Abstract
The iron-regulatory hormone, hepcidin, regulates systemic iron homeostasis by interacting with the iron export protein ferroportin (FPN1) to adjust iron absorption in enterocytes, iron recycling through reticuloendothelial macrophages, and iron release from storage in hepatocytes. We previously demonstrated that FPN1 was highly expressed in erythroblasts, a cell type that consumes most of the serum iron for use in hemoglobin synthesis. Herein, we have demonstrated that FPN1 localizes to the plasma membrane of erythroblasts, and hepcidin treatment leads to decreased expression of FPN1 and a subsequent increase in intracellular iron concentrations in both erythroblast cell lines and primary erythroblasts. Moreover, injection of exogenous hepcidin decreased FPN1 expression in BM erythroblasts in vivo, whereas iron depletion and associated hepcidin reduction led to increased FPN1 expression in erythroblasts. Taken together, hepcidin decreased FPN1 expression and increased intracellular iron availability of erythroblasts. We hypothesize that FPN1 expression in erythroblasts allows fine-tuning of systemic iron utilization to ensure that erythropoiesis is partially suppressed when nonerythropoietic tissues risk developing iron deficiency. Our results may explain why iron deficiency anemia is the most pronounced early manifestation of mammalian iron deficiency.
Introduction
Ferroportin (FPN1) is the sole iron export protein that has been identified thus far in mammals.1 It is highly expressed on the basolateral membrane of duodenal enterocytes where it facilitates the export of newly absorbed dietary iron into the systemic circulation, in reticuloendothelial macrophages where it exports iron recycled from senescent red blood cells into blood, and in hepatocytes where it releases iron from storage. The expression of FPN1 is regulated posttranslationally by a small peptide hormone, hepcidin, which is secreted mainly by the liver in response to iron overload, inflammation, hypoxia, and erythropoiesis.2 In the circulation, hepcidin binds to FPN1 on the plasma membrane of cells in various tissues and induces its internalization, ubiquitination, and degradation, which blocks the flux of iron into the blood stream.3,4 Iron overload increases hepcidin expression, probably through a pathway that integrates inputs from molecules such as HFE, TfR2, HJV, BMP6, and TMPRSS6 to regulate hepcidin transcription. High-serum hepcidin reduces FPN1 expression and blocks iron flux from tissues into the circulation; conversely, iron deficiency decreases hepcidin expression and allows FPN1 to remain available for transport of iron from tissues into the blood stream.2,5-7 Thus, through a feedback mechanism, the hepcidin-FPN1 interaction regulates systemic iron homeostasis. Dysregulation of hepcidin or FPN1 expression is involved in the pathogenesis of diseases such as hemochromatosis,8-10 anemia of chronic disease,11,12 and β-thalassemia,13,14 underlining the importance of hepcidin and FPN in the regulation of systemic iron homeostasis.
In addition to the regulation by hepcidin at the protein level, FPN1 expression can also be regulated at the posttranscriptional level by the iron-regulatory protein (IRP)/iron-responsive element (IRE) machinery.15-17 The canonical transcript of FPN1 has a IRE in its 5′-untranslated region (UTR), and in iron-deficient conditions IRPs can bind to the IRE and repress the translation of FPN1. This translational repression reduces expression of the FPN1 protein in iron-deficient cells, enabling cells to retain intracellular iron when iron availability is limited. However, in contrast to expectations that are based on regulation by the IRP/IRE machinery, FPN1 protein expression was shown to be up-regulated in the duodenum of animals maintained on a low-iron diet.15,16 Moreover, FPN1 protein expression in enterocytes needs to be up-regulated in iron-deficient animals to provide more iron for systemic iron demands. In the process of investigating the underlying mechanism for up-regulation of duodenal FPN1 protein during iron starvation, we previously identified a FPN1 transcript (FPN1B) that does not have a IRE in its 5′-UTR.18 FPN1B is generated from a promoter located upstream of the promoter of the FPN1A transcript (FPN1 transcript that contains the IRE) and encodes the identical ferroportin protein. FPN1B is specifically expressed in enterocytes and is conserved in both humans and mice.18,19 The expression of FPN1B in enterocytes probably enables the cells to evade translational suppression by the IRP/IRE system and to express FPN1 protein under conditions of iron starvation at the potential risk of generating intracellular iron deficiency.
Most strikingly, we found that FPN1B was also highly expressed in erythroblasts of mouse BM, and the protein levels of FPN1 in erythroblasts exceeded those of BM macrophages.18 The promoter of FPN1B in erythroblasts is regulated through putative GATA and EKLF binding sites, and its expression decreases during differentiation of erythroblasts.18 To explore the physiologic function of FPN1 in erythropoiesis, we investigated the function of FPN1 and its regulation by hepcidin in erythroblasts in vitro and in vivo, and we found that hepcidin regulates FPN1 expression and the intracellular iron homeostasis of erythroblasts.
Methods
Animals
All of the animals used in this study were C57BL/6 mice and were approved by the National Institutes of Health Institutional Review Board of Animal Care Committee. For dietary manipulations, newly weaned mice were maintained on a low-iron diet (2-10 mg/kg chow) or control diet (200 mg/kg chow) for ≥ 3 months before hepcidin injection. For hepcidin treatment, the mice were peritoneally injected with hepcidin (Bachem Americas Inc) 20 μg per mouse in 0.3 mL of PBS, with a second dose injected 20 hours later. The control group was injected with PBS only. The animals were killed 24 hours after the first dose injection, and Ter119-positive erythroblasts were isolated with Dynabead (Invitrogen) as described previously.18 In brief, the BM cells from the femurs were incubated with Dynabeads that had been precoated with biotinylated anti–mouse Ter119 Ab (R&D Systems), and the Dynabead-cell complexes were then isolated, and cells were lysed in RIPA buffer (50mM Tris, 150mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1% Triton X-100) with Protease inhibitor tablet (Roche) for Western blot analysis.
Plasmid preparation, cell culture, and transfection
The pEGFP-N1-FPN1 plasmid was generously provided by Dr Ivana De Domenico (University of Utah); there is no IRE element in its 5′-UTR, and its product is a FPN1GFP (green fluorescent protein) fusion protein.9 The pIRES2EGFP-FPN1 vector was created by cloning mouse FPN1 into the pIRES2EGFP vector (Clontech) with XhoI and KpnI sites. The pDsRed-Monomer-F vector was from Clontech Laboratories, Inc. Erythroblast cell lines K562 and MEL were cultured as described previously.18 The cells were transfected with 2 μg of plasmid per 1 × 106 cells with Neon Transfection system (Invitrogen) according to the manufacturer's protocol, and 30 minutes after transfection 100μM iron-nitrilotriacetate (FeNTA) was added to the medium to counteract the potential intracellular iron starvation induced by FPN1 overexpression.
Primary erythroblast cell culture
The erythroblasts from fetal liver were cultured according to previous protocols with minor modifications.20 Briefly, cells from the fetal liver of embryos at 13.5 embryonic days (E13.5) were cultured in StemPro-34 serum-free medium (Invitrogen) supplemented with 2 U/mL human recombinant erythropoietin, 100 ng/mL murine recombinant stem cell factor, 10μM synthetic glucocorticoid dexamethasone, 40 ng/mL insulin-like growth factor 1, 2mM l-glutamine, 75 μg/mL human holo-transferrin, 140mM monothioglycerol, 100 U/mL penicillin, and 100 μg/mL streptomycin under 37°C with 5% CO2. The cells were maintained at a density of 1-4 × 106 cells/mL, the cell medium was changed daily, and differentiated cells were removed by Ficoll purification every 4-5 days. To induce differentiation, proliferating cells were washed twice with DMEM and then were seeded at ∼ 1-2 × 106 cells/mL in StemPro-34 medium supplemented with 10 U/mL erythropoietin, 4 × 10−4 U/mL insulin, 1 mg/mL holo-transferrin, 2mM l-glutamine, 140mM monothioglycerol, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Microscopy analysis
To study the localization of FPN1 in K562 cells, the cells were cotransfected with 2 μg per 1 × 106 cells of pEGFP-N1-FPN1 and pDsRed-Monomer-F for 24 hours in medium supplemented with 100μM FeNTA and were then checked in a Chambered Coverglass under a confocal microscope. To study the localization of endogenous FPN1 in primary erythroblasts, fetal liver erythroblasts were fixed in 4% paraformaldehyde in PBS and were then permeabilized with 0.1% Triton X-100 in PBS with 0.1% BSA. After blocking with 5% donkey serum in PBS, the slides were incubated with rabbit anti–mouse FPN1 Ab (1:100) that we described previously18 and mouse anti-TfR1 Ab (1:200; Zymed) overnight at 4°C, followed by incubation with FITC-donkey anti–rabbit and Cy3-donkey anti–mouse Abs (Jackson ImmunoResearch Laboratories) for 1 hour at room temperature. The pictures were taken with a Leica SP5 confocal microscope.
Fluorescence-activated cell sorting
The GFP-positive cells and TfR1 expression levels were analyzed by FACS on BD FACSCalibur, and the results were acquired and analyzed with CellQuest. For analysis of TfR1 expression in K562 cells, the cells were incubated with allophycocyanin (APC)–labeled anti–human TfR1 Ab (BD PharMingen) for 30 minutes under 4°C in PBA buffer (1% BSA, 0.1% sodium azide in PBS, pH7.4). To verify cell surface TfR1 expression as an indicator of intracellular iron status, K562 cells were treated with 100μM FeNTA or the iron chelator desferrioxamine (DFO) for 18 hours, followed by incubation with APC-mouse anti–human TfR1 Ab, and were subsequently analyzed with FACS. To analyze TfR1 expression in cells transfected with vectors, a gate was set up to analyze the GFP-positive cells only.
Real-time quantitative PCR
Real-time qPCR experiments were performed with SYBR Green PCR master mixture (Applied Biosystems). Total RNA was prepared with TRIzol reagent (Invitrogen), and cDNA was prepared with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) with 2 μg of total RNA. The primers for FPN1, FPN1A, FPN1B, hemoglobin α adult chain 1, and actin were the same as those described previously.18 The primers for hepcidin were as follows: forward primer, TTGCGATACCAATGCAGAAG, and reverse primer, TGCAACAGATACCACACTGG. The results were normalized against actin levels.
Immunoprecipitation of cell surface proteins, Western blot, and RNA mobility shift assays
Fetal liver erythroblasts were incubated with or without 1 μg/mL hepcidin for 24 hours and were then labeled with or without Sulfo-NHS-LC-Biotin (Pierce) according to the manufacturer's protocol. Biotin-labeled cells were then lysed in RIPA buffer, and cell lysates were incubated with Streptavidin Gel (Pierce) overnight at 4°C. The protein-streptavidin gel complexes were incubated with loading buffer at 65°C for 5 minutes before being loaded to gels for Western blot analysis. Tissues were homogenized in liquid nitrogen, and lysates were prepared in homogenizing buffer (50mM Tris-HCl, pH7.0, 150mM NaCl, 2mM EDTA, 1mM AEBSF, and complete Protease Inhibitor Cocktail). For cultured cells, the cell pellet was resuspended in homogenizing buffer and treated with a sonifier (Sonic Dismembrator model 100) for 10 pulses 1 second each at setting 2. The lysates were centrifuged at 7500g for 10 minutes, supernatants were centrifuged at 100 000g for 60 minutes, and pellets were resuspended in 25mM Tris-HCl, pH7.0, with 5% glycerol and 0.2% NP-40. The protein concentration was measured with BCA protein assay (Pierce). Protein (50 μg for each sample) was separated with 4%-20% Tris-glycine SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The dilution is 1:4000 for the mouse anti–tubulin Ab (Sigma-Aldrich) and 1:5000 for the rabbit anti-GFP Ab. The Abs for anti-FPN1 (Map23 Ab that we described previously), TfR1, L-ferritin, IRP1, and IRP2 were used as previously described.18,21 The membranes were developed with the use of the ECL kit (Pierce). The RNA mobility shift assays were performed as described previously.21
Iron-55 retention experiments
Primary erythroblasts from fetal liver in 1 well of a 6-well plate were loaded with 5μCi (0.185 Bq; 0.2 μg) 55Fe (PerkinElmer) for 2 hours at 37°C in proliferation medium and then were washed with DMEM and cultured in proliferating medium in the presence of hepcidin at the indicated concentration for 24 or 48 hours. After being washed twice with DMEM, 55Fe retained inside the cells was measured with a Liquid Scintillation Counter.
Statistics
All data are presented as mean ± SD, and the statistical analyses were done with the Student t test.
Results
FPN1 is localized to the plasma membrane of erythroblast cell lines and primary erythroblasts
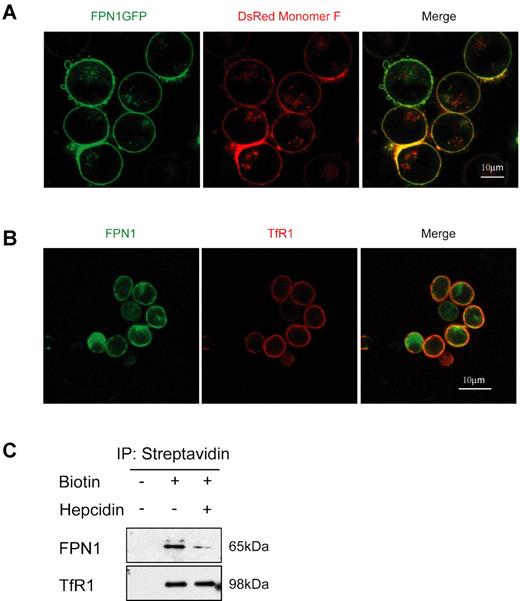
To investigate the localization of FPN1 in erythroblasts, we transiently transfected K562 cells with a plasmid that encodes FPN1GFP (Figure 1A), and we found that FPN1GFP colocalized with the plasma membrane marker DsRed-Monomer-F, suggesting that FPN1 localizes to the plasma membrane of erythroblasts. Transfected FPN1 also localized to the plasma membrane of mouse erythroblast cell line MEL cells (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The localization of endogenous FPN1 was then investigated in fetal liver erythroblasts with the use of a rabbit anti–mouse FPN1 Ab that we described previously (Figure 1B).18 The majority of FPN1 colocalized with TfR1 on the plasma membrane, but some fluorescence observed inside the cells may represent newly synthesized FPN1 that is trafficking through the Golgi/endoplasmic reticulum network (Figure 1B). To verify the localization of FPN1 in primary erythroblasts, cell surface proteins were labeled with biotin and were then pulled down with streptavidin gel, after which FPN1 levels were analyzed by Western blot analysis. As shown in Figure 1C, streptavidin gel immunoprecipitated endogenous FPN1 in the biotin-labeled samples of primary erythroblasts. Hepcidin treatment markedly decreased FPN1 levels on the cell surface. As a positive control, TfR1 was also immunoprecipitated from the biotin-labeled samples. In samples not treated with biotin, neither FPN1 nor TfR1 were detected, confirming that FPN1 localizes to the plasma membrane of erythroblasts (Figure 1C).
FPN1 is localized to the plasma membrane of K562 erythroblast cells and primary erythroblasts. (A) K562 cells were transiently transfected with plasmids to express FPN1GFP and the plasma membrane marker pDsRed-Monomer F for 24 hours to examine their colocalization. (B) Endogenous FPN1 localized mainly to the plasma membrane of primary erythroblasts. Fetal liver erythroblasts were incubated with rabbit anti-FPN1 and mouse anti-TfR1 Abs and were then labeled with FITC-donkey anti–rabbit and Cy3-donkey anti–mouse Abs. Pictures were taken with the Leica SP5 confocal microscope with 63×/1.4 oil objective lens under room temperature. (C) Immunoprecipitations of cell-surface proteins showed that FPN1 localizes on the plasma membrane. Fetal liver erythroblasts were incubated with or without hepcidin for 24 hours, then the cell-surface proteins were labeled with or without Sulfo-NHS-LC-Biotin. The cell lysates were incubated with streptavidin gel, and the immunoprecipitated proteins were immunoblotted with anti-FPN1 and anti-TfR1 Abs.
FPN1 is localized to the plasma membrane of K562 erythroblast cells and primary erythroblasts. (A) K562 cells were transiently transfected with plasmids to express FPN1GFP and the plasma membrane marker pDsRed-Monomer F for 24 hours to examine their colocalization. (B) Endogenous FPN1 localized mainly to the plasma membrane of primary erythroblasts. Fetal liver erythroblasts were incubated with rabbit anti-FPN1 and mouse anti-TfR1 Abs and were then labeled with FITC-donkey anti–rabbit and Cy3-donkey anti–mouse Abs. Pictures were taken with the Leica SP5 confocal microscope with 63×/1.4 oil objective lens under room temperature. (C) Immunoprecipitations of cell-surface proteins showed that FPN1 localizes on the plasma membrane. Fetal liver erythroblasts were incubated with or without hepcidin for 24 hours, then the cell-surface proteins were labeled with or without Sulfo-NHS-LC-Biotin. The cell lysates were incubated with streptavidin gel, and the immunoprecipitated proteins were immunoblotted with anti-FPN1 and anti-TfR1 Abs.
Hepcidin regulates FPN1 expression and intracellular iron levels of erythroblast cells
To assess intracellular iron status, we used FACS analysis to evaluate surface expression of TfR1 in K562 cells. Because the TfR1 transcript has 5 IREs in its 3′-UTR that may be bound and stabilized by IRPs under conditions of iron deficiency, its expression is up-regulated by iron deficiency and down-regulated by iron repletion. As shown in Figure 2A, TfR1 signal intensity was increased by treatment with the iron chelator DFO and decreased by FeNTA supplementation, verifying that TfR1 expression can be used as an indicator of intracellular iron levels in this assay. Expression of the FPN1GFP fusion protein in K562 cells resulted in increased TfR1 expression compared with cells expressing GFP only (Figure 2B), which suggested that FPN1 causes intracellular iron deficiency by exporting iron out of K562 cells. Treatment with 1 μg/mL hepcidin for 24 hours significantly decreased numbers of FPN1GFP-positive cells in both K562 and MEL cells, whereas there was no effect on cells transfected with GFP only, which suggested that hepcidin specifically induces FPN1 degradation in erythroblasts (Figure 2C-D; supplemental Figure 1B-C). Hepcidin at a concentration of 1 μg/mL induced FPN1 degradation in a time-dependent manner (Figure 2E); FPN1 levels significantly decreased after 4 hours of treatment and were almost 50% decreased after 24 hours, whereas there was no effect on GFP levels in cells expressing GFP alone. Hepcidin also induced FPN1 degradation in a dose-dependent manner (Figure 2F). FPN1 expression was significantly decreased by treatment with 50 ng/mL hepcidin for 24 hours, and its expression was decreased further with increased hepcidin concentrations, such that 2 μg/mL hepcidin decreased FPN1 expression by > 50%. Notably, there were fewer positive cells in cells expressing FPN1GFP than cells expressing GFP only, perhaps because the properties of FPN1GFP (encoding a membrane protein with 12 potential trans-membrane domains) differ from those of GFP (soluble protein). The effects of hepcidin on FPN1-induced iron deficiency were also studied in K562 cells transfected with the pIRES2EGFP-FPN1 vector. This vector contains an internal ribosomal entry site and generates a single mRNA from which both FPN1 and GFP can be independently translated into 2 distinct proteins. In transfected cells, GFP expression can thus serve as a marker for cells expressing FPN1, and TfR1 expression can be selectively measured in GFP-expressing cells. Consistent with Figure 2B, expression of FPN1 significantly increased TfR1 expression (Figure 2G), whereas hepcidin treatment decreased TfR1 expression to a level similar to cells transfected with the vector only, suggesting that hepcidin prevents FPN1 from exporting iron in erythroblast cell lines.
FPN1 expression is regulated by hepcidin in transfected erythroblast cell lines in a dose- and time-dependent manner as indicated by FACS analysis. (A) Validation of the use of FACS analysis to measure iron-dependent TfR1 expression. K562 cells were treated with 100μM FeNTA or 100μM DFO for 18 hours, and cells were then labeled by APC-labeled TfR1 Abs and checked by FACS analysis. (B) Expression of FPN1 increases TfR1 expression in K562 cells. K562 cells were transiently transfected with pEGFP-N1-FPN1 to express the fusion protein FPN1GFP for 24 hours, and the expression of TfR1 was then measured by FACS. Expression of FPN1 in K562 (C) and MEL (D) cells was down-regulated by hepcidin. Cells were transiently transfected with pEGFP-N1 or pEGFP-N1-FPN1 and incubated with 1 μg/mL hepcidin for 24 hours, and the GFP-positive cells were then measured by FACS. (E) Time-dependent regulation of FPN1 by hepcidin. K562 cells were transfected with pEGFP-N1-FPN1 or pEGFP-N1 vectors for 12 hours, and then incubated with or without 1 μg/mL hepcidin. The GFP-positive cells were measured by FACS at the indicated time points. (F) Dose-dependent regulation of FPN1 by hepcidin. K562 cells transfected with pEGFP-N1-FPN1 or pEGFP-N1 vectors were incubated with hepcidin of indicated concentrations for 24 hours. The GFP-positive cells were measured by FACS. (G) Hepcidin treatment reverses the FPN1-induced increase of TfR1 in K562 cells. K562 cells were transiently transfected with pIRES2EGFP-FPN1 and incubated with or without 1 μg/mL hepcidin for 24 hours. The expression levels of TfR1 were measured by FACS analysis in GFP-positive cells. All of the experiments (A-G) were independently repeated at least twice with triplicate samples assessed at each time point. a.u. indicates arbitrary unit. **P < .01, ***P < .001.
FPN1 expression is regulated by hepcidin in transfected erythroblast cell lines in a dose- and time-dependent manner as indicated by FACS analysis. (A) Validation of the use of FACS analysis to measure iron-dependent TfR1 expression. K562 cells were treated with 100μM FeNTA or 100μM DFO for 18 hours, and cells were then labeled by APC-labeled TfR1 Abs and checked by FACS analysis. (B) Expression of FPN1 increases TfR1 expression in K562 cells. K562 cells were transiently transfected with pEGFP-N1-FPN1 to express the fusion protein FPN1GFP for 24 hours, and the expression of TfR1 was then measured by FACS. Expression of FPN1 in K562 (C) and MEL (D) cells was down-regulated by hepcidin. Cells were transiently transfected with pEGFP-N1 or pEGFP-N1-FPN1 and incubated with 1 μg/mL hepcidin for 24 hours, and the GFP-positive cells were then measured by FACS. (E) Time-dependent regulation of FPN1 by hepcidin. K562 cells were transfected with pEGFP-N1-FPN1 or pEGFP-N1 vectors for 12 hours, and then incubated with or without 1 μg/mL hepcidin. The GFP-positive cells were measured by FACS at the indicated time points. (F) Dose-dependent regulation of FPN1 by hepcidin. K562 cells transfected with pEGFP-N1-FPN1 or pEGFP-N1 vectors were incubated with hepcidin of indicated concentrations for 24 hours. The GFP-positive cells were measured by FACS. (G) Hepcidin treatment reverses the FPN1-induced increase of TfR1 in K562 cells. K562 cells were transiently transfected with pIRES2EGFP-FPN1 and incubated with or without 1 μg/mL hepcidin for 24 hours. The expression levels of TfR1 were measured by FACS analysis in GFP-positive cells. All of the experiments (A-G) were independently repeated at least twice with triplicate samples assessed at each time point. a.u. indicates arbitrary unit. **P < .01, ***P < .001.
The regulation of hepcidin on FPN1 levels and intracellular iron status was also investigated in K562 cells with Western blot analyses (Figure 3). Overexpression of the FPN1GFP fusion protein in K562 for 24 hours increased IRP2 and TfR1 protein expression, whereas treatment with 1 μg/mL hepcidin decreased FPN1 protein levels (confirmed by both anti-GFP and anti-FPN1 Abs) and restored IRP2 and TfR1 protein levels to levels comparable to those in cells expressing GFP only (Figure 3A). Because IRP1 is a bifunctional enzyme whose activity converts between IRP and cytosolic aconitase in response to changes in intracellular iron concentrations, its protein levels did not change, as expected. Hepcidin induced the degradation of the FPN1GFP fusion protein in a time-dependent manner, and IRP2 and TfR1 protein levels decreased markedly at the 12- and 24-hour time points (Figure 3B). Hepcidin also induced the degradation of FPN1 expression in a dose-dependent manner; treatment with 50 ng/mL hepcidin for 24 hours significantly decreased FPN1 expression, and FPN1 protein decreased further with higher hepcidin levels (≤ 2 μg/mL), with IRP2 and TfR1 protein levels decreasing concomitantly (Figure 3C). Consistent with Figure 2, these results implied that FPN1 exported intracellular iron in erythroblast cell lines, whereas hepcidin treatment prevented FPN1 from depleting intracellular iron and restored intracellular iron homeostasis.
FPN1 protein expression levels in transfected erythroblast cell lines are directly regulated by hepcidin in a dose- and time-dependent manner. (A) Hepcidin regulates FPN1 expression and intracellular iron status of K562 cells. K562 cells transiently transfected with pEGFP-N1 or pEGFP-N1-FPN1 plasmids to generated cells that expressed GFP alone, or the FPNGFP fusion proteins, and were incubated with or without 1 μg/mL hepcidin for 24 hours. (B) Time-dependent regulation of FPN1 by hepcidin in K562 cells. K562 cells were transfected with pEGFP-N1-FPN1 for 12 hours, then incubated with 1 μg/mL hepcidin at the indicated time points. (C) Dose-dependent regulation of FPN1 by hepcidin. K562 cells were transfected with pEGFP-N1-FPN1 and incubated with hepcidin at the indicated concentrations for 24 hours. Total cell lysates were separated with 4%-20% Tris-glycine gels and then were transferred to nitrocellulose membranes. The membranes were then incubated with Abs for GFP, FPN1, TfR1, IRP2, IRP1, and α-tubulin, respectively. All of the experiments were repeated at least twice.
FPN1 protein expression levels in transfected erythroblast cell lines are directly regulated by hepcidin in a dose- and time-dependent manner. (A) Hepcidin regulates FPN1 expression and intracellular iron status of K562 cells. K562 cells transiently transfected with pEGFP-N1 or pEGFP-N1-FPN1 plasmids to generated cells that expressed GFP alone, or the FPNGFP fusion proteins, and were incubated with or without 1 μg/mL hepcidin for 24 hours. (B) Time-dependent regulation of FPN1 by hepcidin in K562 cells. K562 cells were transfected with pEGFP-N1-FPN1 for 12 hours, then incubated with 1 μg/mL hepcidin at the indicated time points. (C) Dose-dependent regulation of FPN1 by hepcidin. K562 cells were transfected with pEGFP-N1-FPN1 and incubated with hepcidin at the indicated concentrations for 24 hours. Total cell lysates were separated with 4%-20% Tris-glycine gels and then were transferred to nitrocellulose membranes. The membranes were then incubated with Abs for GFP, FPN1, TfR1, IRP2, IRP1, and α-tubulin, respectively. All of the experiments were repeated at least twice.
Hepcidin regulates FPN1 expression of primary erythroblasts
The regulation of FPN1 was further investigated in fetal liver erythroblasts, which can be induced to differentiate by changing the medium from proliferation medium to differentiation medium.20 After induction for 48 hours in differentiation medium, expression of the stem cell factor receptor c-kit decreased, whereas the expression of erythroid marker Ter119 and TfR1 increased (Figure 4A). The hemoglobin mRNA levels significantly increased over a 3-day induction period, and the TfR1 mRNA levels initially increased and reached a peak at ∼ 36-48 hours before decreasing (Figure 4B), whereas the size of erythroblasts decreased and hemoglobin protein levels increased (supplemental Figure 2), suggesting that erythroblasts were successfully induced to differentiate. During the differentiation process, expression of the FPN1B transcript decreased significantly over time to reach a baseline level of expression at ∼ 48 hours after differentiation (Figure 4C). In contrast, expression of the FPN1A transcript was quite low in the early stages of differentiation but increased after 60 hours, and the percentage contribution of FPN1B to total FPN1 mRNA levels decreased from ∼ 80% to ∼ 10% as differentiation progressed. The pattern of FPN1B and FPN1A expression in the differentiation of primary erythroblasts was consistent with the pattern that we previously observed in the differentiation of the erythroblast cell line, G1E cells.18 Meanwhile, FPN1 protein levels decreased during differentiation, paralleling decreased expression levels of the FPN1B transcript (Figure 4D), and supporting that FPN1B is the main transcript that encodes FPN1 protein in developing erythroblasts. TfR1 protein expression peaked at 48-60 hours and then decreased during the differentiation process, consistent with the results shown in Figure 4B and the known role of TfR1 in iron uptake during differentiation of erythroblasts.
Distinctive expression patterns of the 2 FPN1 transcripts during differentiation of fetal liver erythroblasts. (A) Expression of c-Kit, TER119, and TfR1 in the process of differentiation of erythroblasts. After being cultured in either proliferation or differentiation medium for 48 hours, erythroblasts were incubated with PE-labeled anti–c-Kit, PE Cy7-labeled anti-TER119, or FITC-labeled anti-TfR1 Abs (BD PharMingen), followed by FACS analysis. (B) mRNA expression of hemoglobin adult α-chain (HBA) and TfR1 during the differentiation of erythroblasts. (C) mRNA expression of FPN1A, FPN1B, and FPN1 during the differentiation of erythroblasts. (D) Protein expression of FPN1, TfR1, and α-tubulin during the differentiation of erythroblasts. At different time points after being cultured in differentiation medium, erythroblasts were harvested, and total RNA or protein was prepared for real-time qPCR (B-C) or Western blot (D) analysis. The real-time qPCR results were normalized with actin mRNA. The real-time qPCR experiments were repeated 3 times with triplicate samples each time; the Western blot analyses were repeated at least twice; a.u. indicates arbitrary units.
Distinctive expression patterns of the 2 FPN1 transcripts during differentiation of fetal liver erythroblasts. (A) Expression of c-Kit, TER119, and TfR1 in the process of differentiation of erythroblasts. After being cultured in either proliferation or differentiation medium for 48 hours, erythroblasts were incubated with PE-labeled anti–c-Kit, PE Cy7-labeled anti-TER119, or FITC-labeled anti-TfR1 Abs (BD PharMingen), followed by FACS analysis. (B) mRNA expression of hemoglobin adult α-chain (HBA) and TfR1 during the differentiation of erythroblasts. (C) mRNA expression of FPN1A, FPN1B, and FPN1 during the differentiation of erythroblasts. (D) Protein expression of FPN1, TfR1, and α-tubulin during the differentiation of erythroblasts. At different time points after being cultured in differentiation medium, erythroblasts were harvested, and total RNA or protein was prepared for real-time qPCR (B-C) or Western blot (D) analysis. The real-time qPCR results were normalized with actin mRNA. The real-time qPCR experiments were repeated 3 times with triplicate samples each time; the Western blot analyses were repeated at least twice; a.u. indicates arbitrary units.
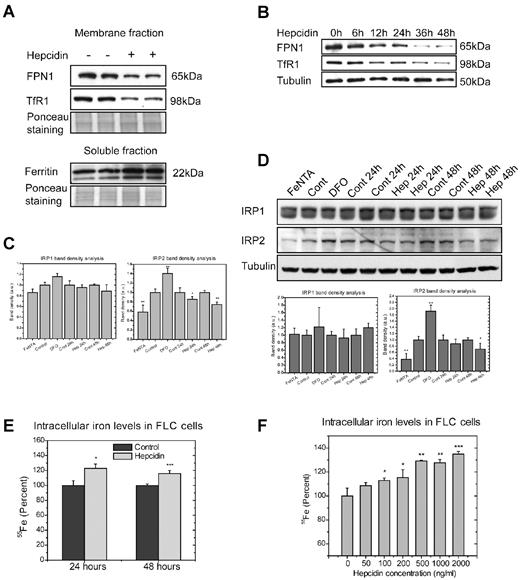
In the primary erythroblasts cultured in proliferation medium, endogenous FPN1 protein levels decreased on treatment with 1 μg/mL hepcidin for 24 hours (Figure 5A), and the TfR1 levels decreased simultaneously, whereas ferritin levels increased, suggesting that hepcidin treatment increased intracellular iron availability by blocking iron export through FPN1. The effect of hepcidin on FPN1 expression was studied at different time points during growth in proliferation medium. FPN1 protein expression decreased slightly after 6 hours of incubation with hepcidin, and it gradually decreased further before reaching a stable baseline after 36 hours. TfR1 expression decreased in parallel (Figure 5B). To confirm the consequences of hepcidin treatment on the intracellular iron concentration of erythroblasts, we measured the activity of IRPs with RNA mobility shift assays. When iron concentrations decreased in samples treated with the iron chelator DFO, IRP2 activity significantly increased, and IRP1 activity increased slightly (Figure 5C); however, treatment with hepcidin markedly reduced IRP2 activity on the gel-shift assay. The protein levels of IRPs were checked with Western blot analyses as well, and IRP2 protein levels were increased by DFO treatment and decreased by hepcidin treatment (Figure 5D), whereas IRP1 protein levels did not change in response to iron or hepcidin treatments. The IRP2 activity and protein levels decreased in response to hepcidin treatment, suggesting that hepcidin treatment increases intracellular iron availability. In addition, the intracellular iron availability of erythroblasts treated by hepcidin was measured by an iron-55 retention experiment (Figure 5E). Treatment with 1 μg/mL hepcidin for 24 and 48 hours significantly increased intracellular 55Fe contents, confirming that a hepcidin-mediated decrease in FPN1 expression led to increased intracellular iron concentrations (Figure 5E). Consistent with the FPN1 levels in Figures 2F and 3C, the intracellular 55Fe activity increased in a hepcidin dose-dependent manner; 100 ng/mL hepcidin significantly increased the intracellular 55Fe concentration, and 2 μg/mL hepcidin increased the 55Fe concentration > 30% (Figure 5F), suggesting that physiologically relevant levels of hepcidin (50 ng/mL to 2 μg/mL) can block iron export by FPN1 and increase intracellular iron concentrations of erythroblasts.22
Hepcidin regulates endogenous FPN1 expression and intracellular iron levels of fetal liver erythroblasts. (A) Expression of endogenous FPN1 in erythroblasts was decreased after a 24-hour hepcidin treatment. Erythroblasts in proliferation medium were incubated with 1 μg/mL hepcidin for 24 hours, and protein expression was then measured by Western blot analysis for FPN1 and TfR1 in membrane fractions of sample lysates and for L-ferritin in soluble fractions of sample lysates. Ponceau S staining of a specific portion of the gel was used as a loading control. (B) Expression of FPN1 at different time points of incubation with hepcidin during growth in proliferation medium. (C) IRP2 activity was decreased by hepcidin in erythroblasts. Erythroblasts were incubated with 1 μg/mL hepcidin for 24 or 48 hours or with 100μM FeNTA or DFO for 18 hours, and samples were then prepared with or without β-mercaptoethanol to measure IRP activity by electrophoresis mobility shift assay. The density of the bands was analyzed with ImageJ (http://imagej.nih.gov/ij/) and normalized to IRP1 activity after β-mercaptoethanol treatment and statistical comparisons were performed. (D) Western blot analyses showed decreased IRP2 protein levels in samples treated by hepcidin. The cells were treated as described in panel C. (E) Hepcidin increases intracellular iron contents of erythroblasts. Erythroblasts in 1 well of a 6-well plate in proliferation medium were loaded with 55Fe by incubation with 5 μCi 55Fe (0.185 Bq; 0.2 μg) for 2 hours and were then washed twice and incubated with 1 μg/mL hepcidin for 24 or 48 hours, after which time intracellular 55Fe levels were measured with a Liquid Scintillation Counter. (F) Hepcidin increases intracellular iron levels in a dose-dependent manner. Erythroblasts in proliferation medium were loaded with 55Fe as described in panel E and were then incubated with hepcidin of indicated concentrations for 24 hours, after which time intracellular 55Fe levels were measured with Liquid Scintillation Counter. a.u. indicates arbitrary unit. Experiments in panels A through D were repeated ≥ 3 times, and experiments in E and F were repeated twice with triplicate samples for each time point. *P < .05, **P < .01, and ***P < .001.
Hepcidin regulates endogenous FPN1 expression and intracellular iron levels of fetal liver erythroblasts. (A) Expression of endogenous FPN1 in erythroblasts was decreased after a 24-hour hepcidin treatment. Erythroblasts in proliferation medium were incubated with 1 μg/mL hepcidin for 24 hours, and protein expression was then measured by Western blot analysis for FPN1 and TfR1 in membrane fractions of sample lysates and for L-ferritin in soluble fractions of sample lysates. Ponceau S staining of a specific portion of the gel was used as a loading control. (B) Expression of FPN1 at different time points of incubation with hepcidin during growth in proliferation medium. (C) IRP2 activity was decreased by hepcidin in erythroblasts. Erythroblasts were incubated with 1 μg/mL hepcidin for 24 or 48 hours or with 100μM FeNTA or DFO for 18 hours, and samples were then prepared with or without β-mercaptoethanol to measure IRP activity by electrophoresis mobility shift assay. The density of the bands was analyzed with ImageJ (http://imagej.nih.gov/ij/) and normalized to IRP1 activity after β-mercaptoethanol treatment and statistical comparisons were performed. (D) Western blot analyses showed decreased IRP2 protein levels in samples treated by hepcidin. The cells were treated as described in panel C. (E) Hepcidin increases intracellular iron contents of erythroblasts. Erythroblasts in 1 well of a 6-well plate in proliferation medium were loaded with 55Fe by incubation with 5 μCi 55Fe (0.185 Bq; 0.2 μg) for 2 hours and were then washed twice and incubated with 1 μg/mL hepcidin for 24 or 48 hours, after which time intracellular 55Fe levels were measured with a Liquid Scintillation Counter. (F) Hepcidin increases intracellular iron levels in a dose-dependent manner. Erythroblasts in proliferation medium were loaded with 55Fe as described in panel E and were then incubated with hepcidin of indicated concentrations for 24 hours, after which time intracellular 55Fe levels were measured with Liquid Scintillation Counter. a.u. indicates arbitrary unit. Experiments in panels A through D were repeated ≥ 3 times, and experiments in E and F were repeated twice with triplicate samples for each time point. *P < .05, **P < .01, and ***P < .001.
Hepcidin regulates FPN1 expression of erythroblasts in vivo
To study the effect of hepcidin on FPN1 expression in erythroblasts in vivo, we attempted to decrease endogenous levels of hepcidin by maintaining the mice on a low-iron diet. Because hepcidin is mainly regulated at the transcriptional level, the hepcidin mRNA levels in liver were measured with real-time qPCR. The hepcidin mRNA levels in liver tissues from mice maintained on the low-iron diet were decreased to barely detectable levels compared with mice on the control diet (Figure 6A). The L-ferritin levels in Ter119-positive erythroblasts of BM (Figure 6B) and in spleen (Figure 6C) were significantly lower in mice on the low-iron diet, and TfR1 expression in the spleen of mice on the low-iron diet increased (Figure 6C), confirming that iron diet treatments were successful in inducing systemic iron deficiency. FPN1 protein levels significantly increased in erythroblasts of mice on the low-iron diet compared with mice on the control diet (Figure 6B), and the increased FPN1 expression in erythroblasts was consistent with the low hepcidin levels, suggesting that hepcidin induced degradation of FPN1 in erythroblasts in vivo. FPN1 protein expression was also increased in the spleens of mice maintained on the low-iron diet (Figure 6C). In primary erythroblasts, we also checked the effect of manipulating iron status on FPN1 protein levels (Figure 6D), but we did not find significant iron-dependent changes, suggesting that FPN1 expression is not regulated by intracellular iron status in erythroblasts, whereas TfR1 was still clearly regulated by iron levels.
Expression of FPN1 in erythroblasts of mouse BM increased in mice maintained on a low-iron diet. (A) Expression of hepcidin mRNA was decreased to barely detectable levels in the livers of mice on the low-iron diet. Total RNA was prepared, and hepcidin expression was measured by real-time qPCR and normalized to actin mRNA levels. a.u. indicates arbitrary unit. (B) FPN1 expression increased in Ter119-positive erythroblasts from the BM of mice maintained on a low-iron diet compared with mice on the control diet. Expression of FPN1 and L-ferritin was measured by Western blot analyses in total cell lysates. A distinctive band in Ponceau S staining was used as a loading control. (C) FPN1 expression increased in the spleens of mice maintained on the low-iron diet. Expression levels of FPN1, TfR1, L-ferritin, and α-tubulin were measured by Western blot analyses. (D) FPN1 expression, unlike TfR1, is not regulated by iron status in cultured primary erythroblasts. Fetal liver erythroblasts were cultured in proliferation medium and incubated with 100μM FeNTA or DFO for 18 hours. Then the membrane fractions were prepared, and the expression levels of FPN1, TfR1, and α-tubulin was measured by Western blot analyses. The experiments in panels A through C were repeated in 3 animals in each group and panel D was repeated twice.
Expression of FPN1 in erythroblasts of mouse BM increased in mice maintained on a low-iron diet. (A) Expression of hepcidin mRNA was decreased to barely detectable levels in the livers of mice on the low-iron diet. Total RNA was prepared, and hepcidin expression was measured by real-time qPCR and normalized to actin mRNA levels. a.u. indicates arbitrary unit. (B) FPN1 expression increased in Ter119-positive erythroblasts from the BM of mice maintained on a low-iron diet compared with mice on the control diet. Expression of FPN1 and L-ferritin was measured by Western blot analyses in total cell lysates. A distinctive band in Ponceau S staining was used as a loading control. (C) FPN1 expression increased in the spleens of mice maintained on the low-iron diet. Expression levels of FPN1, TfR1, L-ferritin, and α-tubulin were measured by Western blot analyses. (D) FPN1 expression, unlike TfR1, is not regulated by iron status in cultured primary erythroblasts. Fetal liver erythroblasts were cultured in proliferation medium and incubated with 100μM FeNTA or DFO for 18 hours. Then the membrane fractions were prepared, and the expression levels of FPN1, TfR1, and α-tubulin was measured by Western blot analyses. The experiments in panels A through C were repeated in 3 animals in each group and panel D was repeated twice.
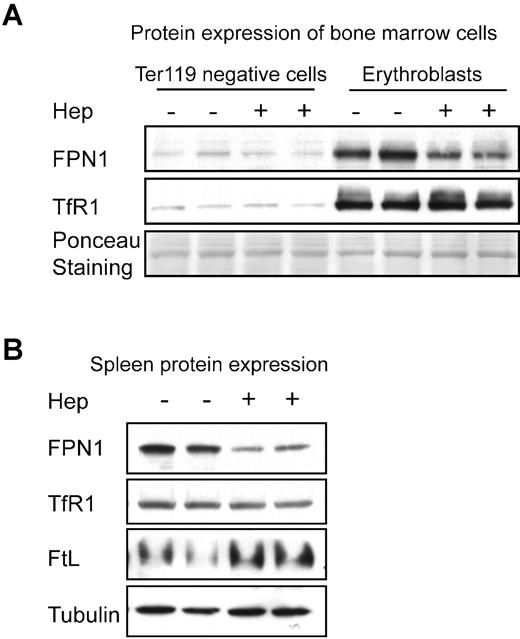
To exclude the possibility that increased FPN1 expression in erythroblasts of mice on the low-iron diet was caused by factors other than from decreased hepcidin-mediated degradation, we injected exogenous hepcidin into mice to check the in vivo effects of hepcidin on FPN1 expression in erythroblasts. Hepcidin injection significantly decreased FPN1 expression in erythroblasts (Figure 7A). Hepcidin treatment significantly decreased FPN1 expression in the spleen as well (Figure 7B), whereas L-ferritin expression increased with hepcidin treatment, suggesting that hepcidin increased intracellular iron availability in vivo (although TfR1 expression did not significantly change on hepcidin treatment in both spleen and erythroblasts). There was little effect of hepcidin injection on FPN1 protein levels in either spleen or erythroblasts when hepcidin was injected into mice maintained on a control diet (supplemental Figure 3), presumably because endogenous hepcidin levels were already high in those mice (Figure 6A).
Expression levels of FPN1 in erythroblasts of mouse BM and in mouse spleen were decreased by hepcidin injection. (A) Expression levels of FPN1 and TfR1 in Ter119-positive erythroblasts or the Ter119-negative cells from BM were compared by Western blot analyses after injection of either hepcidin or PBS for 24 hours. A band from Ponceau S staining of the gel was used as a loading control. (B) Hepcidin injections decreased FPN1 expression in mouse spleen. Expression levels of FPN1, TfR1, L-ferritin, and α-tubulin were measured by Western blot analyses. The experiments were performed in mice maintained on a low-iron diet as described in “Methods.” Experiments in panels A and B were repeated in 3 animals in each group.
Expression levels of FPN1 in erythroblasts of mouse BM and in mouse spleen were decreased by hepcidin injection. (A) Expression levels of FPN1 and TfR1 in Ter119-positive erythroblasts or the Ter119-negative cells from BM were compared by Western blot analyses after injection of either hepcidin or PBS for 24 hours. A band from Ponceau S staining of the gel was used as a loading control. (B) Hepcidin injections decreased FPN1 expression in mouse spleen. Expression levels of FPN1, TfR1, L-ferritin, and α-tubulin were measured by Western blot analyses. The experiments were performed in mice maintained on a low-iron diet as described in “Methods.” Experiments in panels A and B were repeated in 3 animals in each group.
Discussion
Subcellular localization on the plasma membrane is essential for the iron export function of FPN1, and some mutations of FPN1 that lead to sequestration within intracellular vesicles cause a specific type of hemochromatosis.23,24 The endogenous FPN1 in primary human erythroblasts derived from CD34+ cells was previously reported to be localized in cytosol,19 raising the possibility that FPN1 in erythroblasts might not have the capacity to export iron and to be regulated by hepcidin. Here, our results with both transiently transfected FPN1 in erythroblast cell lines and endogenous FPN1 in primary fetal liver erythroblasts showed that FPN1 trafficks to the plasma membrane, the subcellular location required for FPN1 to export iron and to be regulated by hepcidin. Furthermore, our results in both erythroblast cell lines and primary erythroblasts showed that hepcidin treatment within a physiologic concentration range (50-2000 ng/mL) decreased FPN1 expression (Figures 2,3,5) and increased intracellular iron availability, consistent with the finding that FPN1 localizes on the plasma membrane of erythroblasts.22
There are 2 FPN1 transcripts, including FPN1A, which is ubiquitously expressed and is subject to translational repression by the IRP/IRE machinery, and FPN1B, which is specifically expressed in enterocytes and erythroblasts and is not regulated by IRPs.18 Consistent with our previous results in the erythroblast cell line, G1E cells, FPN1B is the main transcript in cultured primary erythroblasts (Figure 2), and its levels decreased during the differentiation of erythroblasts, whereas expression of the FPN1A transcript increased during differentiation. Levels of FPN1 protein decreased and correlated with FPN1B transcript levels during differentiation, confirming that FPN1 protein is mainly generated from FPN1B in developing erythroblasts.18 In cell types such as macrophages and hepatocytes, FPN1 protein levels are positively regulated by intracellular iron concentrations.17 In contrast, FPN1 protein expression remains constant in erythroblasts after incubation with FeNTA or DFO for 18 hours, consistent with the observations that FPN1B (the FPN1 transcript without a 5′-IRE) is the main FPN1 transcript in erythroblasts and suggesting that expression of FPN1 protein in developing erythroblasts is regulated mainly at the systemic level by hepcidin.
Because erythropoiesis is the main iron-consuming process in vertebrates, it is not surprising that expression of hepcidin is regulated by erythropoiesis, and erythropoietic signals can override some other hepcidin-regulatory signals such as iron overload or inflammation.2 Erythropoiesis induced by phlebotomy or hypoxia has been shown to repress hepcidin expression, which allows more iron to flow into the circulating transferrin pool to support red blood cell production.25 Moreover, erythropoiesis can inhibit both inflammatory and iron-sensing pathways, probably by the suppression of the STAT3 and SMAD4 pathways in vivo, to repress hepcidin transcription.26 In β-thalassemia, anemia caused by reduced expression of β-globin chains results in suppression of hepcidin synthesis despite severe iron overload.13 The exact mechanism by which erythropoiesis regulates hepcidin expression is not known yet, but in β-thalassemia it might be through GDF15 and TWSG1, 2 molecules of the TGF-β superfamily, which are remarkably up-regulated in erythroblasts of patients with β-thalassemia.27,28 In any case, the mechanism by which erythropoiesis regulates hepcidin expression remains to be fully elucidated.
Here, we have shown that hepcidin regulates FPN1 expression and intracellular iron homeostasis of erythroblasts, raising the possibility that hepcidin may be directly involved in regulation of erythropoiesis. The involvement of FPN1 and hepcidin in regulation of erythropoiesis was observed in several previous studies. In polycythemic mice, a 58-bp microdeletion in the FPN1A promoter alters the canonical transcriptional start sites and eliminates the IRE in the 5′-UTR of the FPN1A transcript, resulting in increased FPN1 protein expression in some tissues and leading to polycythemia in heterozygous mice and microcytic anemia in young homozygous mice.29,30 Both the polycythemia of heterozygous mice and the microcytic anemia of homozygous mice improved spontaneously in adults, and the improvement correlated with increased hepcidin expression, which supports the possible role of hepcidin-FPN1 interactions in the regulation of erythropoiesis. In another study, hepcidin treatment reduced erythroid colony formation when cultures were performed in medium that contained low erythropoietin concentrations, providing direct evidence for hepcidin involvement in erythropoiesis.31 Anemia of chronic diseases (ACD) is a common form of anemia associated with chronic inflammatory disorders which is characterized by high hepcidin expression.32 Hepcidin is strikingly high in patients with ACD, probably because inflammatory factors, including IL-6 levels, increase hepcidin transcription. Hepcidin subsequently blocks iron absorption in the duodenum and iron recycling in reticuloendothelial macrophages, leading to systemic iron deficiency. In contrast to iron deficiency anemia (IDA) in which patients have low iron saturations (< 10%) and low ferritin levels (< 30 ng/mL) in serum, patients with ACD have low iron saturations (< 15%) but high serum ferritin levels (> 200 ng/mL, perhaps because macrophages, the main source of serum ferritin, are iron overloaded33 ). It has been a mystery that the mean corpuscular volume of patients with ACD is mostly normal compared with patients with IDA. Considering that both patients with ACD and with IDA have limited iron and one of the biggest distinctions between ACD and IDA is the higher hepcidin levels in ACD, the nearly normal mean corpuscular volume of patients with ACD suggests that the interaction of hepcidin with FPN1 in erythroblasts may increase erythroid iron stores at an important developmental stage.34
The discovery of FPN1B expression and hepcidin binding in erythroblasts may have important physiologic implications. Because FPN1 is blocked by hepcidin, FPN1 in erythroblasts is active only in iron-deficient conditions when hepcidin is low. FPN1B expression in erythroblasts probably represents a fine-tuning mechanism to prevent misallocation of iron. In iron-replete conditions, down-regulation of FPN1 ensures that erythroblasts retain iron to produce hemoglobin, helping to prevent complete saturation of circulating transferrin. Conversely, in iron-deficient conditions, erythroblasts allow other tissues to take priority by exporting their iron, similar to a process that probably occurs in duodenal enterocytes, and thereby helping other tissues avoid severe iron depletion. Considering that erythroblasts have efficient iron uptake machinery (TfR1 is highly expressed on erythroblasts and > 80% of total TfR1 in vivo is expressed on erythroblasts) and only ∼ 1% of red blood cells in humans need to be replaced daily, it is probably better for the organism to decrease erythropoietic activity during a period of iron deficiency than to risk developing iron deficiency in vital organs such as the brain and heart. The connection between hepcidin and ferroportin expression on erythroblasts probably explains why anemia is usually the first prominent symptom of iron deficiency. Although hepcidin levels are extremely low in hemochromatosis, the blood cell indices are normal in mouse models of hemochromatosis, and the mean corpuscular volume in patients with hemochromatosis is slightly higher than in control groups, most probably because high transferrin saturations can support high iron influx into erythroblasts, enabling them to override the export capacity of FPN1.35,36
In conclusion, the results of our study show that the iron-regulatory hormone hepcidin can regulate FPN1 expression of the erythroid precursors at a point in development when FPN1 expression is not yet regulated by the IRE/IRP system. FPN1 expression in erythroblasts may represent a fine-tuning mechanism that maintains systemic iron homeostasis and ensures that erythroblasts titrate the amount of iron they retain according to organismal iron status. We predict that mice with a conditional deletion of FPN1 in erythroblasts will not be able to maintain systemic iron homeostasis; in iron-deficient conditions, the animals may maintain normal hematocrit values or develop polycythemia, even while they are experiencing iron starvation in nonerythropoietic tissues. Thus, it may be possible to address the physiologic significance of FPN1 and hepcidin binding in erythropoiesis with the FPN1 conditional knockout mice.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Ivana De Domenico for providing them with the pEGFP-N1-FPN1 plasmid and Dr Jeffery Miller for constructive comments on the paper.
This work was supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
National Institutes of Health
Authorship
Contribution: D.-L.Z. designed and performed experiments, analyzed data, and wrote the paper; T.S., H.O.-W., and T.T. performed experiments; M.C.G. performed experiments and analyzed data; and T.A.R. oversaw the study, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tracey A. Rouault, Molecular Medicine Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD 20892; e-mail: rouault@mail.nih.gov.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal