Abstract
Interferon regulatory factor 4 (IRF4) is a member of the interferon regulatory factor family of transcription factors and has been shown to have critical functions at several stages of B-cell development. Genome-wide association study identified a polymorphism in the 3′ untranslated region of IRF4 as a chronic lymphocytic leukemia risk locus. In this study, we report a recurrent heterozygous somatic mutation in the DNA-binding domain of IRF4 detected in 7 of 457 chronic lymphocytic leukemia patients (1.5%). Patients with IRF4 mutation have a good prognosis, and 4 of 6 have a trisomy 12. We also found that IRF4 mRNA expression is higher in the patients with the mutation.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most common form of lymphoid neoplasia in the Western countries. Genetic aberrations can be identified in > 80% of CLL patients. The most frequent recurrent alterations include deletion/inactivation of 13q14 (> 50%), 11q22–23 (18%), trisomy of 12 (15%-18%), and deletion 17p (7%-10%).1 Genome-wide association studies allowed finding out the susceptibility loci for CLL.2 Among them, single nucleotide polymorphism in the 3′-untranslated region of IRF4 was strongly associated with an increased risk of CLL.2 The corresponding gene, interferon regulatory factor 4 (IRF4), is a lymphocyte-restricted member of the interferon regulatory factor family of transcription factors.3-5 Members of the IRF family are involved in important cellular processes, such as pathogen response, cytokine signaling, and apoptosis.6 IRF4 has been shown to have critical functions at several stages of B-cell development.7 The role of IRF4 has been clearly established in multiple myeloma but is still an enigma in CLL.8 Recently, a whole genome sequencing study of 38 multiple myeloma patients showed a mutation in the IRF4 DNA-binding domain in 2 patients (K123R) and by sequencing in 2 more multiple myeloma patients (4 of 200; 2%).9
Methods
Sequencing
The study was carried out in accordance with the Institutional Review Board protocol approved by the Ohio State University. CLL samples were obtained from 457 untreated CLL patients: 184 aggressive CLL samples, 106 indolent CLL samples, and 167 CLL samples, which could not be classified as aggressive or indolent, were analyzed. Aggressive status was defined as unmutated imunoglobulin variable heavy chain (IgVH; > 98% of homology to the germline), and > 20% of ZAP70-positive cells. Indolent status was defined as mutated IgVH (< 98% of homology to the germline) and < 20% of ZAP70-positive cells. Patients were enrolled in the CLL Research Consortium on written informed consent. DNA was extracted with the DNeasy Blood & Tissue Kit (QIAGEN). IRF4 mutations were determined by PCR amplification of the 8 coding exons with the high-fidelity Advantage 2 polymerase master mix (Clontech). The primers for exon 2 were: 2dir, aactcaggagttcgatgctgc; 2rev, aactcaggagttcgatgctgc. Clinical and prognostic data were obtained for 6 of the 7 patients with the mutation. IRF4 exon 2 was sequenced in 200 normal DNA samples (Sigma-Aldrich).
Functional studies
IRF4 wild-type (WT) and mutants were PCR amplified and cloned in the p3XFLAG-CMV-10 vector (Sigma-Aldrich). This construct was used to generate the mutants using Quick Change site-directed mutagenesis kit (Stratagene). HEK 293 cells were transfected with 1 μg of the WT/mutant plasmids using lipofectamine 2000 (Invitrogen). For gene-expression profiling, RNA samples were analyzed with the use of Affymetrix U133 plus 2.0 GeneChips. IRF4 mRNA expression was quantified by quantitative reverse-transcribed polymerase chain reaction using ABI Prism 7900HT sequence detection systems with Applied Biosystems TaqMan gene-expression assays (IRF4 Hs01056534_m1). Normalization was performed with GAPHD. Apoptosis was measured by annexin V/propidium iodide stain (BD Biosciences PharMingen) performed at 24 and 48 hours after transfection. Caspase-3 and -7 activity was measured at 48 hours using the Caspase-Glo 3/7 Assay (Promega).
Results and discussion
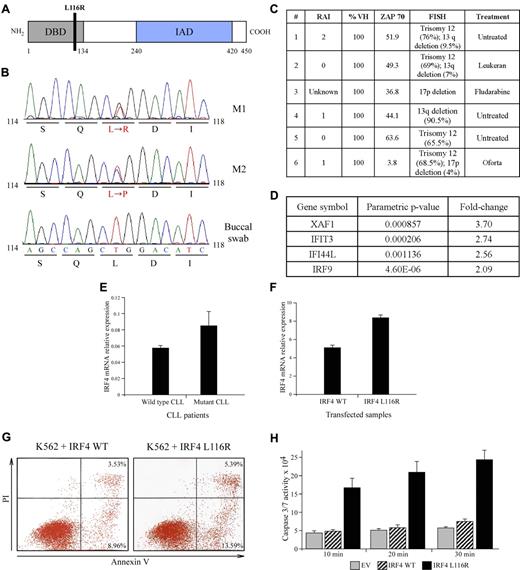
Sequencing of IRF4 coding exons in 93 CLL patients revealed a heterozygous missense somatic substitution localized in the exon 2 of the gene in 2 patients. The substitution (leucine → arginine–L116R–M1) was located in the DNA-binding domain of the gene (codons 20-137; Figure 1A-B). Sequencing of the exon 2 in other 364 additional CLL identified 4 more patients with the same heterozygous mutation (1.4%), and one other patient had another substitution (leucine → proline–L116P–M2) affecting the same base. The mutation was not found in the buccal swabs analyzed (Figure 1B). Thus, it could not represent a polymorphism. To further eliminate the possibility that we found a rare polymorphism, we sequenced 200 DNA samples from normal persons and no mutations in the exon 2 of IRF4 were detected (not shown). This mutation was not reported as a polymorphism in the single nucleotide polymorphism databases analyzed (University of California Santa Cruz browser and National Cancer Institute single nucleotide polymorphism database). Interestingly, 4 of the 6 patients had a trisomy 12 detected by fluorescence in situ hybridization (Figure 1C). Trisomy 12 was considered the third abnormality in frequency and to have an intermediate prognosis.1 The molecular consequences of trisomy 12 are unknown. However, all patients with the IRF4 mutation have an unmutated IgVH status (> 98% of homology to the germline), which predicts an inferior clinical course in CLL.10 CLL with IRF4 expression seems to have a more favorable clinical course and is more likely to have disease at low Rai stage.11
IRF4 mutations in CLL. (A). Schematic representation of the L116R mutation. (B) Chromatographs of L116R and L116P mutations. (C) Clinical characteristics of patients showing IRF4 mutations. (D) Significantly overexpressed genes in HEK 293 cells transfected with IRF4 L116R versus WT IRF4. (E-F) Mutant and WT IRF4 mRNA levels in CLL samples and transfected HEK 293 cells. (G) Expression of IRF4 L116R mutant increases apoptosis in K562 cells (representative experiment is shown). (H) Expression of IRF4 L116R mutant increases caspase-3/caspase-7 activity in K562 cells. Reaction was stopped at the indicated time points.
IRF4 mutations in CLL. (A). Schematic representation of the L116R mutation. (B) Chromatographs of L116R and L116P mutations. (C) Clinical characteristics of patients showing IRF4 mutations. (D) Significantly overexpressed genes in HEK 293 cells transfected with IRF4 L116R versus WT IRF4. (E-F) Mutant and WT IRF4 mRNA levels in CLL samples and transfected HEK 293 cells. (G) Expression of IRF4 L116R mutant increases apoptosis in K562 cells (representative experiment is shown). (H) Expression of IRF4 L116R mutant increases caspase-3/caspase-7 activity in K562 cells. Reaction was stopped at the indicated time points.
To investigate functional consequences of L116R mutation, we transfected HEK 293 cells with WT and mutant IRF4 constructs and carried out gene expression analysis using Affymetrix chips. We found several genes differentially expressed between HEK 293 cells transfected with the IRF4 mutant compared with IRF4 WT (Figure 1D). XAF1, a proapoptotic gene, was the most significantly overexpressed gene in IRF4 mutant transfected HEK 293 cells. To explore the possibility that the effect of the mutation might be mediated through differential IRF4 expression, we investigated the mRNA expression of IRF4 in 4 mutant samples compared with 2 patients without mutation and found higher IRF4 expression in the mutant patients (with a fold-change of 1.5; Figure 1E). The same ratio was found when HEK 293 cells were transfected with mutant (M1) versus WT IRF4 plasmid (Figure 1F). Transfection of IRF4 mutant plasmid in K562 cells resulted in an increase in apoptosis (∼ 18.0% vs ∼ 12.2%, Figure 1G) and the caspase-3/caspase-7 activity (∼ 3.5-fold, Figure 1H).
Mittrücker et al generated Irf4-deficient mice by replacing exons 2 and 3 of the gene with a neomycin resistance gene.12 The mice displayed profound defects in mature T-cell function but also a lymphoproliferative disorder with a progressive accumulation of T and B cells in the spleen and lymph nodes. This phenotype suggests that Irf4 may have an important role in regulating the activation but also in controlling the apoptosis of the lymphocytes. Moreover, expression of IRF4 in Jurkat T cells enhances their sensitivity to Fas-mediated apoptosis by the activation of the initiator caspase-8.13 Of note, in multiple myeloma cell lines, IRF4 was shown to target a number of genes involved in different cellular processes, such as cell cycle, metabolism, general transcription, membrane biogenesis, cell death, and plasma cell functions.14 IRF4 inhibition by shRNA was toxic for multiple myeloma cells.14
IRF4 is an essential regulator of multiple steps in B-cell differentiation. Spleens of Irf4-deficient mice showed B lymphocytes blocked at a late stage of B-cell maturation and consistently no germinal centers in B-cell follicles.12 The unmutated IgVH status of our IRF4 mutants seems to be in concordance with this finding. Consequently, Irf4-deficient mice do not produce antigen-specific antibody on immunization. The inability of Irf4-deficient B cells to proliferate and survive on B-cell receptor stimulation may be sufficient to prevent germinal center B-cell differentiation, but IRF4 may also regulate a key gene(s) required to promote germinal center differentiation.14 Saito et al recently showed that IRF4 binds to BCL6 promoter and represses BCL6 transcription.15 No difference in BCL6 protein expression was observed after transfection of IRF4 mutant or WT in HEK 293 cells or in Raji cell line, a Burkitt line with high expression of BCL6 (not shown). Future work is needed to understand the relevance and functional consequence of this recurrent mutation in the IRF4 DNA-binding domain and to better understand its impact in CLL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
V.H. was supported by the Belgian Fonds National de la Recherche Scientifique (grant Télévie). This work was also supported by the American Cancer Society (Research Scholar Grant, Y.P.) and National Institutes of Health CLL Research Consortium (grant PO1-CA81534, L.R., T.K., and C.M.C.).
National Institutes of Health
Authorship
Contribution: V.H., T.N., Y.P., and C.M.C. designed research; L.R. and T.K. provided patient samples and clinical data; V.H., T.N., Y.P., A.P., and H.A. performed research and analyzed data; V.H., Y.P., and C.M.C. wrote the paper; and all authors critically reviewed and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlo M. Croce, The Ohio State University, Comprehensive Cancer Center, Biomedical Research Tower, Rm 460, West 12th Ave, Columbus, OH 43210; e-mail: carlo.croce@osumc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal