | + | GC | NA |
| 10 | BFM | DLBCL, cb | 4/female | IGH/IRF4 | − | − | − | - | ++ | ++ | + | + | - | ++ | GC | NA |
| 11 | BFM | DLBCL, cb | 10/male | IGL/IRF4 | − | − | − | - | ++ | + | ++ | - | - | ++ | GC | NA |
| 12 | BFM | FL3B/DLBCL | 5/male | IGH/IRF4 | − | − | − | - | ++ | ++ | ++ | + | NA | + | GC | NA |
| 13 | BFM | DLBCL, cb | 9/male | IGH/IRF4 | Split | − | − | - | ++ | ++ | ++ | - | - | ++ | GC | NA |
| 14 | BFM | FL3A/DLBCL | 12/male | IGH/IRF4 | − | Split | − | + | + | ++ | + | + | - | ++ | Non-GC | NA |
| 15 | Routine | B-cell lymphoma, NOS‡ | 72/male | IGK/IRF4 | − | − | − | + | - | ++ | NA | NA | NA | NA | Non-GC | NA |
| 16 | DSHNHL | FL3B/DLBCL | 22/male | IGH/IRF4 | − | − | − | - | + | ++ | ++ | - | - | ++ | GC | NA |
| 17 | DSHNHL | DLBCL, cb | 20/male | IGH/IRF4 | Split | − | − | ++ | - | ++ | + | ++ | - | + | Non-GC | NA |
| 18 | DSHNHL | FL3B | 42/female | IGH/IRF4 | Split | − | − | - | - | ++ | NA | ++ | NA | ++ | Non-GC | NA |
| 19 | DSHNHL | DLBCL, cb | 28/male | IGH/IRF4 | Split | − | − | - | NA | + | - | - | - | ++ | NA | NA |
| 20 | BFM | FL3B/DLBCL | 8/female | IGH/IRF4 | − | − | − | + | + | ++ | + | ++ | - | ++ | GC | NA |
| Predicted 1 MPI059 | MMML | DLBCL, NOS | 31/male | Atypical IRF4 break (no IG) | − | − | − | - | ++ | ++ | ++ | - | NA | + | GC | GCB |
| Predicted 2 MPI514 | MMML | DLBCL, cb | 79/female | Atypical IGH/IRF4§ | − | − | − | ++ | - | ++ | + | ++ | NA | ++ | Non-GC | GCB |
| Predicted 3 MPI823 | MMML | DLBCL, poly | 49/male | IGH/IRF4 | Split | − | − | - | - | ++ | ++ | ++ | + | ++ | Non-GC | Unclassified |
| Extra+ | BFM | FL3A | 14/male | IRF4 break, IGH break | NA | NA | NA | - | + | NA | + | NA | NA | ++ | GC | NA |
| Case no. . | Study . | Diagnosis . | FISH . | Immunohistochemistry . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, y/sex . | IRF4 translocation . | BCL6 break . | MYC break . | t(14;18)/BCL2 break . | CD5 . | CD10 . | MUM1 . | BCL6 . | BCL2 . | BLIMP . | ki67 . | Hans* classifier16 . | GEP . | |||
| 1-MPI093 | MMML | DLBCL, NOS | 12/female | IGL/IRF4 | − | − | − | - | NA | ++ | ++ | + | NA | + | NA | GCB |
| 2-MPI124 | MMML | DLBCL, cb | 62/female | IGH/IRF4 | Split | − | − | ++ | - | ++ | ++ | ++ | - | ++ | Non-GC | Unclassified |
| 3-MPI233 | MMML | DLBCL, cb | 8/female | IGH/IRF4† | − | − | − | ++ | - | ++ | ++ | ++ | NA | ++ | Non-GC | GCB |
| 4-MPI276 | MMML | DLBCL, cb | 79/female | IGH/IRF4 | Split | − | − | - | - | ++ | + | - | NA | ++ | Non-GC | GCB |
| 5-MPI571 | MMML | DLBCL, cb | 7/female | IGH/IRF4† | − | − | − | - | ++ | ++ | + | ++ | NA | + | GC | GCB |
| 6-MPI581 | MMML | DLBCL, NOS | 6/female | IGH/IRF4 | − | − | − | - | ++ | ++ | ++ | ++ | - | + | GC | GCB |
| 7-MPI584 | MMML | DLBCL, cb | 15/male | IGH/IRF4 | − | − | − | - | ++ | ++ | ++ | - | NA | ++ | GC | GCB |
| 8 | BFM | FL3B | 7/male | IGH/IRF4 | − | − | − | - | + | + | ++ | ++ | NA | + | GC | NA |
| 9 | BFM | DLBCL, cb | 15/male | IGH/IRF4 | Split | − | − | - | + | ++ | ++ | - | NA | + | GC | NA |
| 10 | BFM | DLBCL, cb | 4/female | IGH/IRF4 | − | − | − | - | ++ | ++ | + | + | - | ++ | GC | NA |
| 11 | BFM | DLBCL, cb | 10/male | IGL/IRF4 | − | − | − | - | ++ | + | ++ | - | - | ++ | GC | NA |
| 12 | BFM | FL3B/DLBCL | 5/male | IGH/IRF4 | − | − | − | - | ++ | ++ | ++ | + | NA | + | GC | NA |
| 13 | BFM | DLBCL, cb | 9/male | IGH/IRF4 | Split | − | − | - | ++ | ++ | ++ | - | - | ++ | GC | NA |
| 14 | BFM | FL3A/DLBCL | 12/male | IGH/IRF4 | − | Split | − | + | + | ++ | + | + | - | ++ | Non-GC | NA |
| 15 | Routine | B-cell lymphoma, NOS‡ | 72/male | IGK/IRF4 | − | − | − | + | - | ++ | NA | NA | NA | NA | Non-GC | NA |
| 16 | DSHNHL | FL3B/DLBCL | 22/male | IGH/IRF4 | − | − | − | - | + | ++ | ++ | - | - | ++ | GC | NA |
| 17 | DSHNHL | DLBCL, cb | 20/male | IGH/IRF4 | Split | − | − | ++ | - | ++ | + | ++ | - | + | Non-GC | NA |
| 18 | DSHNHL | FL3B | 42/female | IGH/IRF4 | Split | − | − | - | - | ++ | NA | ++ | NA | ++ | Non-GC | NA |
| 19 | DSHNHL | DLBCL, cb | 28/male | IGH/IRF4 | Split | − | − | - | NA | + | - | - | - | ++ | NA | NA |
| 20 | BFM | FL3B/DLBCL | 8/female | IGH/IRF4 | − | − | − | + | + | ++ | + | ++ | - | ++ | GC | NA |
| Predicted 1 MPI059 | MMML | DLBCL, NOS | 31/male | Atypical IRF4 break (no IG) | − | − | − | - | ++ | ++ | ++ | - | NA | + | GC | GCB |
| Predicted 2 MPI514 | MMML | DLBCL, cb | 79/female | Atypical IGH/IRF4§ | − | − | − | ++ | - | ++ | + | ++ | NA | ++ | Non-GC | GCB |
| Predicted 3 MPI823 | MMML | DLBCL, poly | 49/male | IGH/IRF4 | Split | − | − | - | - | ++ | ++ | ++ | + | ++ | Non-GC | Unclassified |
| Extra+ | BFM | FL3A | 14/male | IRF4 break, IGH break | NA | NA | NA | - | + | NA | + | NA | NA | ++ | GC | NA |
Extra+ indicates additional IRF4-break-positive case; DSHNHL, German high-grade Lymphoma Study Group; BFM, Berlin-Frankfurt-Münster NHL trials; FL 3A/B, follicular lymphoma grade 3A or 3B; NOS, not otherwise specified; cb, centroblastic; poly, polymorphic; −: negative; ++, high expression (> 50%); +, moderate expression (25%-50%); -, no expression (< 25%); NA, not available; and GEP, gene expression profiling.
Hans classifier was applied to lymphomas other that DLBCLs, though not developed originally for those.
Translocation cloned.
Transformation, transition in a secondary malignant B-cell lymphoma.
Lack of der(6)t(6;14)(p25;q32).
To investigate the frequency of IG/IRF4 fusion in a population-representative cohort, we next screened 235 B-cell lymphomas from the trials of the German pediatric (Berlin-Frankfurt-Münster-NHL, n = 74) and adult (German High-Grade NHL Study Group, NHL-B trials, n = 161) aggressive lymphoma study groups by FISH for IRF4 breaks. This led to the identification of 13 additional cases with IRF4 breakpoints. Finally, 1 of 9 additional lymphomas with unknown IG translocation analyzed in a diagnostic setting was shown to have an IGK/IRF4 translocation (Figure 1C).
In summary, FISH screening identified a total of 23 lymphomas with IRF4 breakpoints (Table 1): 17 cases with IGH/IRF4, 2 with IGL/IRF4, and 1 with IGK/IRF4 fusion (Figure 1C; supplemental Figure 3A-B). In the 3 additional cases, the partner remained unproven or ambiguous. Thus, these cases were classified as “atypical IRF4-positive” cases but excluded from further analyses as the nature of the translocation could not be resolved. Among the 20 proven IG/IRF4-positive cases, 7 carried a BCL6 break, 1 carried a MYC break, and none a BCL2 break and/or t(14;18).
Compared with 118 lymphomas of the MMML cohort with copy number data available lacking IRF4 breakpoints, the 7 IG/IRF4-positive lymphomas had fewer chromosomal imbalances (9.2 vs 4.9 alterations, P = .049), suggesting that the IRF4 translocation is an early event in lymphomagenesis (supplemental Figure 4A-D).
Histopathology and immunohistochemistry of IG/IRF4-positive lymphomas
Thirteen of the 20 IG/IRF4-positive lymphomas were classified as DLBCL, 2 as FL 3B, 4 as composite FL grade 3/DLBCL, and 1 as B-cell non-Hodgkin lymphoma, not further classified. All 11 subclassifiable DLBCLs were of the centroblastic variant.
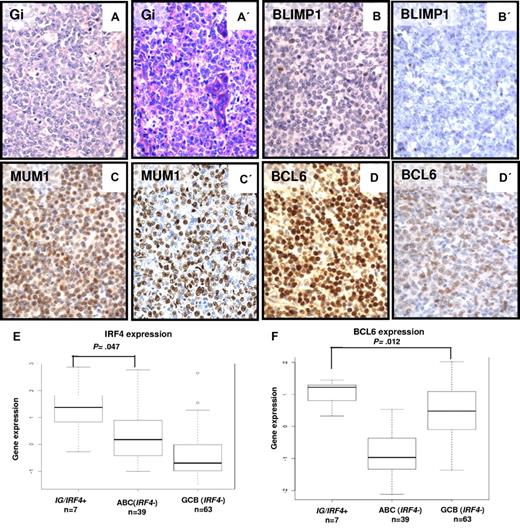
The immunophenotypes of the IG/IRF4-positive lymphomas were mostly characterized by a MUM1+/BCL6+/BLIMP1− pattern (Table 1; Figure 2). MUM1 protein encoded by the IRF4 gene was expressed in all 20 cases with IG/IRF4 fusion, with mostly strong staining intensity (17 of 20). BCL6 was expressed in 17 of 18 (94%) cases, whereas all 10 cases studied were negative for BLIMP1. CD10 was expressed in 12 of 18 (66%), CD5 in 6 of 20 (30%; including 4 of 6 cases lacking CD10 expression), and BCL2 in 12 of 19 (63%) evaluable cases. The expression of Ki-67 was variable but overall high. According to the Hans algorithm,16 11 of 18 (61%) cases were classified as GCB and 7 (39%) as non-GCB.
Morphologic and immunohistochemical features of IG/IRF4-positive cases. (A-D) Morphology and immunophenotype of 2 IG/IRF4-positive cases (A-D, case 14; A′-D′, case 20; original magnification ×400): DLBCL component with GC immunophenotype, strong nuclear staining for MUM1, and negativity for BLIMP1. The follicular component of both cases showed an identical immunophenotype (data not shown). Gi indicates Giemsa staining. (E) Differential expression levels of IRF4 (probe set U133A 216986_s_at) between IG/IRF4-positive cases and the other DLBCL subtypes, specifically with the ABC subtype (P = .047). (F) Comparison of the expression levels of BCL6 (2 probes encoding BCL6, 203140_at and 215990_s_at, summarized to BCL6 index) between IG/IRF4-positive cases and the other DLBCL subtypes. Specifically, IG/IRF4-positive cases presented higher expression of BCL6 than the GCB-DLBCLs (P = .012).
Morphologic and immunohistochemical features of IG/IRF4-positive cases. (A-D) Morphology and immunophenotype of 2 IG/IRF4-positive cases (A-D, case 14; A′-D′, case 20; original magnification ×400): DLBCL component with GC immunophenotype, strong nuclear staining for MUM1, and negativity for BLIMP1. The follicular component of both cases showed an identical immunophenotype (data not shown). Gi indicates Giemsa staining. (E) Differential expression levels of IRF4 (probe set U133A 216986_s_at) between IG/IRF4-positive cases and the other DLBCL subtypes, specifically with the ABC subtype (P = .047). (F) Comparison of the expression levels of BCL6 (2 probes encoding BCL6, 203140_at and 215990_s_at, summarized to BCL6 index) between IG/IRF4-positive cases and the other DLBCL subtypes. Specifically, IG/IRF4-positive cases presented higher expression of BCL6 than the GCB-DLBCLs (P = .012).
IG/IRF4-positive lymphomas show characteristics of GCB cells
Gene expression data from 7 lymphomas with typical IG/IRF4 fusion were available through the MMML consortium. One case could not be classified as either GCB or ABC, and 6 had the GCB signature. All 6 interpretable cases had highly mutated IGHV genes. Identity to published germline sequences ranged from 86.1% to 93.9% with ongoing mutation detected in 4 of 4 cases (supplemental Table 6).
Gene expression profiling of IG/IRF4-positive lymphomas
IRF4 (P = .011) and BCL6 (P = .013) were both also expressed at significantly higher transcript levels in IG/IRF4-positive than in IRF4-negative lymphomas (supplemental Figure 5A-B). Moreover, allelic expression analysis of 3 IG/IRF4-positive lymphomas heterozygous for an IRF4 SNP rs872071 suggested that transcription is heavily skewed toward the translocated allele (supplemental Figure 6). Although IRF4 and BCL6 are hallmark genes of the ABC and GCB signatures, respectively, in IG/IRF4-positive cases the transcript levels of IRF4 were significantly higher than in ABC-DLBCL (P = .047; Figure 2E) and the levels of BCL6 significantly higher than in GCB-DLBCL (P = .012; Figure 2F). Therefore, we investigated whether these lymphomas show gene expression features different from both GCB- and ABC-DLBCL. Compared with negative cases (n = 136), lymphomas with IG/IRF4 fusion (n = 7) showed differential expression of 132 genes (161 probe sets), including 123 up-regulated and 9 down-regulated in IG/IRF4-positive cases (supplemental Figure 7; supplemental Table 7).
As these findings suggest IG/IRF4-positive lymphomas to be different from GCB and ABC lymphomas, we used a supervised learning algorithm to derive a gene expression classifier allowing prediction of IG/IRF4-positive lymphomas. A linear classifier based on 27 genes (32 probe sets), which predicted the 7 IG/IRF4-positive cases with 96.5% accuracy in cross-validation, was identified (Figure 3; supplemental Table 8). Application of this classifier to an independent cohort of 293 MMML cases not investigated by FISH as well as the 2 atypical cases identified by FISH (MPI-059 and MPI-514) identified 3 lymphomas with a gene expression profiling typical for IG/IRF4 fusion. These included 2 “atypical IRF4-positive cases” described in the preceding paragraph and 1 additional case, which by FISH subsequently showed a signal pattern indicative of IG/IRF4 fusion. These 3 predicted cases shared the typical morphologic, immunohistochemical, and somatic hypermutation features with the other IG/IRF4-positive cases, except 1 of them that expressed weakly PRDM1/BLIMP1 (Table 1).
Gene expression profiling-based classifier of IG/IRF4-positive cases. This figure represents a heatmap of an IG/IRF4-positive classifier consisting of 32 probe sets (27 genes). This classifier distinguishes IG/IRF4-positive cases from the rest of DLBCL subtypes.
Gene expression profiling-based classifier of IG/IRF4-positive cases. This figure represents a heatmap of an IG/IRF4-positive classifier consisting of 32 probe sets (27 genes). This classifier distinguishes IG/IRF4-positive cases from the rest of DLBCL subtypes.
Alterations of the BCL6-IRF4-PRDM1/BLIMP1 regulatory pathway
Under physiologic conditions, IRF4 suppresses expression of BCL6 and activates PRDM1/BLIMP1. Because the IG/IRF4-positive cases in contrast showed strong expression of BCL6 but lacked PRDM1/BLIMP1 expression, we searched for potential genomic defects in the IRF4-mediated pathway. Sequence and copy number analyses of PRDM1/BLIMP1 in 10 IG/IRF4-positive lymphomas failed to provide evidence for mutational inactivation of this gene (supplemental Table 9). In contrast, 8 of 23 lymphomas (35%) with proven IRF4 breaks showed chromosomal breakpoints affecting the BCL6 locus, and 1 case carried a gain of BCL6. In addition, all 7 IG/IRF4-positive lymphomas investigated carried somatic BCL6 mutations, from which one even directly affected a predicted IRF4-binding site (supplemental Table 10).18 No differential expression of IRF4 target genes and BCL6 target genes described by Shaffer et al24 and Polo et al25 between cases with and without the translocation was observed (data not shown).
Clinical features of IG/IRF4-positive lymphomas
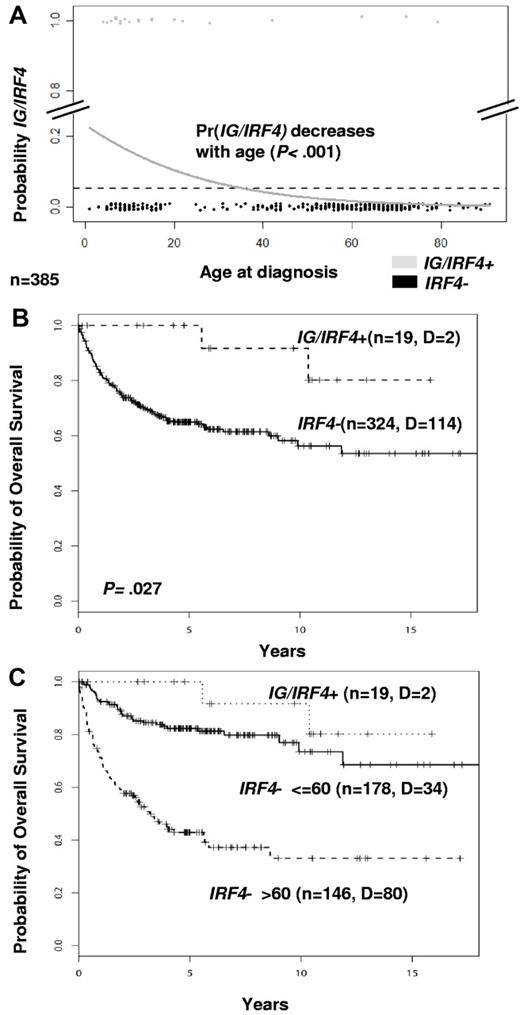
There were 9 female and 11 male patients with IG/IRF4-positive lymphomas with a median age of 12 years (range, 4-79 years). Clinical and follow-up data are summarized in supplemental Table 11. Among the 385 cases studied by FISH with available age data, IG/IRF4-positive lymphomas were significantly more frequent in children (≤ 18 years) than in adults (> 18 years; 15% vs 2%; P < .001). Logistic regression analysis showed a significant decrease in the probability for IG/IRF4 fusion with age (P < .001; Figure 4A). Clinical presentation was significantly skewed toward the involvement of the head and neck region, including Waldeyer ring (80%, P = .011) and limited disease stages (84%, P < .001). In the whole population analyzed by FISH in which clinical data were available (n = 343), IG/IRF4-positive cases were associated with a significantly better prognosis (5-year overall survival, 100% vs 64.9%; P = .027; Figure 4B). Adjusting for age in a COX regression (age, relative risk: 4.26; P < .001; IG/IRF4, relative risk: 0.39; P = .19) showed that this effect was predominantly associated with the low age of the positive cases (Figure 4C; supplemental Figure 8).
Age and survival analyses. (A) Logistic regression analysis of age distribution. The diagram shows the decreasing probability of having IG/IRF4 translocations in association with increasing age (n = 385; P < .001). Gray dots represent IG/IRF4-positive cases; and black dots, IRF4-break-negative cases. (B) Kaplan-Meier curves show a better survival of IG/IRF4-positive cases (P = .027). (C) Kaplan-Meier curves stratifying IRF4-break-negative cases by age (≤ 60 years and > 60 years).
Age and survival analyses. (A) Logistic regression analysis of age distribution. The diagram shows the decreasing probability of having IG/IRF4 translocations in association with increasing age (n = 385; P < .001). Gray dots represent IG/IRF4-positive cases; and black dots, IRF4-break-negative cases. (B) Kaplan-Meier curves show a better survival of IG/IRF4-positive cases (P = .027). (C) Kaplan-Meier curves stratifying IRF4-break-negative cases by age (≤ 60 years and > 60 years).
Discussion
Recurrent translocations juxtaposing oncogenes and IG genes are hallmarks of subtypes of B-cell lymphomas and thus add to their definition and aid their diagnosis.26 Here we show that recurrent IG/IRF4 translocations activate the transcription factor IRF4 in a subtype of mature B-cell lymphomas. IG/IRF4 fusions are associated with a hitherto unrecognized subgroup of GC B-cell lymphomas composing FL grade 3 or (centroblastic) DLBCL characterized by coexpression of MUM1 and BCL6 in the absence of PRDM1/BLIMP1, a specific gene expression profile, and a disease onset predominantly in childhood or young adulthood.
Remarkably, despite that all 6 classifiable lymphomas with IG/IRF4 fusion were assigned to the GCB subtype by the gold standard gene expression profiling, 2 of them lacked CD10 expression. In the presence of strong MUM1 positivity, these 2 CD10-negative cases are assigned to the non-GCB subtype according to the immunohistochemistry-based Hans algorithm yielding conflicting results between both classifiers.16 Interestingly, the expressions of CD10 and CD5 were mutually exclusive, and 5 of 8 CD10-negative cases were positive for CD5, a marker discussed to identify a distinct subgroup of DLBCL.27 Both features, the lacking correlation of gene expression and immunohistochemical classification and the high rate of CD5 positivity in CD10-negative lymphomas, might indicate that IG/IRF4 positive lymphomas indeed constitute a defined subgroup of mature B-cell lymphomas.
Translocations affecting the IRF4 locus have not yet been described as recurrent aberrations in GC B-cell lymphomas, although at least 2 DLBCLs with IRF4 translocations have been published (supplemental Table 12).28,29 The reason might be that t(6;14)(p25;q32) is cytogenetically cryptic as shown by Tamura et al.28 The only case from our series with karyotype available was case 15, which showed der(6)t(2;6)(p12;p25), but light chain variants are obviously rare. In contrast, recurrent aberrations affecting the IRF4 locus have been described in T-lineage anaplastic large cell lymphomas30-33 and multiple myeloma.23,34,35 In the latter, IRF4 is similarly juxtaposed by an illegitimate IG switch recombination to the IG loci.23,34 Moreover, in multiple myeloma, expression of IRF4 is not only driving those cases with IG/IRF4-fusion but is also essential for survival in cases lacking this translocation.24 Whether a subset of GC-derived lymphomas is similarly “addicted” to the expression of the IRF4 oncogene remains elusive. In contrast to plasma cell neoplasms, the IG/IRF4-positive B-cell lymphomas strongly express the GC master regulator BCL6 and lack expression of PRDM1/BLIMP1, which is necessary to drive plasma cell differentiation (supplemental Figure 9).36 Physiologically, IRF4 suppresses BCL6 expression. The simultaneous high expression of both proteins in IG/IRF4-positive cases suggests that this regulatory loop is interrupted by the IG/IRF4 juxtaposition and the mutations/translocations at the BCL6 locus. This would explain why attempts to identify a significantly differential expression of IRF4 target genes and BCL6 target genes described by Shaffer et al24 and Polo et al25 between cases with and without the translocation failed.
The IG/IRF4-positive B-cell lymphomas were FL grade 3, DLBCL, or a composite of both. Interestingly, the IG/IRF4-positive lymphomas composed 4 FLs grade 3 (with or without additional DLBCL component) in children. Giving the overall rarity of pediatric FL and the overlap in IG/IRF4 fusion in both FL grade 3 and DLBCL, these findings raise the question of whether pediatric FL might represent part of the spectrum of DLBCL in childhood rather than a distinct FL subtype. Our results also suggest that DLBCL in childhood and young adulthood may be biologically different from older adult DLBCL. Although there is no sharp age border for the presence of IG/IRF4-positive lymphomas, the probability of IG/IRF4 positivity decreases as a function of age. Although age is the strongest predictor of outcome in mature aggressive B-cell lymphoma, it is possible that the prevalence of favorable versus unfavorable genetic alterations in different age groups accounts for some of this difference, as has been described for B-cell acute lymphoblastic leukemia. Future studies have to show whether, similar to the situation in acute lymphoblastic leukemia, treatment according to genetic rather than age-based risk stratification might improve outcome also in mature B-cell neoplasms.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Reina Zühlke-Jenisch, Magret Ratjen, Claudia Becher, and Olivera Batic for their excellent technical support.
This work was supported by the Deutsche Krebshilfe through the network project Molecular Mechanisms in Malignant Lymphomas (70-3173-Tr3), the Kinderkrebsinitative Buchholz/Holm-Seppensen (W.K., R. Siebert), and the Alexander von Humboldt Foundation (I.S.).
Authorship
Contribution: I.S. performed molecular analysis, analyzed data, and wrote the manuscript; I.O. provided samples, performed pathology review, and wrote the manuscript; C. Philipp, S.G., H.T., M.S., M.A., J.R., F.M., C. Pott, I.M.-G., S.P., M.F., L.H., M.H., C.S., and S.W. performed molecular analysis and analyzed data; C.W.K., M.K., H.B., D.H., M.R., M.L., and R. Spang performed statistical analysis; B.B., L.T., and A.R. provided samples, clinical data, and administrative support; M.P., H.S., and W.K. provided samples and performed pathology review; and R.K., W.K., and R. Siebert designed the project, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The company Abbott/Vysis discounts the Deutsche Krebshilfe (70-3173-Tr3) project Molecular Mechanisms in Malignant Lymphomas for FISH probes. R. Siebert received speakers fees from Abbott/Vysis. H.S. received royalties for antibody development. The remaining authors declare no competing financial interests.
For a complete list of the members of the Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe, see the online supplemental Appendix.
Correspondence: Reiner Siebert, Institute of Human Genetics, University Hospital Schleswig-Holstein, Campus Kiel/Christian-Albrechts University Kiel, Schwanenweg 24, D-24105 Kiel, Germany; e-mail: rsiebert@medgen.uni-kiel.de; and Itziar Salaverria, Institute of Human Genetics, University Hospital Schleswig-Holstein, Campus Kiel/Christian-Albrechts University Kiel, Schwanenweg 24, D-24105 Kiel, Germany; e-mail: isalaverria@medgen.uni-kiel.de.
References
Author notes
I.S., C. Philipp, I.O., C.W.K., and M.K. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal