Abstract
Primary mediastinal large B-cell lymphoma (PMBCL) is a recognized non-Hodgkin lymphoma entity with unique pathologic, clinical, and molecular characteristics distinct from those of other diffuse large B-cell lymphomas. Immunohistochemical characterization and molecular studies strongly suggest that PMBCL is of germinal center or postgerminal center origin. Pivotal gene expression profiling work defined major deregulated pathway activities that overlap with Hodgkin lymphoma and prompted a more detailed analysis of candidate genes. In particular, the nuclear factor-κB and the Janus Kinase-Signal Transducer and Activator of Transcription signaling pathways are targeted by multiple genomic hits, and constitutive activity of both pathways can be considered molecular hallmark alterations of PMBCL. Moreover, data are emerging giving unique insight into remodeling of the epigenome that affects transcriptional regulation of a multitude of genes. More recently, the tumor microenvironment of PMBCL has shifted into focus based on a number of gene perturbations altering expression of surface molecules that contribute to immune escape. These findings highlight the importance of immune privilege in the pathogenesis of PMBCL and suggest that disrupting crosstalk between the tumor cells and the microenvironment might be a rational new therapeutic target in conjunction with traditional treatment strategies.
Introduction
Primary mediastinal large B-cell lymphoma (PMBCL) represents approximately 2% to 4% of all non-Hodgkin lymphomas.1 PMBCL is now a fully recognized entity in the World Health Organization (WHO) classification of lymphoid neoplasms. Previously, in the revised European-American classification of lymphoid neoplasms2 and frequently in clinical practice, PMBCL was considered a subtype of diffuse large B-cell lymphoma (DLBCL), arising in the mediastinum, reflected in study designs that did not distinguish PMBCL from DLBCL patients in earlier clinical trials. In 2001, the WHO classification listed PMBCL as a separate entity. Currently, with characteristic clinical and immunophenotypic features, and recent molecular characterization of PMBCL, there is now conclusive evidence that PMBCL represents a distinct clinicopathologic entity.
Distinguishing PMBCL from other forms of DLBCL can be viewed as a paradigm for reclassification of cancer subtypes based on molecular features, as the molecular characterization of PMBCL highlighted an overlap with the nodular sclerosis subtype of classic Hodgkin lymphoma (cHL) sharing a number of genetic and gene expression features.3,4 Interestingly, this association had been long suspected based on similarities in clinical features and reports of sequential or composite occurrences of cHL and PMBCL.5,6 Functional analyses of both lymphomas confirmed that the malignant cells rely on the same principal signaling pathways, namely, Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) and nuclear factor-κB (NF-κB) signaling, both providing these cells with a proliferative advantage.7-9 However, more recent lines of evidence also suggest that acquired immune privilege by the malignant cells contributes to the cancer phenotype of PMBCL in a cooperative way.10,11 The specific composition of the inflammatory infiltrates discovered through histologic study of primary PMBCL biopsies further supports the significance of the interactions of the malignant cells with the surrounding non-neoplastic immune cells. Unraveling the underlying genetic mechanism(s) of these tumor-microenvironment interactions might be important for future therapies targeting the immune privilege phenotype of the malignant cells.
From a clinical viewpoint, PMBCL patients treated with multiagent chemotherapy appear to have a better survival compared with DLBCL based on retrospective analyses.12,13 However, the intensity of primary chemotherapy, the role of radiotherapy, and the effectiveness of added rituximab are still the subject of debate, and definitive conclusions are hampered by the paucity of prospective studies. Equally, meaningful biomarker studies to inform risk stratification and outcome prediction of PMBCL are largely unavailable.
With a number of important biologic insights in PMBCL reported in the last 12 months, a critical review of the molecular pathogenesis of this disease was warranted. We first focus on the pathologic and clinical features of PMBCL to define in detail the disease entity compared with related lymphoid cancers. We then characterize the genetic alterations and deregulated signaling pathways that underlie the pathogenesis and review the recent data that highlight the importance of the microenvironment. Finally, we pay special attention to the potential synergy of the underlying genetic alterations that cooperate to develop the full malignant phenotype, and briefly outline avenues for clinical translation of these novel findings.
Pathology and clinical features
PMBCL characteristically presents with a bulky anterior mediastinal mass and affects predominantly female patients in their third or fourth decade of life (Figure 1).14 PMBCL can also occur in children and adolescents showing similar clinical and pathologic characteristics with adult cases.15 In contrast to DLBCL,16 the bone marrow is rarely involved and the tumor extension is typically local, with pleural, pericardial effusions and superior vena cava obstruction as typical clinical complications in a significant number of patients. Systemic involvement beyond the thorax and Ann Arbor stage III are uncommon at initial diagnosis,12,17 and the absence of infradiaphragmatic lymph node or bone marrow involvement was recommended as a prerequisite for diagnosis of PMBCL in the WHO classification, to exclude systemic DLBCL with secondary mediastinal involvement.1 However, particularly at relapse, the disease has a very high incidence of extranodal spread with a tendency to spare lymph nodes.18 Prospective clinical trials specifically enrolling PMBCL are lacking in the literature, in part because of small patient numbers and the recent recognition of PMBCL as a distinct entity. Therefore, outcome studies and treatment recommendations are based on retrospective studies or subgroup analysis of randomized trials of DLBCL. As relapses after first-line therapies usually occur early and salvage therapies have been reported to have high failure rates, successful primary treatment has been considered critical. In particular, more dose-intense regimens have been suggested and shown to be effective, especially compared with DLBCL.12,19,20 Consolidating radiation therapy has also been studied in noncomparator studies.21 However, it remains to be determined whether the addition of rituximab immunotherapy is making radiation and high-dose therapies obsolete. Very recent reports of combining rituximab with dose-adjusted etoposide, prednisone, vincristine, cyclophosphamide, and doxorubicin have shown very promising results,20 and equally, PMBCL subgroup analysis of a randomized DLBCL trial (Mabthera International Trial group study) showed increased response rates and event-free survival in patients receiving rituximab that was similar to the benefit seen in DLBCL patients.13
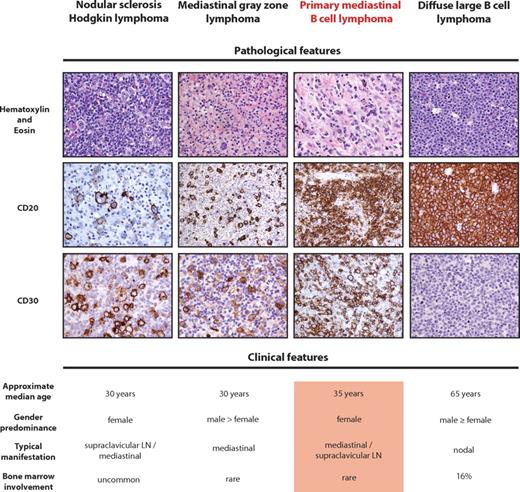
Pathologic and clinical features of PMBCL compared with related diseases. (Top panel) Representative H&E and immunohistochemical stains. Positivity of CD20 and CD30 in PMBCL is characteristic, although not mandatory, for the diagnosis of PMBCL. The H&E stain of PMBCL shows typical sheets of medium-sized to large cells containing abundant pale cytoplasm in a background of fine sclerosis. Of note, CD20 staining in nodular sclerosis subtype of cHL is typically heterogeneous, and CD30 is usually absent in DLBCL. (Bottom panel) The typical clinical features of PMBCL are given and contrasted with related entities.
Pathologic and clinical features of PMBCL compared with related diseases. (Top panel) Representative H&E and immunohistochemical stains. Positivity of CD20 and CD30 in PMBCL is characteristic, although not mandatory, for the diagnosis of PMBCL. The H&E stain of PMBCL shows typical sheets of medium-sized to large cells containing abundant pale cytoplasm in a background of fine sclerosis. Of note, CD20 staining in nodular sclerosis subtype of cHL is typically heterogeneous, and CD30 is usually absent in DLBCL. (Bottom panel) The typical clinical features of PMBCL are given and contrasted with related entities.
Of note, most of the prognostic factors that were established in DLBCL proved to be of limited value in PMBCL, and individual clinical parameters such as poor performance status or elevated LDH have been inconsistently reported to be associated with treatment failure.12,22 Importantly, the age-adjusted International Prognostic Index did not predict treatment outcome in most series.12,17 This lack of prognostic factors highlights the need for molecular biomarkers and an improved understanding of the underlying pathobiology.
Although PMBCL is a distinct clinicopathologic entity, the histomorphologic features and phenotype often vary and underscore the relatedness to other B-cell lymphomas (BCLs), namely, DLBCL and nodular sclerosis cHL.1 The histologic appearance of PMBCL is characterized by sheets of medium- to large-sized cells containing abundant pale cytoplasm resembling other large BCL subtypes. Occasionally, some neoplastic cells are multinucleated and resemble Hodgkin-Reed-Sternberg cells, and a fine background sclerosis, distinct from broad collagen bands seen in nodular sclerosis cHL, often leads to compartmentalization of the malignant cells (Figure 1). This relatedness of HL and PMBCL is also reflected by the revised WHO classification recognizing mediastinal gray zone lymphomas that are defined as unclassifiable BCLs with features intermediate between DLBCL and cHL.1,23 Although histopathologic and genetic data are limited, recent molecular studies confirm the transitional nature of mediastinal gray zone lymphoma existing in the borderland between PMBCL and cHL.24-26 Figure 1 highlights the most prominent clinical, morphologic, and molecular features recognized in PMBCL, cHL, DLBCL, and mediastinal gray zone lymphoma demonstrating the biologic continuum of these BCLs. However, the main focus of this review is PMBCL, and reference to the related entities will only be made if relevant to the molecular pathogenesis of PMBCL.
Based on immunophenotypic data and mutational analysis of various loci that are typical targets of somatic hypermutation (SHM), PMBCL has been suggested to be of germinal center or postgerminal center origin.27,28 The B-cell immunophenotype shows general positivity for surface CD19, CD20, CD22, and CD79a and frequent nuclear positivity of the transcriptional regulators BOB1, PU.1, OCT2, PAX5, BCL6, and IRF4.27,29 However, the mature B-cell phenotype is incomplete as human leukocyte antigen (HLA) class I and II molecules are expressed at various levels and are often completely lacking and, equally, surface immunoglobulin (Ig) expression is frequently absent.30-32 Of note, most PMBCL cases also stain positively for CD30, but the staining intensity is often weaker compared with HL or anaplastic large cell lymphoma33,34 (Figure 1). The malignant cells typically lack expression of CD15 and do not harbor latent Epstein-Barr virus. SHM of the immunoglobulin (IG) genes and BCL6 are considered reliable markers indicating transit of a B cell through the germinal center and, in agreement with the proposed (post-) germinal center origin of PMBCL, isotype-switched IG genes and BCL6 gene mutations unequivocally show the pattern of SHM.28,35,36 Of note, the spatial distribution and targeted motifs of hypermutation in the BCL6 5′ noncoding region were found to be distinct from DLBCL and follicular lymphoma, suggesting specificity of the SHM process and a distinct clonal evolution in PMBCL. However, the significance of this finding remains unclear.37 Interestingly, activation-induced cytidine deaminase (AID), a gene that plays a key role in SHM and class switch recombination, has been shown to be constitutively expressed in PMBCL and the PMBCL-derived cell line KARPAS1106P38 showed ongoing SHM, but not class switch recombination, involving IG genes and BCL6.39 Although, taken together, these data are consistent with a germinal center origin of PMBCL and migration into the thymus, another potential explanation is that PMBCL is derived from AID-expressing non-germinal center B lymphocytes residing in the medullary thymus.40
Genetic alterations and deregulated signaling pathways
Since recognition of PMBCL as a distinct entity, the number of studies describing the specific molecular features and genetic underpinnings of the disease is increasing. Whereas earlier studies used target gene approaches, seminal genome-wide gene expression profiling work defined major deregulated pathway activities and boosted investigation of candidate genes, some of which overlapped with cHL.3,41 Table 1 summarizes the most recurrent gene alterations involved in the pathogenesis of PMBCL. Among the first studied genes was CDKN2A (p16/INK), which was found to be mutated or hypermethylated in a small number of cases. Other gene alterations encompass MYC rearrangements, MYC promoter sequence variations, which might be because of SHM, and coding sequence mutations in TP53.42,43 In contrast to DLBCL, BCL6 translocations are only sporadically reported. Of importance, genetic heterogeneity in some of these older studies almost certainly reflects the inclusion of cases of typical DLBCL with secondary mediastinal lymph node localization that may have little in common with PMBCL.
Recurrent gene alterations involved in the pathogenesis of primary mediastinal B-cell lymphoma
| Gene . | Pathway/function . | Frequency of alteration in PMBCL, % (primary reference) . |
|---|---|---|
| Copy number gain | ||
| REL | NF-κB pathway | 75 (45) |
| PDL1/PDL2 | Induction of T-cell exhaustion/apoptosis | 63 (10) |
| JAK2 | IL/JAK-STAT pathway/histone modification | 63 (10) |
| JMJD2C | Histone modification | 63 (61) |
| Chromosomal translocation/rearrangement | ||
| CIITA | Transcriptional regulation of HLA class II/antigen presentation | 38 (11) |
| Coding sequence mutation | ||
| SOCS1 | IL/JAK-STAT pathway | 45 (64) |
| STAT6 | IL/JAK-STAT pathway | 36 (68) |
| TNFAIP3 | NF-κB pathway | 36 (4) |
| MYC | Transcriptional regulation/chromatin remodeling | 25 (42) |
| TP53 | p53 pathway | 13 (42) |
| Promoter hypermethylation | ||
| p16/INK | Cell-cycle progression, p53 pathway | 9 (42) |
| Gene . | Pathway/function . | Frequency of alteration in PMBCL, % (primary reference) . |
|---|---|---|
| Copy number gain | ||
| REL | NF-κB pathway | 75 (45) |
| PDL1/PDL2 | Induction of T-cell exhaustion/apoptosis | 63 (10) |
| JAK2 | IL/JAK-STAT pathway/histone modification | 63 (10) |
| JMJD2C | Histone modification | 63 (61) |
| Chromosomal translocation/rearrangement | ||
| CIITA | Transcriptional regulation of HLA class II/antigen presentation | 38 (11) |
| Coding sequence mutation | ||
| SOCS1 | IL/JAK-STAT pathway | 45 (64) |
| STAT6 | IL/JAK-STAT pathway | 36 (68) |
| TNFAIP3 | NF-κB pathway | 36 (4) |
| MYC | Transcriptional regulation/chromatin remodeling | 25 (42) |
| TP53 | p53 pathway | 13 (42) |
| Promoter hypermethylation | ||
| p16/INK | Cell-cycle progression, p53 pathway | 9 (42) |
NF-κB signaling pathway
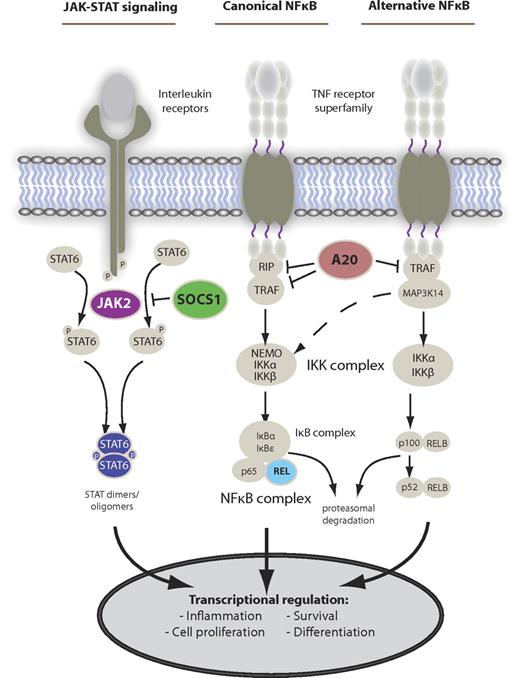
The NF-κB signaling pathway has been described as one of the most important dysregulated pathways in the malignant pathogenesis of BCLs, and of cHL and PMBCL in particular (for detailed review of HL see Kuppers7 ). In normal B-cell development, the activity of the NF-κB transcription factor complex is influenced and regulated by several extrinsic and intrinsic factors, of which stimulation through tumor necrosis factor (TNF) receptors plays a major role for pathway activation (Figure 2). In PMBCL, early gene expression profiling studies revealed that several genes of the NF-κB signaling pathway, such as TNF family members, TNF receptor-associated factors, and NF-κB complex members, were overexpressed compared with DLBCL and that this gene expression pattern more closely resembled cHL.3,41 Subsequent studies confirmed overexpression of an NF-κB activation signature and nuclear localization of NF-κB transcription factor complexes in PMBCL-derived cell line KAPRAS1106P compared with the activated B cell and germinal center B type DLBCL.9 Moreover, small molecule inhibition of the IκB kinase complex, which functions as an activator of NF-κB signaling, was toxic to KARPAS1106P cells demonstrating dependence on this pathway.44 The underlying genetic causes for this overexpression and pathway activation are not yet fully understood. More recently, specific gene mutations and structural alterations of pathway genes have been described, leading to constitutive activity. Chromosomal gains and amplifications of the REL gene locus on chromosome 2p16.1 have been consistently found in 50% or more cases using array comparative genomic hybridization and fluorescence in situ hybridization.45-47 Although amplification of the REL gene locus did not correlate with elevated mRNA levels, genomic amplification was associated with nuclear localization of REL protein, indicating increased pathway activity in these cases. Other chromosomal imbalances involving NF-κB regulators encompass amplification of BCL10 (1p22) and MALT1 (18q21).46
Main deregulated signaling pathways in PMBCL. The main activation cascades of JAK-STAT and NF-κB signaling. Alternative pathway activation exists. Known gene alterations leading to constitutive pathway activity in PMBCL are shown in color.
Main deregulated signaling pathways in PMBCL. The main activation cascades of JAK-STAT and NF-κB signaling. Alternative pathway activation exists. Known gene alterations leading to constitutive pathway activity in PMBCL are shown in color.
Very recently, TNFAIP3 (encoding A20) has been reported as a novel tumor suppressor gene in HL and PMBCL. The A20 protein acts as a ubiquitin-modifying enzyme that inhibits NF-κB signaling downstream of TNF receptor engagement by interacting with RIP1, TRAF1, and TRAF2.48 Specifically, A20 adds polyubiquitins to RIP1 (NF-κB essential modulator), targeting it for proteasomal degradation and thereby negatively regulates the IκB kinase complex and NF-κB activity.49 Destructive and biallelic gene mutations were found in 36% of PMBCL cases leading to constitutive NF-κB activity.4 One study included cell line KARPAS1106P that was found to be null for A20 protein expression; and, importantly, earlier array comparative genomic hybridization study of primary PMBCL cases revealed chromosomal deletion of the genomic TNFAIP3 locus on chromosome 6q23.3 in ∼ 30% of patients.46,50 These data provide further evidence for complete inactivation of the gene in a significant proportion of cases. Somatic TNFAIP3 mutations and/or chromosomal 6q deletion have also been described in several other B-cell malignancies, such as mantle cell lymphoma,51 activated B cell type DLBCL,51 marginal zone BCL,52 mucosa-associated lymphoid tissue lymphomas53,54 and AIDS-related BCLs,55 highlighting the frequency and overall importance of TNFAIP3 in BCL pathogenesis.
Moreover, unlike cHL, NFKBIA mutations were absent in PMBCL, suggesting a different mutational landscape of gene mutations in the 2 diseases leading to NF-κB activation.56 Genome-wide discovery studies using next-generation sequencing will clarify the mutational landscape in the very near future.
JAK-STAT signaling
Similar to NF-κB signaling, the JAK-STAT pathway is a major regulator of gene transcription involved in cellular proliferation, apoptosis, and angiogenesis and has been implicated in the pathogenesis of multiple malignancies, including lymphoid cancers.57 In normal B cells, the pathway is typically activated by interleukins or interferons binding to a variety of receptors, which lead to auto-phosphorylation of JAK molecules, subsequent STAT phosphorylation, and translocation of STAT dimers into the nucleus where they act as DNA-binding transcription factors (Figure 2). In PMBCL, gene expression profiling revealed high levels of IL13 receptor expression and downstream effectors, such as JAK2 and STAT1, indicating up-regulation of pathway genes.3,41,58 Moreover, constitutive STAT6 activation has been identified as a characteristic feature of PMBCL compared with DLBCL and, in contrast to cHL, this is probably not the result of autocrine IL-4 or IL-13 production.8,59
Genomic amplification of a subtelomeric region of chromosome 9 (9p24.1) involving several genes, including JAK2, has been consistently reported in more than half of cases and can be considered as one of the hallmark genetic alterations in PMBCL10,46,60,61 (Table 1). In these high-resolution copy number studies, a minimally amplified region of approximately 3.5 Mb could be identified, leading to coamplification of the JAK2, CD274, PDCD1LG2, and JMJD2C genes that probably cooperate in PMBCL pathogenesis (Table 2). Wessendorf et al further delineated a high-level amplicon of ∼ 1.6 Mb harboring the aforementioned genes.46 Focusing on JAK-STAT signaling, Green et al demonstrated a significant correlation between JAK2 genomic gains and mRNA expression.10
Proposed synergy of genetic alterations
| Genomic event . | Gene components . | Proposed synergy . |
|---|---|---|
| Unbalanced CIITA rearrangement | SOCS1, CIITA, CIITA fusion partners (eg, PDL1 and PDL2) | Impaired CIITA function, overexpression of fusion partners, and deletion of tumor suppressor genes might be simultaneous consequences of a single unbalanced translocation event: (1) proliferative advantage through increased JAK-STAT signaling in the malignant cells by deletion of SOCS1 cooperates with escape from immune surveillance by (2) overexpression of PD-1 ligands and (3) down-regulation of HLA class II |
| Chromosomal gain of 9p | JAK2, JMJD2C, PDL1, PDL2 | Overexpression of JAK2, JMJD2C, PDL1, and PDL2 is linked as these genes are part of the same amplicon involving cytoband 9p24.1: (1) JAK2 overexpression increases JAK-STAT signaling promoting proliferation; (2) JAK2 transcriptionally augments PD-1 ligand expression inhibiting T-cell activation in the tumor microenvironment and (3) cooperates with JMJD2C to alter the epigenome in PMBCL by histone H3 modifications promoting proliferation and survival |
| Genomic event . | Gene components . | Proposed synergy . |
|---|---|---|
| Unbalanced CIITA rearrangement | SOCS1, CIITA, CIITA fusion partners (eg, PDL1 and PDL2) | Impaired CIITA function, overexpression of fusion partners, and deletion of tumor suppressor genes might be simultaneous consequences of a single unbalanced translocation event: (1) proliferative advantage through increased JAK-STAT signaling in the malignant cells by deletion of SOCS1 cooperates with escape from immune surveillance by (2) overexpression of PD-1 ligands and (3) down-regulation of HLA class II |
| Chromosomal gain of 9p | JAK2, JMJD2C, PDL1, PDL2 | Overexpression of JAK2, JMJD2C, PDL1, and PDL2 is linked as these genes are part of the same amplicon involving cytoband 9p24.1: (1) JAK2 overexpression increases JAK-STAT signaling promoting proliferation; (2) JAK2 transcriptionally augments PD-1 ligand expression inhibiting T-cell activation in the tumor microenvironment and (3) cooperates with JMJD2C to alter the epigenome in PMBCL by histone H3 modifications promoting proliferation and survival |
Furthermore, suppressor of cytokine signaling 1 (SOCS1) has been identified as a recurrently mutated tumor suppressor gene in PMBCL. SOCS1 can be viewed as the key effector of a classic negative feedback loop in which expression of SOCS1 is induced by activated STATs, inhibits JAK phosphorylation, and targets phosphorylated JAK for proteasomal degradation.62 In the MedB-1 mediastinal B-cell line63 biallelic SOCS1 mutations were found abrogating the SOCS box function of the protein and leading to constitutive phosphorylation of JAK2 and STAT5.64 This constitutive pathway activation was abolished by ectopic wild-type overexpression of SOCS1 in MedB-1 cells, demonstrating the tumor-suppressor gene function of SOCS1 in a PMBCL context. In a study of 20 patients with PMBCL, SOCS1 deletion mutations were detected in 45% of cases. Notably, SOCS1 mutations and genomic JAK2 amplification were not mutually exclusive, suggesting an additive effect of these 2 alterations. Interestingly, SOCS1 mutations have been found to be consistent with aberrant SHM in nodular lymphocyte-predominant HL, PMBCL, and other germinal center-derived lymphomas. However, further study of the mutational patterns, specifically in PMBCL, is needed.65,66 In addition, a copy number study revealed homozygous deletion of the SOCS1 locus in cell line KARPAS1106P with similar functional consequences as in MedB-1.67 However, recurrent copy number losses of the SOCS1 locus could not be definitively detected by array comparative genomic hybridization in a study of 37 PMBCL patients.46
Recurrent somatic mutations of STAT6 have been described in 36% of PMBCL cases, contrasting with DLBCL, in which no mutations could be detected, adding further evidence that the JAK-STAT signaling pathway is the target of multiple genomic hits, specifically in PMBCL.68 Of note, concurrent SOCS1 gene alterations and JAK2 gene amplification were not significantly more prevalent in either the STAT6 mutated or nonmutated cases, further suggesting an additive effect of the signaling pathway hits. Investigating the STAT6 DNA binding domain, all gene mutations were found to be heterozygous, including 2 mutations in MedB1 that were on the same allele (“CIS” configuration). Moreover, the mutational pattern of predominantly base pair transversions (A → C or G → T) suggested a genesis other than SHM. However, questions remain on the mechanistic level how the identified mutations in the STAT6 DNA binding domain contribute to the malignant phenotype. As mutated STAT6 protein appears to have diminished DNA binding properties and decreased ability to activate transcription in HEK293 cells, these results are unexpected given the overall pathway activation characteristic of PMBCL. Furthermore, expression of known STAT6-regulated target genes remained unaltered, which has led to the hypothesis that STAT6 mutations might be “passenger” mutations.69 Additional experiments using alternative in vitro and in vivo model systems are needed to clarify the functional role of these heterozygous mutations.
Histone modification
Gene expression regulation and aberrant gene expression by histone modification in normal tissue development and carcinogenesis are increasingly recognized, and altered regulation of the histone modification process itself by mutations in key enzymes have been described in the most current literature regarding BCLs.70,71 In PMBCL, specifically JAK2 and JMJD2C have been shown to be key players in histone 3 modifications essential to tumor cell proliferation and survival using an RNA interference screen.61 This finding was unique to PMBCL and HL cell lines with the 9p24 amplicon compared with DLBCL cell lines without 9p amplification, highlighting the specificity of this observation linked to overexpression of both genes. JMJD2C functions as a demethylase for the trimethylation mark at lysine 9 of histone 3 and impedes the recruitment of HP1α and heterochromatin formation.72 Furthermore, activated phospho-JAK2 cooperates in this process by phosphorylating tyrosine 41 of histone 3 and phosphorylation of HP1α, thereby inactivating HP1α and further contributing to increased formation of active chromatin.73 Interestingly, on RNA interference with JAK2 and JMJD2C, MYC was identified as one of the most regulated genes targeted by altering the chromatin structure of the MYC locus.61 These studies for the first time give unique insight into remodeling of the PMBCL epigenome and open avenues for further investigations and potentially therapeutic intervention with JAK2 and JMJD2C as potential drug targets.
The tumor microenvironment of PMBCL
The number of lymphoma subtypes for which interactions of the malignant cells with non-neoplastic cells of the microenvironment are described is constantly increasing. However, the underlying genetic events harbored by the malignant cells that influence the immune microenvironment remain largely unknown. Only recently, a handful of specific mechanisms have been described in PMBCL. By contrast, the microenvironment in cHL has been extensively studied and serves as a paradigm of tumor-microenvironmental interactions. In the following two paragraphs, we review the most important microenvironment-related features of PMBCL known to date, including the composition of the cellular infiltrate, overexpression of surface molecules, and the underlying genetic changes contributing to an immune escape phenotype of the malignant cells (Figure 3).
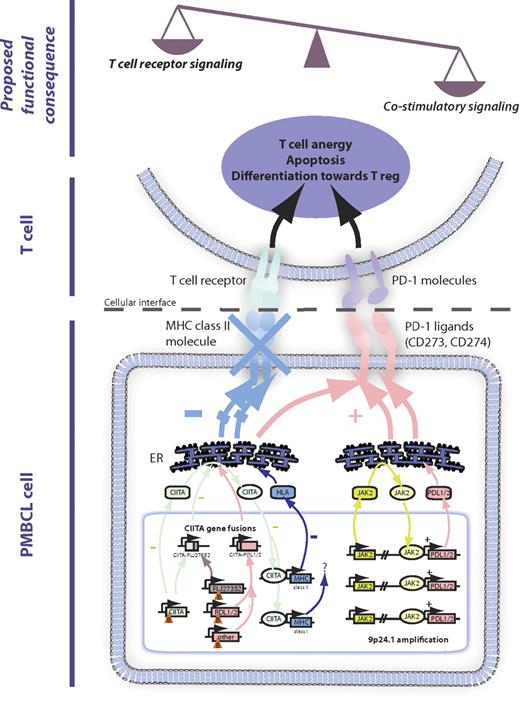
Impact of genetic alterations on the tumor microenvironment. The major mechanisms of 16p13.13 gene rearrangements leading to CIITA gene fusions and 9p24.1 amplification. These mechanisms involve down-regulation of MHC class II and up-regulation of PD-1 ligands, leading to an imbalance of T-cell receptor versus costimulatory signaling. Functional consequences for the tumor microenvironment encompass induction of T-cell anergy, apoptosis, and skewed differentiation toward regulatory T cells. Orange triangles indicate chromosomal break (B). ER indicates endoplasmic reticulum.
Impact of genetic alterations on the tumor microenvironment. The major mechanisms of 16p13.13 gene rearrangements leading to CIITA gene fusions and 9p24.1 amplification. These mechanisms involve down-regulation of MHC class II and up-regulation of PD-1 ligands, leading to an imbalance of T-cell receptor versus costimulatory signaling. Functional consequences for the tumor microenvironment encompass induction of T-cell anergy, apoptosis, and skewed differentiation toward regulatory T cells. Orange triangles indicate chromosomal break (B). ER indicates endoplasmic reticulum.
Down-regulation of HLA class II expression
As a defining feature of the immunophenotype of PMBCL, early studies described reduced expression of major histocompatibility (MHC) class II genes in a substantial number of cases.31 In subsequent work, decreased expression of MHC class II molecules has been consistently linked to inferior outcome,32 similar to observations in DLBCL74,75 and cHL.76 Furthermore, MHC class II loss was found to be correlated with a decreased number of infiltrating cytotoxic T cells in PMBCL,77 and a similar correlation was previously described in DLBCL, suggesting a possible mechanism of escape from immune surveillance.78 Specifically, of 92 PMBCL cases, 65% were negative for HLA-DR protein expression using immunohistochemistry.77 Lack of HLA-DR expression correlated significantly with diminished numbers of cytotoxic T cells (CD8+, TIA1+), but not with CD68+ macrophages, and both loss of HLA-DR expression and decreased TIA1+ cells were correlated with disease-specific and progression-free survival. In aggregate, these data provide the first evidence that immune escape of malignant cells by down-regulation of HLA class II molecules might be an important part of the malignant phenotype. However, the exact functional link between HLA class II loss and reduced numbers of CD8+ cytotoxic T cells still needs to be established, as a CD8+ immune response is usually linked to MHC class I expression. Involvement of T helper cells to assist or modulate the cytotoxic T-cell response as the final common pathway of cell-mediated death appears likely. Interestingly, the density of a distinct T-cell subset, CD4+CD25+FOXP3+ cells (so-called Treg cells), has been found to have high variability (average FOXP3+ proportion 2.6%) compared with other B- and T-cell lymphomas, warranting further investigations focusing on the role of this T-cell subset in the microenvironment of PMBCL.79
The cause for down-regulation of HLA class II in PMBCL and other lymphomas is still not well understood. Gene expression array data revealed a strong correlation between expression of the master transcriptional regulator of class II expression CIITA (MHC2TA) and HLA class II expression.32 These data suggest a distinct mechanism compared with immune-privileged site-associated BCLs,80,81 such as testicular or primary central nervous system lymphomas, in which copy number loss as a result of deletion at the HLA locus on chromosome 6p21.3 has been recurrently described in > 60% of cases.82 By contrast, in PMBCL, chromosomal loss of a distinct region on 6p21 harboring the MHC class I and II genes has only been seen in a small number of cases.46
More evidence for an important role of CIITA in the regulation of MHC class II expression in PMBCL comes from the recent finding of recurrent genomic CIITA rearrangements.11 Evaluation of 263 BCLs by fluorescence in situ hybridization demonstrated that genomic CIITA breaks were recurrent in PMBCL (38%) and cHL (15%) but only rarely found in DLBCL (3%). In PMBCL, these breaks were also linked to inferior disease-specific survival. Moreover, CIITA was found as a promiscuous gene with various partners, resulting in novel in-frame gene fusions; and strikingly, in these cases with gene fusions, a cluster breakpoint region of 1.6 kb in CIITA intron 1 could be identified, disrupting normal CIITA transcription of this allele. Interestingly, in some cases, CIITA breaks were shown to occur in more than one allele, raising the possibility that CIITA rearrangements might lead to complete loss of function. Of note, CIITA coding sequence mutations have been described in a small number of DLBCLs,83 but the functional consequences remain unclear. The CIITA gene was initially found in studies of patients with bare lymphocyte syndrome (complementation group A), a rare autosomal recessive disease characterized by mutations in CIITA leading to loss of MHC class II expression and clinical manifestations because of an immunodeficiency phenotype.84 Several groups found that CIITA functions as a transactivator in a complex of transcription factors (RFX, NFY, and X2BP) that bind to class II MHC promoters.85 Previous studies also describe deletion mutants that inhibit wild-type CIITA function.86-89 Consistent with these findings, in vitro functional experiments in the cHL-derived cell line KM-H2 and DLBCL-derived cell line SUDHL-4 demonstrated that a specific CIITA fusion (CIITA-FLJ27352) suppresses HLA class II expression.11 These data suggest that, in addition to haploinsufficiency of CIITA, MHC class II expression may be suppressed in a dominant-negative manner (Figure 3). In summary, CIITA breaks probably lead to reduced MHC class II expression and might explain, in part, the previously identified clinical correlations discussed in this section.
Overexpression of PD-1 ligands (PDL1/CD274, PDL2/PDCD1LG2)
The surface molecule PDCD1LG2 (CD273, PDL2) has been identified as a highly overexpressed PMBCL-specific gene by gene expression profiling. It is harbored in the common amplicon on chromosome 9p discussed in “JAK-STAT signaling,”3,41,60 and a direct correlation between 9p copy number changes and mRNA expression of both PD-1 ligands, CD274 and PDCD1LG2, has been demonstrated by integrative analysis.10 Both CD274 and PDCD1LG2 have also been found as recurrent gene fusion partners of CIITA,11 and both genes are highly expressed under control of CIITA promoter III by translocation t(9;16)(p24.1;p13.13), resulting in the formation of in-frame CIITA-CD274 and CIITA-PDCD1LG2 gene fusions, respectively.90,91 CD274 and PDCD1LG2 belong to the CD28 costimulatory receptor superfamily and regulate T-cell activity by providing an additional signal to T cells via the PD1 receptor in conjunction with T-cell receptor signaling (Figure 3).92 In DLBCL-derived U2932 cells93 that do not express wild-type PD1 ligands, forced expression of the CIITA-CD274 and CIITA-PDCD1LG2 fusions inhibits activation of Jurkat T cells in coculture, a mechanism that is mediated by PD-1 receptor signaling. These observations were in agreement with findings using U2940,94 a cell line with PMBCL features that expresses high wild-type levels of PD-1 ligands, inducing anergy in cocultured Jurkat T cells.11 A similar mechanism has been described for cHL, in which PD1 ligand overexpression has been shown to contribute to T-cell exhaustion of the infiltrating T cells.95 These data demonstrate that PD-1 ligands can be overexpressed by either gene amplification or translocation and strongly suggest that in a subset of PMBCL the malignant B cells escape immunosurveillance by overexpression of surface molecules inactivating infiltrating effector T cells. Of note, elevated surface expression of galectin-1 (encoded by LGALS1) has been described in HL influencing the composition of the microenvironment toward increased T helper 2 and T regulatory cells,96 providing further support for the hypothesis that the malignant cells uniquely orchestrate their microenvironment in certain lymphoma subtypes.
Synergy of alterations and perspective
Similar to other cancers, the pathogenesis of PMBCL is presumably a multistep process with accumulation of multiple genetic alterations. The JAK-STAT signaling pathway is illustrative, demonstrating the coincidence of multiple pathway hits that probably cooperate in conferring an additive survival advantage to the malignant cells. However, recent studies also provide evidence that multiple pathways can be affected by a single underlying event on the genomic level.10,11,61 This process can be viewed as a comparably economic way of cancer cells gaining a survival advantage in contrast to accumulation of multiple independent events.
In the following 2 paragraphs, we discuss the simultaneous consequences of unbalanced chromosome 16p13.13 rearrangements and chromosomal gain of the 9p24 amplicon (Table 2). Finally, we outline how the dependence of PMBCL on some of the previously described key pathways might be translated into novel therapeutic approaches.
Unbalanced 16p13.13 rearrangements
Based on cytogenetic studies of chromosomal rearrangements of the CIITA locus (16p13.13), a substantial number of cases were found to harbor unbalanced rearrangements with loss of genetic material centromeric of the breakpoint.11 This finding is in keeping with previous reports describing deletions of the tumor-suppressor gene SOCS1 that closely maps to CIITA on chromosome 16, leading to downstream activation of JAK-STAT signaling.67 As these rearrangements coincide with disruptive CIITA breaks and overexpression of CIITA fusion partners, one can reasonably hypothesize that deletion of tumor suppressor genes, such as SOCS1, overexpression of oncogenes resulting from gene fusion and CIITA loss of function might be concurrent consequences of a single genetic event. The functional consequences might be diverse as numerous CIITA fusion partners have been described; however, it appears probable that, at least in a subset of PMBCL cases, a proliferative advantage through JAK-STAT signaling is combined with an immune escape mechanism through down-regulation of MHC class II and up-regulation of PD-1 ligands inducing T-cell anergy (Figure 3).
Chromosomal gain of 9p24
As discussed in “JAK-STAT signaling,” amplification of a well-defined region on chromosome 9p24 has been identified in the majority of cases and can be considered as one of the genetic hallmark events of PMBCL pathogenesis. However, only recently have systematic approaches identified the target genes in the amplicon that are overexpressed and prove to be functionally most relevant. Remarkably, these studies did not identify a single target gene but revealed that multiple genes are coamplified and cooperate in the pathogenesis.10,61 Integrative study of copy number and mRNA expression level changes confirmed overexpression of JAK2 and PD-1 ligands in cases with 9p amplification and demonstrated that JAK2 further augments PD-1 ligand expression by transcriptional up-regulation.10 Moreover, JAK2, JMJD2C, and RANBP6 were identified as critical genes required for proliferation and survival by an RNA interference screen, of which JAK2 and JMJ2D2C were shown to cooperate in modifying the epigenome in these lymphomas.61 In aggregate, increased JAK-STAT signaling through JAK2 amplification, PD-1 ligand surface expression sculpting a favorable microenvironment, and epigenetic modifications changing gene expression of a wide range of genes, such as MYC, probably cooperate to develop the full malignant phenotype of PMBCL.
Novel therapeutic approaches
Dose-intensified treatments have been suggested to improve clinical outcomes in patients with PMBCL in a retrospective study.20 However, no randomized clinical trials have been reported to date. With the exception of rituximab (#NCT00001337), there are currently no registered trials that are actively recruiting (www.clinicaltrials.gov, accessed April 26, 2011) that use targeted therapy approaches to provide an alternative to dose escalation and to minimize treatment-related sequelae. As treatment recommendations are historically linked to DLBCL, the relative specificity of JAK-STAT,8 NF-κB pathway activation,9 overexpression of PD-1 ligands,3 and the histone-modifying genes JAK2 and JMJD2C61 stand out as the most rational targets for specifically tailored future therapies in PMBCL. In vitro studies in MedB-1 cells demonstrate proof of principle that JAK pathway inhibition and reduction of STAT5 phosphorylation are toxic to the cells,58 raising hope that similar approaches of JAK-STAT pathway inhibition might be a promising approach. Similarly, NF-κB pathway inhibition has yielded promising results in preclinical models.44,97 Specifically, the histone deacetylase inhibitor vorinostat has also been described to specifically inhibit STAT6 phosphorylation in HL.98 Furthermore, targeting the tumor cell microenvironment interaction by inhibition of T-cell accessory signaling might be very promising based on encouraging data in melanoma, in which a combined PD-1/CTLA-4 blockade led to increased infiltration of effector T cells promoting tumor cell kill.99 These data highlight the importance of host immunity and that immune escape mechanisms of tumor cells can be targeted in conjunction with cytostatic therapy; a paradigm shift in treatment approaches that might be fruitful in a subset of PMBCLs and related lymphomas.
Acknowledgments
The authors thank Dr David Scott for critical review of the manuscript.
This work was supported by the Cancer Research Society (Steven E. Drabin postdoctoral fellowship; C.S.). R.D.G. is supported by the Canadian Institutes for Health Research (grant 178536) and the Terry Fox Foundation (Program Project grant 019001).
Authorship
Contribution: C.S. and R.D.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Steidl, Department of Pathology, Centre for Lymphoid Cancer, British Columbia Cancer Agency and Experimental Therapeutics, British Columbia Cancer Research Centre, 675 West 10th Avenue, Room 5-114, Vancouver, BC V5Z 1L3, Canada; e-mail: csteidl@bccancer.bc.ca.