Abstract
Gene expression profiling (GEP) of purified plasma cells 48 hours after thalidomide and dexamethasone test doses showed these agents' mechanisms of action and provided prognostic information for untreated myeloma patients on Total Therapy 2 (TT2). Bortezomib was added in Total Therapy 3 (TT3), and 48 hours after bortezomib GEP analysis identified 80 highly survival-discriminatory genes in a training set of 142 TT3A patients that were validated in 128 patients receiving TT3B. The 80-gene GEP model (GEP80) also distinguished outcomes when applied at baseline in both TT3 and TT2 protocols. In context of our validated 70-gene model (GEP70), the GEP80 model identified 9% of patients with a grave prognosis among those with GEP70-defined low-risk disease and 41% of patients with favorable prognosis among those with GEP70-defined high-risk disease. PMSD4 was 1 of 3 genes common to both models. Residing on chromosome 1q21, PSMD4 expression is highly sensitive to copy number. Both higher PSMD4 expression levels and higher 1q21 copy numbers affected clinical outcome adversely. GEP80 baseline-defined high risk, high lactate dehydrogenase, and low albumin were the only independent adverse variables surviving multivariate survival model. We are investigating whether second-generation proteasome inhibitors (eg, carfilzomib) can overcome resistance associated with high PSMD4 levels.
Introduction
Much of the recent progress in the treatment of multiple myeloma has been attributed to the introduction of several novel agents which, when combined with standard cytotoxic agents and each other, have imparted a high frequency of clinical responses.1-4 We have previously reported on the superior survival outcomes in Total Therapy 3A (TT3A), with added bortezomib (Bz),5 compared with Total Therapy 2 (TT2), which randomly assigned patients to a control arm or an experimental thalidomide arm of a multiple agent chemotherapy and tandem autograft-supported high-dose melphalan program.6 The major advance with TT3A was observed in the 85% of patients presenting with gene expression profiling (GEP)–defined low-risk myeloma7,8 and, considering molecular subgroup designations,9 in the 13% exhibiting MMSET/FGFR3 translocations t(4;14) (the MS subgroup).10 As part of TT3A, pharmacogenomic investigations of Bz, using GEP analysis before and after 48-hour test-dosing (1.0 mg/m2), were performed in an attempt to further delineate low- versus high-risk disease. Strong leads obtained in 142 of 303 patients receiving TT3A motivated the conduct of a successor trial, Total Therapy 3B (TT3B), for validation purposes. The clinical results of both trials have been recently updated.10 Herein, we report on the Bz-associated gene expression alterations observed in the TT3 trials toward unraveling the mechanisms of action of this first-in-class proteasome inhibitor, the addition of which profoundly improved the outcome of patients with low-risk myeloma as defined by a previously published and validated 70-gene GEP model (GEP70).7
Methods
Patient population
All patients met the criteria of symptomatic or progressive multiple myeloma according to International Myeloma Working Group 2006 criteria.11 Standard prognostic parameters included β2-microglobulin, albumin, C-reactive protein, lactate dehydrogenase (LDH), M-protein quantifications in serum and urine for response assessment, BM examinations with metaphase karyotyping to determine the presence of cytogenetic abnormalities, and GEP.
Characteristics of patients with GEP data both at baseline and after Bz in the 2 TT3 trials were comparable, except for overrepresentation of low albumin and CD-1 and CD-2 subgroup classifications in patients in the TT3B trial (Table 1). Despite these less favorable features, clinical outcomes were comparable to those of patients enrolled in TT3A, whether examined among all 177 and 303 patients, respectively,7 or in the subset of 128 and 142 patients with GEP data both at baseline and after Bz.
Baseline characteristics of patients enrolled on TT3A (protocol UARK2003-33) and TT3B (protocol UARK2006-66)
| Variable . | TT3A (2003-33) . | TT3B (2006-66) . | P* . |
|---|---|---|---|
| Age ≥ 65 y | 36/142 (25) | 35/128 (27) | .711 |
| Albumin level < 3.5 g/dL | 31/142 (22) | 58/127 (46) | < .001 |
| B2M level ≥ 3.5 mg/L | 63/142 (44) | 70/125 (56) | .058 |
| ISS Stage I | 66/142 (46) | 37/125 (30) | .005 |
| ISS Stage II | 43/142 (30) | 53/125 (42) | .039 |
| ISS Stage III | 33/142 (23) | 35/125 (28) | .373 |
| Creatinine level ≥ 2.0 mg/dL | 13/142 (9) | 9/127 (7) | .537 |
| CRP level ≥ 8 mg/L | 54/142 (38) | 44/127 (35) | .565 |
| Hb level < 10 g/dL | 40/142 (28) | 39/127 (31) | .648 |
| LDH level ≥ 190 U/L | 38/142 (27) | 32/127 (25) | .770 |
| Cytogenetic abnormalities | 55/141 (39) | 49/128 (39) | .984 |
| Baseline 70-gene high risk | 22/142 (15) | 27/128 (21) | .233 |
| GEP CD-1 subgroup | 3/142 (2) | 13/128 (10) | .005 |
| GEP CD-2 subgroup | 11/142 (8) | 21/128 (16) | .028 |
| GEP HY subgroup | 40/142 (28) | 34/128 (27) | .768 |
| GEP LB subgroup | 18/142 (13) | 10/128 (8) | .191 |
| GEP MF subgroup | 10/142 (7) | 8/128 (6) | .794 |
| GEP MS subgroup | 13/142 (9) | 20/128 (16) | .105 |
| GEP PR subgroup | 15/142 (11) | 19/128 (15) | .290 |
| GEP Proliferation Index ≥ 10 | 20/142 (14) | 14/128 (11) | .436 |
| GEP TP53 deletion | 17/142 (12) | 12/128 (9) | .491 |
| Variable . | TT3A (2003-33) . | TT3B (2006-66) . | P* . |
|---|---|---|---|
| Age ≥ 65 y | 36/142 (25) | 35/128 (27) | .711 |
| Albumin level < 3.5 g/dL | 31/142 (22) | 58/127 (46) | < .001 |
| B2M level ≥ 3.5 mg/L | 63/142 (44) | 70/125 (56) | .058 |
| ISS Stage I | 66/142 (46) | 37/125 (30) | .005 |
| ISS Stage II | 43/142 (30) | 53/125 (42) | .039 |
| ISS Stage III | 33/142 (23) | 35/125 (28) | .373 |
| Creatinine level ≥ 2.0 mg/dL | 13/142 (9) | 9/127 (7) | .537 |
| CRP level ≥ 8 mg/L | 54/142 (38) | 44/127 (35) | .565 |
| Hb level < 10 g/dL | 40/142 (28) | 39/127 (31) | .648 |
| LDH level ≥ 190 U/L | 38/142 (27) | 32/127 (25) | .770 |
| Cytogenetic abnormalities | 55/141 (39) | 49/128 (39) | .984 |
| Baseline 70-gene high risk | 22/142 (15) | 27/128 (21) | .233 |
| GEP CD-1 subgroup | 3/142 (2) | 13/128 (10) | .005 |
| GEP CD-2 subgroup | 11/142 (8) | 21/128 (16) | .028 |
| GEP HY subgroup | 40/142 (28) | 34/128 (27) | .768 |
| GEP LB subgroup | 18/142 (13) | 10/128 (8) | .191 |
| GEP MF subgroup | 10/142 (7) | 8/128 (6) | .794 |
| GEP MS subgroup | 13/142 (9) | 20/128 (16) | .105 |
| GEP PR subgroup | 15/142 (11) | 19/128 (15) | .290 |
| GEP Proliferation Index ≥ 10 | 20/142 (14) | 14/128 (11) | .436 |
| GEP TP53 deletion | 17/142 (12) | 12/128 (9) | .491 |
Values are expressed as number with Factor/number with valid data for factor (%).
ISS indicates International Staging System; and Hb, hemoglobin.
Determined with chi-square test.
Progression-free survival (PFS) and overall survival (OS) durations were measured from the time of initiation of protocol therapy; events included relapse or death from any cause in the former and death from any cause in the latter. OS and PFS curves were estimated by the product-limit method12 and were compared by the log-rank test.13 Univariate and multivariate regression analyses of covariates and time-to-event outcomes were performed by Cox regression.14
In keeping with institutional, federal, and Helsinki Declaration guidelines, all patients signed a written informed consent both for receiving protocol-based therapy and for undergoing repeated BM sampling for GEP analyses. Both protocols and their revisions had been reviewed and approved by the Institutional Review Board at the University of Arkansas for Medical Sciences, and annual progress reports on translational research investigations have been submitted. According to National Cancer Institute requirements, ∼ 80% of patient records have been audited by an external team of experienced independent auditors.
TT protocols
The details of the 2 TT regimens (TT3A, 2003-33; TT3B, 2006-66) have been published.5,10 Briefly, each protocol called for 2 cycles of induction therapy with VTD-PACE, comprising Bz (Velcade), thalidomide, and dexamethasone and 4-day continuous infusions of cisplatin, doxorubicin, cyclophosphamide, and etoposide, before melphalan-based high-dose therapy with autologous hematopoietic progenitor support, followed by 2 consolidation cycles of dose-attenuated VTD-PACE. Maintenance regimens differed in that TT3A called for VTD in year 1 and TD in years 2 and 3, whereas TT3B used VD and lenalidomide (Revlimid) instead of thalidomide for all 3 years. TT2 data were used for validation purposes.6,15
GEP and sample preparation
GEP sample procurement and processing, as well as analyses for the derivation of GEP70-based high-risk designation,7 molecular subgroup classification,9 and delTP53 assignment,16 have been reported previously. For Bz test-dosing, consenting patients received a bolus dose of Bz 1.0 mg/m2 intravenously and, 48 hours later, a further BM sample was obtained under local anesthesia from the posterior iliac crest. Plasma cell (PC) isolation from mononuclear cell fractions was performed by immunomagnetic bead selection with monoclonal mouse anti–human CD138 antibody with the use of the AutoMACS automated separation system (Miltenyi-Biotec). PC purity of > 95% homogeneity was confirmed by 2-color flow cytometry with the use of CD138+/CD45− criteria (Becton Dickinson), immunocytochemistry for cytoplasmic light-chain immunoglobulin, and morphology by Wright-Giemsa staining. RNA was extracted with the QIAGEN RNeasy kit. cDNA was prepared and biotinylated with the Affymetrix GeneChip HT 3′ IVT Express Kit. Samples were hybridized to an Affymetrix U133Plus2.0 microarray according to the manufacturer's recommendations and then read on a GeneChip Scanner 3000 (Affymetrix). The data files were deposited in the ArrayExpress archive (http://www.ebi.ac.uk/arrayexpress) under the accession number E-TABM-1138.
FISH analysis
Tricolor interphase FISH analysis was performed so that we could correlate GEP data with FISH-derived 1q21 copy number.17 BM aspirates were first subjected to Ficoll-Hypaque gradient-centrifugation separation to remove erythrocytes. The cells were immediately fixed in 100% ethanol and stored at −20°C. FISH probes were labeled by a nick translation reaction. Briefly, 1 μg of BAC DNA (RP11-180N18 for the AHCYL1 locus [1p13.3], RP11-498A2 for the CKS1B locus [1q21.3]) was mixed with 10μM fluorescent Green/Red-dUTP, 5μM dTTP, and 15μM each of dATP, dCTP, and dGTP in a nick translation buffer system including NT enzymes (Abbot Molecular Inc). After coprecipitation with 10 μg of human Cot-1 DNA (Invitrogen Corp), the labeled DNA probes were suspended in 100 μL of DNA in an in situ hybridization solution (Dako Corp). The 2 probes were mixed 1:1 before the hybridization. At least 100 tumor cells each were counted for the copies of 1p13.3 (green) and 1q21.3 (red). The threshold of significance abnormality (gain or loss) of each probe was set at ≥ 90%, as we previously described.17
Ingenuity pathway analysis
Data were analyzed with the Ingenuity Pathways Analysis online tool (Ingenuity Systems, www.ingenuity.com). All 80 genes from the GEP80 model were considered for analysis. Canonical pathways analysis identified the pathways from the Ingenuity Pathways Analysis library of canonical pathways that were most significant to the dataset. The significance of the association between the dataset and the canonical pathway was measured in 2 ways. First, a ratio was calculated by dividing the number of molecules from the dataset that mapped to the canonical pathway by the total number of molecules that mapped to the pathway. Second, the Fisher exact test was used to calculate a P value.
Results
Development and validation of the GEP80 model
Affymetrix U133Plus2.0 microarrays were preprocessed and normalized with the MAS5.0 method. With the use of TT3A protocol data as a training set (n = 142 patients with GEP data both at baseline and after Bz), we identified 1051 genes whose expression levels significantly changed 48 hours after the Bz test dose relative to baseline by applying a paired t test on each gene and a .005 P-value cutoff with a false discovery rate (q value) of 0.22.18 We then correlated each of these 1051 genes with PFS in a Cox regression model and ranked the genes by P values. To explore the various potential survival implications, 2 sets of covariates were considered in the Cox models for each gene: model a (percentage of change at 48 hours compared with baseline expression) and model b (48-hour expression).
Fitting model a on each of the 1051 genes produced a minimum q-value of 0.94,18 which means that, even if we selected only the most significant gene, the chance of it being a false positive was 94%. Fitting model b produced much smaller q-values than model a, with a minimum of 0.005, indicating that the 48-hour expression, compared with the percentage of change, was more strongly associated with PFS. Accordingly, we chose model b over model a and ranked genes by the P values of the 48-hour expression levels.
Once genes were ranked, PFS was predicted by computing a summary score on the basis of the top-ranked genes for each patient as described previously.7 The score for a particular patient was defined as the mean differential expression of prognosis-unfavorable genes and prognosis-favorable genes, whereby the latter were those associated with a hazard ratio (HR) < 1 (“good” genes) and the former with a HR ≥ 1 (“bad” genes). To produce a robust model, we used a 10-fold cross-validation approach (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) to simultaneously select the number of genes to calculate the gene score and an optimal cut point to dichotomize that score. In brief, we examined a range of top genes (from the top 5 to the top 150) to calculate a score and a range of percentiles (from the 50th to the 90th) to dichotomize that score. For each top gene set and percentile, a log-rank test was conducted comparing the predicted high-/low-risk groups by cross validation. The optimal score with the lowest log-rank P value used 80 genes to compute the score and dichotomized at the 82nd percentile. After cross validation, the 80 genes and 82nd percentile were applied to the entire training set to find the optimal cut point of 2.48. Patients having scores greater than the cut point were deemed to have high-risk disease and patients with scores below the cut point low-risk disease.
Once model selection was completed, we then examined the independent prognostic power of the post-Bz GEP80 [GEP80(PB)] in multivariate analysis of PFS and OS, along with other prognostic factors. This score was then applied to the test set of the TT3B protocol (n = 128 patients with GEP data both at baseline and after Bz, the latter of which had also been preprocessed and normalized with the Affymetrix MAS5.0 algorithm, followed by log2 transformation). The risk score was again calculated as the mean expression of the prognosis-unfavorable genes minus the mean expression of prognosis-favorable genes (Table 2), as with the training set. Each patient of the test set was then classified as having high-risk disease if the score was ≥ 2.48 and low-risk disease otherwise.
List of 80 genes altered 48 hours after Bz test-dosing and linked to PFS
| Probe set . | Gene symbol . | Chromosomal location . | No. probe sets on array . | P for PFS . | PFS HR . | Mean log2 expression change (all patients) . | Mean log2 expression change (GEP80 low-risk) . | Mean log2 expression change (GEP80 high-risk) . |
|---|---|---|---|---|---|---|---|---|
| Unfavorable genes | ||||||||
| 201754_at | COX6C | 8q22-q23 | 1 | .00027 | 4.4 | 0.12 | 0.11 | 0.15 |
| 219110_at | NOLA1 | 4q25 | 1 | .0014 | 3.4 | 0.11 | 0.07 | 0.32 |
| 201652_at | COPS5 | 8q13.2 | 1 | .00093 | 3.2 | 0.12 | 0.08 | 0.31 |
| 200642_at | SOD1 | 21q22.1 21q22.11 | 1 | .0014 | 3.1 | 0.15 | 0.14 | 0.21 |
| 209251_x_at | TUBA6 | 12q12-q14 | 2 | .0017 | 3.0 | 0.12 | 0.10 | 0.19 |
| 200014_s_at | HNRPC | 14q11.2 | 7 | .012 | 2.9 | 0.11 | 0.09 | 0.23 |
| 200039_s_at | PSMB2 | 1p34.2 | 4 | .0037 | 2.8 | 0.17 | 0.13 | 0.33 |
| 201252_at | PSMC4 | 19q13.11-q13.13 | 1 | .0082 | 2.7 | 0.16 | 0.11 | 0.37 |
| 226924_at | LOC400657 | 18q22.3 | 1 | .0043 | 2.6 | 0.11 | 0.10 | 0.16 |
| 225638_at | C1orf31 | 1q42.2 | 1 | .000 22 | 2.6 | 0.18 | 0.13 | 0.40 |
| 235346_at | FUNDC1 | Xp11.3 | 1 | .0037 | 2.6 | 0.13 | 0.10 | 0.22 |
| 211069_s_at | SUMO1 | 2q33 | 3 | .012 | 2.6 | 0.10 | 0.08 | 0.18 |
| 202244_at | PSMB4 | 1q21 | 3 | .016 | 2.5 | 0.15 | 0.14 | 0.23 |
| 201400_at | PSMB3 | 17q12 | 1 | .0081 | 2.5 | 0.13 | 0.09 | 0.30 |
| 202596_at | ENSA | 1q21.2 | 5 | .0091 | 2.4 | 0.12 | 0.09 | 0.26 |
| 202243_s_at | PSMB4 | 1q21 | 3 | .0098 | 2.4 | 0.19 | 0.17 | 0.28 |
| 218351_at | COMMD8 | 4p12 | 1 | .0044 | 2.4 | 0.16 | 0.15 | 0.19 |
| 223480_s_at | MRPL47 | 3q26.33 | 2 | .022 | 2.4 | 0.09 | 0.08 | 0.14 |
| 209503_s_at | PSMC5 | 17q23-q25 | 1 | .0062 | 2.3 | 0.17 | 0.11 | 0.45 |
| 212626_x_at | HNRPC | 14q11.2 | 7 | .019 | 2.3 | 0.16 | 0.14 | 0.27 |
| 203396_at | PSMA4 | 15q25.1 | 1 | .013 | 2.3 | 0.14 | 0.12 | 0.18 |
| 200882_s_at | PSMD4 | 1q21.2 | 4 | .00046 | 2.3 | 0.19 | 0.14 | 0.42 |
| 201157_s_at | NMT1 | 17q21.31 | 3 | .016 | 2.3 | 0.12 | 0.09 | 0.24 |
| 200786_at | PSMB7 | 9q34.11-q34.12 | 4 | .0083 | 2.3 | 0.15 | 0.11 | 0.34 |
| 209628_at | NXT2 | Xq22.3 | 2 | .000083 | 2.2 | 0.13 | 0.09 | 0.32 |
| 204587_at | SLC25A14 | Xq24 | 2 | .0049 | 2.2 | −0.10 | −0.12 | −0.03 |
| 210460_s_at | PSMD4 | 1q21.2 | 4 | .0012 | 2.2 | 0.19 | 0.14 | 0.44 |
| 200830_at | PSMD2 | 3q27.1 | 1 | .011 | 2.2 | 0.13 | 0.11 | 0.24 |
| 202690_s_at | SNRPD1 | 18q11.2 | 2 | .024 | 2.2 | 0.11 | 0.07 | 0.30 |
| 211609_x_at | PSMD4 | 1q21.2 | 4 | .0018 | 2.2 | 0.20 | 0.15 | 0.40 |
| 218566_s_at | CHORDC1 | 11q14.3 | 2 | .024 | 2.1 | 0.15 | 0.15 | 0.14 |
| 212296_at | PSMD14 | 2q24.2 | 1 | .015 | 2.1 | 0.23 | 0.21 | 0.31 |
| 217933_s_at | LAP3 | 4p15.32 | 1 | .0079 | 2.0 | 0.15 | 0.13 | 0.24 |
| 201114_x_at | PSMA7 | 20q13.33 | 2 | .023 | 1.9 | 0.21 | 0.18 | 0.37 |
| 205687_at | UBPH | 16p12 | 3 | .014 | 1.8 | 0.16 | 0.14 | 0.23 |
| 210334_x_at | BIRC5 | 17q25 | 4 | .00029 | 1.8 | −0.35 | −0.43 | 0.02 |
| 229417_at | STAU2 | — | 9 | .0044 | 1.7 | 0.22 | 0.18 | 0.38 |
| 238996_x_at | ALDOA | 16q22-q24 | 3 | .02 | 1.7 | 0.36 | 0.34 | 0.48 |
| 236554_x_at | TMC8 | 17q25.3 | 2 | .0015 | 1.7 | 0.24 | 0.26 | 0.12 |
| 229856_s_at | C1orf128 | 1p36.11 | 3 | .022 | 1.6 | −0.32 | −0.36 | −0.16 |
| 214752_x_at | FLNA | Xq28 | 3 | .01 | 1.5 | 0.18 | 0.13 | 0.41 |
| 208576_s_at | HIST1H3B | 6p21.3 | 1 | .018 | 1.5 | 0.40 | 0.27 | 0.98 |
| Average | 0.13 | 0.10 | 0.28 | |||||
| Favorable genes | ||||||||
| 202768_at | FOSB | 19q13.32 | 1 | .025 | 0.8 | −0.54 | −0.48 | −0.81 |
| 230292_at | LOC644250 | — | 1 | .012 | 0.8 | −0.34 | −0.24 | −0.78 |
| 217513_at | C17orf60 | 17q23.3 | 1 | .022 | 0.8 | 0.32 | 0.26 | 0.59 |
| 1564423_a_at | LZTR2 | 1q25.2 | 7 | .019 | 0.7 | −0.23 | −0.11 | −0.75 |
| 211302_s_at | PDE4B | 1p31 | 4 | .018 | 0.7 | −0.28 | −0.23 | −0.49 |
| 233909_at | STAU2 | — | 9 | .023 | 0.7 | −0.40 | −0.39 | −0.41 |
| 215671_at | PDE4B | 1p31 | 4 | .0023 | 0.7 | −0.42 | −0.43 | −0.37 |
| 208869_s_at | GABARAPL1 | 12p13.2 | 2 | .023 | 0.7 | 0.20 | 0.22 | 0.14 |
| 1552542_s_at | TAGAP | 6q25.3 | 4 | .0093 | 0.7 | −0.19 | −0.19 | −0.19 |
| 1561060_at | — | — | 1 | .017 | 0.7 | −0.27 | −0.18 | −0.65 |
| 1557633_at | LOC643318 | 22q11.2 | 1 | .025 | 0.7 | −0.29 | −0.19 | −0.77 |
| 223961_s_at | CISH | 3p21.3 | 3 | .00015 | 0.7 | −0.32 | −0.30 | −0.45 |
| 202340_x_at | NR4A1 | 12q13 | 3 | .00027 | 0.7 | −0.79 | −0.67 | −1.33 |
| 226499_at | MGC61598 | 9q34.3 | 1 | .017 | 0.7 | −0.29 | −0.28 | −0.34 |
| 242548_x_at | ANKRD37 | 4q35.1 | 2 | .022 | 0.7 | −0.39 | −0.28 | −0.91 |
| 228984_at | KIAA1394 | 11q13.1 | 1 | .0049 | 0.6 | −0.29 | −0.22 | −0.61 |
| 229388_at | — | — | 1 | .012 | 0.6 | 0.22 | 0.21 | 0.25 |
| 1552519_at | ACVR1C | 2q24.1 | 1 | .0018 | 0.6 | −0.17 | −0.18 | −0.12 |
| 239343_at | — | 12q23.3 | 1 | 0.008 | 0.6 | 0.19 | 0.14 | 0.43 |
| 212960_at | TBC1D9 | 4q31.21 | 2 | 0.0083 | 0.6 | −0.19 | −0.18 | −0.24 |
| 236986_at | — | — | 1 | 0.0025 | 0.6 | −0.13 | −0.07 | −0.39 |
| 223643_at | CRYGS | 3q25-qter | 2 | 0.023 | 0.6 | −0.18 | −0.10 | −0.52 |
| 203708_at | PDE4B | 1p31 | 4 | 0.0018 | 0.6 | −0.25 | −0.20 | −0.44 |
| 221223_x_at | CISH | 3p21.3 | 3 | 7.20E-06 | 0.5 | −0.20 | −0.16 | −0.37 |
| 213658_at | ZNF710 | — | 9 | 0.0097 | 0.5 | −0.12 | −0.08 | −0.34 |
| 238801_at | RBM33 | 7q36.3 | 3 | 0.004 | 0.5 | −0.26 | −0.28 | −0.19 |
| 235670_at | STX11 | — | 2 | 0.018 | 0.5 | −0.14 | −0.14 | −0.13 |
| 225582_at | KIAA1754 | 10q25.1 | 1 | 0.0074 | 0.5 | −0.14 | −0.17 | −0.04 |
| 227524_at | — | — | 1 | 0.01 | 0.5 | −0.17 | −0.14 | −0.33 |
| 239082_at | — | — | 1 | 0.0016 | 0.5 | −0.11 | −0.09 | −0.17 |
| 223377_x_at | CISH | 3p21.3 | 3 | 0.000 24 | 0.5 | −0.18 | −0.16 | −0.31 |
| 216215_s_at | RPL41 | 22q13.1 | 4 | 0.000 13 | 0.5 | −0.16 | −0.14 | −0.27 |
| 212050_at | WIRE | 17q21.2 | 5 | 0.0079 | 0.5 | −0.10 | −0.10 | −0.11 |
| 200673_at | LAPTM4A | 2p24.1 | 1 | 0.021 | 0.4 | 0.12 | 0.12 | 0.13 |
| 223250_at | KLHL7 | 7p15.3 | 4 | 0.023 | 0.4 | 0.09 | 0.10 | 0.07 |
| 227893_at | C9orf130 | 9q22.32 | 4 | 0.018 | 0.4 | 0.16 | 0.18 | 0.03 |
| 223215_s_at | C14orf100 | 14q23.1 | 2 | 0.013 | 0.4 | 0.12 | 0.12 | 0.11 |
| 226399_at | — | — | 1 | 0.0092 | 0.4 | 0.09 | 0.08 | 0.17 |
| Average | −0.16 | −0.13 | −0.29 |
| Probe set . | Gene symbol . | Chromosomal location . | No. probe sets on array . | P for PFS . | PFS HR . | Mean log2 expression change (all patients) . | Mean log2 expression change (GEP80 low-risk) . | Mean log2 expression change (GEP80 high-risk) . |
|---|---|---|---|---|---|---|---|---|
| Unfavorable genes | ||||||||
| 201754_at | COX6C | 8q22-q23 | 1 | .00027 | 4.4 | 0.12 | 0.11 | 0.15 |
| 219110_at | NOLA1 | 4q25 | 1 | .0014 | 3.4 | 0.11 | 0.07 | 0.32 |
| 201652_at | COPS5 | 8q13.2 | 1 | .00093 | 3.2 | 0.12 | 0.08 | 0.31 |
| 200642_at | SOD1 | 21q22.1 21q22.11 | 1 | .0014 | 3.1 | 0.15 | 0.14 | 0.21 |
| 209251_x_at | TUBA6 | 12q12-q14 | 2 | .0017 | 3.0 | 0.12 | 0.10 | 0.19 |
| 200014_s_at | HNRPC | 14q11.2 | 7 | .012 | 2.9 | 0.11 | 0.09 | 0.23 |
| 200039_s_at | PSMB2 | 1p34.2 | 4 | .0037 | 2.8 | 0.17 | 0.13 | 0.33 |
| 201252_at | PSMC4 | 19q13.11-q13.13 | 1 | .0082 | 2.7 | 0.16 | 0.11 | 0.37 |
| 226924_at | LOC400657 | 18q22.3 | 1 | .0043 | 2.6 | 0.11 | 0.10 | 0.16 |
| 225638_at | C1orf31 | 1q42.2 | 1 | .000 22 | 2.6 | 0.18 | 0.13 | 0.40 |
| 235346_at | FUNDC1 | Xp11.3 | 1 | .0037 | 2.6 | 0.13 | 0.10 | 0.22 |
| 211069_s_at | SUMO1 | 2q33 | 3 | .012 | 2.6 | 0.10 | 0.08 | 0.18 |
| 202244_at | PSMB4 | 1q21 | 3 | .016 | 2.5 | 0.15 | 0.14 | 0.23 |
| 201400_at | PSMB3 | 17q12 | 1 | .0081 | 2.5 | 0.13 | 0.09 | 0.30 |
| 202596_at | ENSA | 1q21.2 | 5 | .0091 | 2.4 | 0.12 | 0.09 | 0.26 |
| 202243_s_at | PSMB4 | 1q21 | 3 | .0098 | 2.4 | 0.19 | 0.17 | 0.28 |
| 218351_at | COMMD8 | 4p12 | 1 | .0044 | 2.4 | 0.16 | 0.15 | 0.19 |
| 223480_s_at | MRPL47 | 3q26.33 | 2 | .022 | 2.4 | 0.09 | 0.08 | 0.14 |
| 209503_s_at | PSMC5 | 17q23-q25 | 1 | .0062 | 2.3 | 0.17 | 0.11 | 0.45 |
| 212626_x_at | HNRPC | 14q11.2 | 7 | .019 | 2.3 | 0.16 | 0.14 | 0.27 |
| 203396_at | PSMA4 | 15q25.1 | 1 | .013 | 2.3 | 0.14 | 0.12 | 0.18 |
| 200882_s_at | PSMD4 | 1q21.2 | 4 | .00046 | 2.3 | 0.19 | 0.14 | 0.42 |
| 201157_s_at | NMT1 | 17q21.31 | 3 | .016 | 2.3 | 0.12 | 0.09 | 0.24 |
| 200786_at | PSMB7 | 9q34.11-q34.12 | 4 | .0083 | 2.3 | 0.15 | 0.11 | 0.34 |
| 209628_at | NXT2 | Xq22.3 | 2 | .000083 | 2.2 | 0.13 | 0.09 | 0.32 |
| 204587_at | SLC25A14 | Xq24 | 2 | .0049 | 2.2 | −0.10 | −0.12 | −0.03 |
| 210460_s_at | PSMD4 | 1q21.2 | 4 | .0012 | 2.2 | 0.19 | 0.14 | 0.44 |
| 200830_at | PSMD2 | 3q27.1 | 1 | .011 | 2.2 | 0.13 | 0.11 | 0.24 |
| 202690_s_at | SNRPD1 | 18q11.2 | 2 | .024 | 2.2 | 0.11 | 0.07 | 0.30 |
| 211609_x_at | PSMD4 | 1q21.2 | 4 | .0018 | 2.2 | 0.20 | 0.15 | 0.40 |
| 218566_s_at | CHORDC1 | 11q14.3 | 2 | .024 | 2.1 | 0.15 | 0.15 | 0.14 |
| 212296_at | PSMD14 | 2q24.2 | 1 | .015 | 2.1 | 0.23 | 0.21 | 0.31 |
| 217933_s_at | LAP3 | 4p15.32 | 1 | .0079 | 2.0 | 0.15 | 0.13 | 0.24 |
| 201114_x_at | PSMA7 | 20q13.33 | 2 | .023 | 1.9 | 0.21 | 0.18 | 0.37 |
| 205687_at | UBPH | 16p12 | 3 | .014 | 1.8 | 0.16 | 0.14 | 0.23 |
| 210334_x_at | BIRC5 | 17q25 | 4 | .00029 | 1.8 | −0.35 | −0.43 | 0.02 |
| 229417_at | STAU2 | — | 9 | .0044 | 1.7 | 0.22 | 0.18 | 0.38 |
| 238996_x_at | ALDOA | 16q22-q24 | 3 | .02 | 1.7 | 0.36 | 0.34 | 0.48 |
| 236554_x_at | TMC8 | 17q25.3 | 2 | .0015 | 1.7 | 0.24 | 0.26 | 0.12 |
| 229856_s_at | C1orf128 | 1p36.11 | 3 | .022 | 1.6 | −0.32 | −0.36 | −0.16 |
| 214752_x_at | FLNA | Xq28 | 3 | .01 | 1.5 | 0.18 | 0.13 | 0.41 |
| 208576_s_at | HIST1H3B | 6p21.3 | 1 | .018 | 1.5 | 0.40 | 0.27 | 0.98 |
| Average | 0.13 | 0.10 | 0.28 | |||||
| Favorable genes | ||||||||
| 202768_at | FOSB | 19q13.32 | 1 | .025 | 0.8 | −0.54 | −0.48 | −0.81 |
| 230292_at | LOC644250 | — | 1 | .012 | 0.8 | −0.34 | −0.24 | −0.78 |
| 217513_at | C17orf60 | 17q23.3 | 1 | .022 | 0.8 | 0.32 | 0.26 | 0.59 |
| 1564423_a_at | LZTR2 | 1q25.2 | 7 | .019 | 0.7 | −0.23 | −0.11 | −0.75 |
| 211302_s_at | PDE4B | 1p31 | 4 | .018 | 0.7 | −0.28 | −0.23 | −0.49 |
| 233909_at | STAU2 | — | 9 | .023 | 0.7 | −0.40 | −0.39 | −0.41 |
| 215671_at | PDE4B | 1p31 | 4 | .0023 | 0.7 | −0.42 | −0.43 | −0.37 |
| 208869_s_at | GABARAPL1 | 12p13.2 | 2 | .023 | 0.7 | 0.20 | 0.22 | 0.14 |
| 1552542_s_at | TAGAP | 6q25.3 | 4 | .0093 | 0.7 | −0.19 | −0.19 | −0.19 |
| 1561060_at | — | — | 1 | .017 | 0.7 | −0.27 | −0.18 | −0.65 |
| 1557633_at | LOC643318 | 22q11.2 | 1 | .025 | 0.7 | −0.29 | −0.19 | −0.77 |
| 223961_s_at | CISH | 3p21.3 | 3 | .00015 | 0.7 | −0.32 | −0.30 | −0.45 |
| 202340_x_at | NR4A1 | 12q13 | 3 | .00027 | 0.7 | −0.79 | −0.67 | −1.33 |
| 226499_at | MGC61598 | 9q34.3 | 1 | .017 | 0.7 | −0.29 | −0.28 | −0.34 |
| 242548_x_at | ANKRD37 | 4q35.1 | 2 | .022 | 0.7 | −0.39 | −0.28 | −0.91 |
| 228984_at | KIAA1394 | 11q13.1 | 1 | .0049 | 0.6 | −0.29 | −0.22 | −0.61 |
| 229388_at | — | — | 1 | .012 | 0.6 | 0.22 | 0.21 | 0.25 |
| 1552519_at | ACVR1C | 2q24.1 | 1 | .0018 | 0.6 | −0.17 | −0.18 | −0.12 |
| 239343_at | — | 12q23.3 | 1 | 0.008 | 0.6 | 0.19 | 0.14 | 0.43 |
| 212960_at | TBC1D9 | 4q31.21 | 2 | 0.0083 | 0.6 | −0.19 | −0.18 | −0.24 |
| 236986_at | — | — | 1 | 0.0025 | 0.6 | −0.13 | −0.07 | −0.39 |
| 223643_at | CRYGS | 3q25-qter | 2 | 0.023 | 0.6 | −0.18 | −0.10 | −0.52 |
| 203708_at | PDE4B | 1p31 | 4 | 0.0018 | 0.6 | −0.25 | −0.20 | −0.44 |
| 221223_x_at | CISH | 3p21.3 | 3 | 7.20E-06 | 0.5 | −0.20 | −0.16 | −0.37 |
| 213658_at | ZNF710 | — | 9 | 0.0097 | 0.5 | −0.12 | −0.08 | −0.34 |
| 238801_at | RBM33 | 7q36.3 | 3 | 0.004 | 0.5 | −0.26 | −0.28 | −0.19 |
| 235670_at | STX11 | — | 2 | 0.018 | 0.5 | −0.14 | −0.14 | −0.13 |
| 225582_at | KIAA1754 | 10q25.1 | 1 | 0.0074 | 0.5 | −0.14 | −0.17 | −0.04 |
| 227524_at | — | — | 1 | 0.01 | 0.5 | −0.17 | −0.14 | −0.33 |
| 239082_at | — | — | 1 | 0.0016 | 0.5 | −0.11 | −0.09 | −0.17 |
| 223377_x_at | CISH | 3p21.3 | 3 | 0.000 24 | 0.5 | −0.18 | −0.16 | −0.31 |
| 216215_s_at | RPL41 | 22q13.1 | 4 | 0.000 13 | 0.5 | −0.16 | −0.14 | −0.27 |
| 212050_at | WIRE | 17q21.2 | 5 | 0.0079 | 0.5 | −0.10 | −0.10 | −0.11 |
| 200673_at | LAPTM4A | 2p24.1 | 1 | 0.021 | 0.4 | 0.12 | 0.12 | 0.13 |
| 223250_at | KLHL7 | 7p15.3 | 4 | 0.023 | 0.4 | 0.09 | 0.10 | 0.07 |
| 227893_at | C9orf130 | 9q22.32 | 4 | 0.018 | 0.4 | 0.16 | 0.18 | 0.03 |
| 223215_s_at | C14orf100 | 14q23.1 | 2 | 0.013 | 0.4 | 0.12 | 0.12 | 0.11 |
| 226399_at | — | — | 1 | 0.0092 | 0.4 | 0.09 | 0.08 | 0.17 |
| Average | −0.16 | −0.13 | −0.29 |
Genes are ordered by descending hazard ratio (HR).
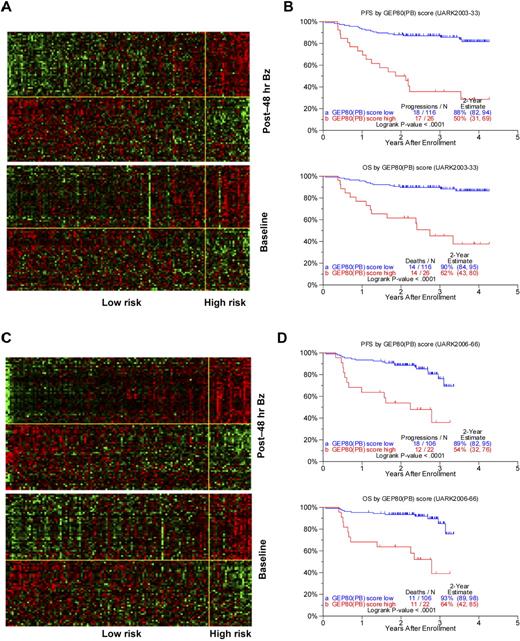
Using the training set of 142 patients in the TT3A trial, we identified 80 differentially expressed genes (Table 2) whose 48-hour expression levels were highly associated with PFS. The heat maps of these 80 genes are portrayed in Figure 1A, in which patients are grouped according to ascending GEP80 scores at 48 hours (upper panel) and at baseline (lower panel). The heat map of the 48-hour expression shows 2 major gene clusters (separated by the yellow horizontal line) with the upper gene cluster comprising many genes coding for subunits of the proteasome. The 26 patients in the high-risk group showed a concerted up-regulation of 42 bad genes (including genes coding for the proteasome) and down-regulation of 38 good genes. The clinical course for these patients was characterized by highly significantly inferior PFS and OS (Figure 1B). Results were validated in the test set of 128 patients receiving TT3B, showing similar gene expression patterns (Figure 1C) and survival differences (Figure 1D).
Development and validation of post-Bz–based GEP80 predictive model. (A) Heat map of GEP80 expression levels 48 hours after Bz administration (top) and at baseline (bottom) in training set of 142 patients enrolled in the UARK2003-33 TT3A protocol. In both panels, samples are represented in columns and genes in rows. Each column represents one patient, and each row represents one gene. Samples are ordered by ascending GEP80(PB) score, which is indicated by the green bar on the top, and genes are ordered by hierarchical cluster analysis. The horizontal yellow line separates the unfavorable genes (above yellow line) from the favorable genes (below yellow line) in each panel. The vertical yellow line separates the low-risk (left of yellow line) from the high-risk patients (right of yellow line). (B) The GEP80(PB) model distinguishes between high-risk and low-risk patients in the training set, reflected in significantly different PFS (top) and OS (bottom). (C) Heat map of GEP80 expression levels 48 hours after Bz administration (top) and at baseline (bottom) in test set of 128 patients enrolled in the UARK2006-66 TT3B protocol. In both panels, samples are represented in columns and genes in rows. Each column represents one patient, and each row represents one gene. Samples are ordered by ascending GEP80(PB) score, which is indicated by the green bar on the top, and genes are ordered by hierarchical cluster analysis. The horizontal yellow line separates the unfavorable genes (above yellow line) from the favorable genes (below yellow line) in each panel. The vertical yellow line separates the low-risk (left of yellow line) from the high-risk patients (right of yellow line). (D) The GEP80(PB) model distinguishes between high-risk and low-risk patients in the test set, reflected in significantly different PFS (top) and OS (bottom).
Development and validation of post-Bz–based GEP80 predictive model. (A) Heat map of GEP80 expression levels 48 hours after Bz administration (top) and at baseline (bottom) in training set of 142 patients enrolled in the UARK2003-33 TT3A protocol. In both panels, samples are represented in columns and genes in rows. Each column represents one patient, and each row represents one gene. Samples are ordered by ascending GEP80(PB) score, which is indicated by the green bar on the top, and genes are ordered by hierarchical cluster analysis. The horizontal yellow line separates the unfavorable genes (above yellow line) from the favorable genes (below yellow line) in each panel. The vertical yellow line separates the low-risk (left of yellow line) from the high-risk patients (right of yellow line). (B) The GEP80(PB) model distinguishes between high-risk and low-risk patients in the training set, reflected in significantly different PFS (top) and OS (bottom). (C) Heat map of GEP80 expression levels 48 hours after Bz administration (top) and at baseline (bottom) in test set of 128 patients enrolled in the UARK2006-66 TT3B protocol. In both panels, samples are represented in columns and genes in rows. Each column represents one patient, and each row represents one gene. Samples are ordered by ascending GEP80(PB) score, which is indicated by the green bar on the top, and genes are ordered by hierarchical cluster analysis. The horizontal yellow line separates the unfavorable genes (above yellow line) from the favorable genes (below yellow line) in each panel. The vertical yellow line separates the low-risk (left of yellow line) from the high-risk patients (right of yellow line). (D) The GEP80(PB) model distinguishes between high-risk and low-risk patients in the test set, reflected in significantly different PFS (top) and OS (bottom).
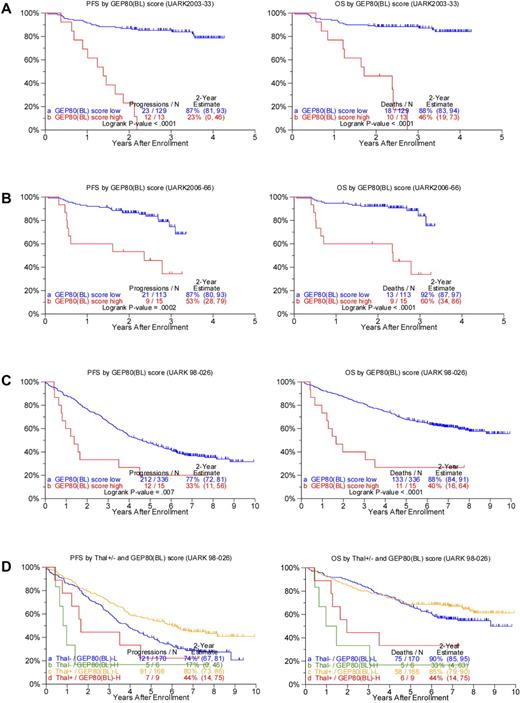
We next examined whether application of the GEP80 model to baseline data [GEP80(BL)] in both TT3 trials had outcome-discriminatory power. Results showed significant survival differences both in the TT3A training set (Figure 2A) and in the TT3B test set (Figure 2B). In the training set, 88% and 87% of low-risk patients, respectively, were alive and progression free at 2 years compared with 46% and 23% of the patients with high-risk disease. Applied to the test set, 2-year OS rates were 92% and 60% and PFS estimates were 87% and 53%, respectively, in low- and high-risk groups. Applying the GEP80 model to the combined TT3 protocols showed no significant differences in OS whether the model was applied to samples after Bz or at baseline (data not shown).
Applying the GEP80 model developed from the 48-hour post-Bz setting to baseline expression levels to divide patients into high- and low-risk groups. (A) Superior PFS (left) and OS (right) were noted in the low-risk group when applied to the training set of 142 patients receiving TT3A (UARK2003-33). (B) Superior PFS (left) and OS (right) were confirmed in the low-risk group when applied to the test set of 128 patients receiving TT3B (UARK2006-66). (C) Superior PFS (left) and OS (right) were noted in the low-risk group when applied to all 351 patients with GEP data before starting TT2 (UARK98-026, both arms combined). (D) Superior PFS (left) and OS (right) were also noted in the low-risk group (GEP80(BL)–L) compared with the high-risk group (GEP80(BL)–H) when examined separately by control arm (Thal−; P = .03 and .0002, respectively) and experimental arm (Thal+) in the TT2 protocol (P = .04 and .007, respectively).
Applying the GEP80 model developed from the 48-hour post-Bz setting to baseline expression levels to divide patients into high- and low-risk groups. (A) Superior PFS (left) and OS (right) were noted in the low-risk group when applied to the training set of 142 patients receiving TT3A (UARK2003-33). (B) Superior PFS (left) and OS (right) were confirmed in the low-risk group when applied to the test set of 128 patients receiving TT3B (UARK2006-66). (C) Superior PFS (left) and OS (right) were noted in the low-risk group when applied to all 351 patients with GEP data before starting TT2 (UARK98-026, both arms combined). (D) Superior PFS (left) and OS (right) were also noted in the low-risk group (GEP80(BL)–L) compared with the high-risk group (GEP80(BL)–H) when examined separately by control arm (Thal−; P = .03 and .0002, respectively) and experimental arm (Thal+) in the TT2 protocol (P = .04 and .007, respectively).
The profound predictive power of the GEP80 model in TT3 led us to investigate its discriminatory power in TT2, which randomly assigned 668 patients between a control arm and an experimental arm with thalidomide, both without Bz. Results showed significant differences in both OS and PFS when both arms of TT2 were considered together (Figure 2C), as well as separately (Figure 2D).
Discriminatory power of the GEP80 model compared with the GEP70 model
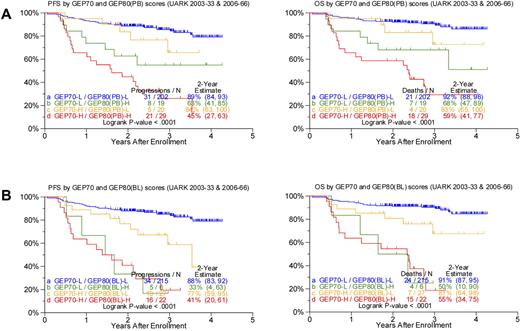
Given the profound outcome differences according to the GEP80 risk model, we next examined its performance in the context of the well-validated GEP70 risk model. Applying the GEP80 model to GEP70-defined low- and high-risk disease showed significant segregation in survival outcomes (Figure 3). Thus, among the 221 patients with GEP70-defined low-risk disease, the GEP80 model, when applied to samples after Bz, identified 19 patients (9%) with a grave prognosis. The 2-year estimates of PFS and OS were 63% and 68%, respectively, in these patients and 89% and 92%, respectively, in the 202 patients with GEP80-defined low-risk status. Conversely, among the 49 patients with GEP70-defined high-risk myeloma, the GEP80 model identified 20 patients (41%) with a favorable prognosis. These patients had 2-year PFS and OS estimates of 84% and 83%, respectively, compared with 45% and 59%, respectively, in the 29 patients in whom GEP80-defined risk was high (Figure 3A).
Comparing survival outcomes in TT3 (UARK2003-33 and 2006-66 combined) according to GEP70, GEP80(PB), and GEP80(BL) scores. (A) PFS (left) and OS (right) in both GEP70 low- and high-risk settings could be further discriminated by the GEP80(PB) model. Best outcomes were observed in patients with GEP70 low-risk and GEP80(PB) low-risk (LL) features, followed by patients with GEP70 high-risk and GEP80(PB) low-risk (HL) features and patients with GEP70 low-risk and GEP80(PB) high-risk (LH) features; worst outcomes were observed in patients with GEP70 high-risk and GEP80(PB) high-risk (HH) characteristics. The LH group exhibited poorer survival than the HL group; however, no significant differences were observed between the 2 groups (P = .216 and .268 for PFS and OS, respectively). (B) PFS (left) and OS (right) in both GEP70 low- and high-risk settings could be further discriminated by the GEP80(BL) model. Best outcomes were observed in patients with GEP70 low-risk and GEP80(BL) low-risk (LL) features, followed by patients with GEP70 high-risk and GEP80(BL) low-risk (HL) features and patients with GEP70 low-risk and GEP80(BL) high-risk (LH) features; worst outcomes were observed in patients with GEP70 high-risk and GEP80(BL) high-risk (HH) characteristics. The LH group exhibited poorer survival than the HL group, and, importantly, significant differences were observed between the 2 groups (P = .05 and .02 for PFS and OS, respectively).
Comparing survival outcomes in TT3 (UARK2003-33 and 2006-66 combined) according to GEP70, GEP80(PB), and GEP80(BL) scores. (A) PFS (left) and OS (right) in both GEP70 low- and high-risk settings could be further discriminated by the GEP80(PB) model. Best outcomes were observed in patients with GEP70 low-risk and GEP80(PB) low-risk (LL) features, followed by patients with GEP70 high-risk and GEP80(PB) low-risk (HL) features and patients with GEP70 low-risk and GEP80(PB) high-risk (LH) features; worst outcomes were observed in patients with GEP70 high-risk and GEP80(PB) high-risk (HH) characteristics. The LH group exhibited poorer survival than the HL group; however, no significant differences were observed between the 2 groups (P = .216 and .268 for PFS and OS, respectively). (B) PFS (left) and OS (right) in both GEP70 low- and high-risk settings could be further discriminated by the GEP80(BL) model. Best outcomes were observed in patients with GEP70 low-risk and GEP80(BL) low-risk (LL) features, followed by patients with GEP70 high-risk and GEP80(BL) low-risk (HL) features and patients with GEP70 low-risk and GEP80(BL) high-risk (LH) features; worst outcomes were observed in patients with GEP70 high-risk and GEP80(BL) high-risk (HH) characteristics. The LH group exhibited poorer survival than the HL group, and, importantly, significant differences were observed between the 2 groups (P = .05 and .02 for PFS and OS, respectively).
Such survival differences were also noted when the GEP80 model was applied to baseline samples (Figure 3B). The GEP80 model identified 6 patients (3%) with grave prognosis among the 221 patients in the GEP70 low-risk group. Thus, 2-year PFS and OS were 88% and 91%, respectively, in the 215 patients with GEP80(BL) low-risk disease, compared with 33% and 50%, respectively, in the 6 patients with GEP80(BL) high-risk status. However, among the 49 patients with GEP70 high-risk disease, 27 (55%) had a favorable prognosis on the basis of the GEP80 model. These patients had 2-year PFS and OS estimates of 77% and 81%, respectively, as opposed to 41% and 55%, respectively, among the 22 patients with GEP80(BL) high-risk disease. Such discriminatory power of the GEP80 model was not observed when applied to GEP70-defined subgroups treated with TT2, whether applied to all patients or examined in the context of the 2 arms of the trial (data not shown).
Gene enrichment analysis
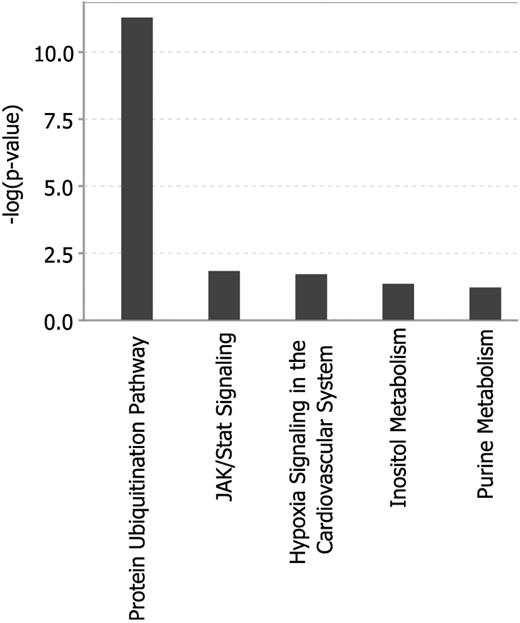
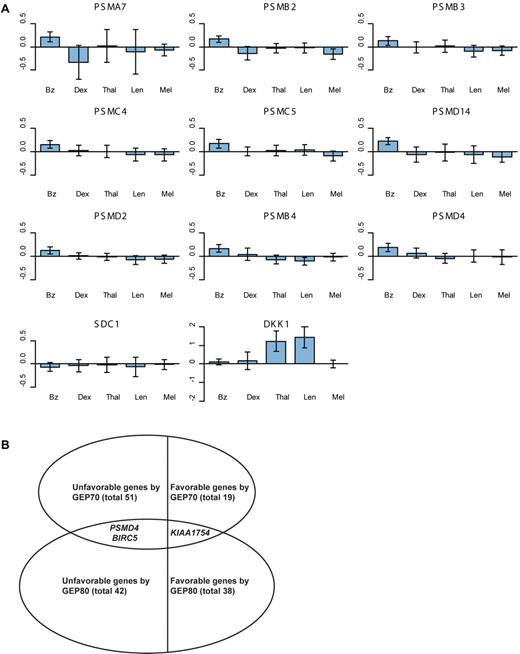
To identify predominantly affected pathways we first used the Ingenuity Pathway Analysis online tool to further explore the 80 selected genes of the GEP80 model. This analysis showed significant enrichment for genes of the protein ubiquitination pathway (Figure 4). We also computed overlaps of our 80 genes with curated gene sets with the use of the Molecular Signature Database of the Gene Set Enrichment Analysis online tool.19 Computing overlaps with a total of 5411 gene sets, the top 4 gene sets thus identified all involved the proteasome20-23 (Table 3). Further analysis showed that the Bz-induced gene expression alterations were unique to this proteasome inhibitor and were not observed in similar pharmacogenomic investigations after test-dosing with dexamethasone, thalidomide, lenalidomide, and melphalan (Figure 5A; supplemental Tables 2.1 and 2.2). Comparing the GEP80 and GEP70 risk models we found only 3 genes, 2 unfavorable and 1 favorable, common to both (Figure 5B). One of these genes, PSMD4, is one of the non-ATPase subunits of the proteosomal 19S regulator and maps to chromosome 1q21, the amplification of which had been linked to inferior clinical outcomes.24 The other 2 genes, BIRC5 and KIAA1754, map to chromosomes 17q25 and 10q25, respectively. The former is a member of the inhibitor of apoptosis gene family whereas KIAA1754, the only favorable gene found in common, encodes an inositol triphosphate receptor interacting protein.
Analysis of the 80 genes of the GEP80 model shows the protein ubiquitination pathway to be primarily affected.
Analysis of the 80 genes of the GEP80 model shows the protein ubiquitination pathway to be primarily affected.
Analysis of the 80 genes of the GEP80 model with the use of the Molecular Signature Database showing significant overlap with proteasome pathway–related gene sets
| Gene set name . | No. genes in gene set, K . | Description . | No. genes in overlap, k . | k/K . | P . |
|---|---|---|---|---|---|
| KEGG_PROTEASOME | 48 | Proteasome | 11 | 0.2292 | 6.52E-14 |
| BIOCARTA_PROTEASOME_PATHWAY | 19 | Proteasome complex | 6 | 0.3158 | 1.59E-13 |
| WONG_PROTEASOME_GENE_MODULE | 46 | Genes that comprise the proteasome gene module | 7 | 0.1522 | 3.77E-13 |
| PROTEASOME_COMPLEX | 23 | Genes annotated by the GO term GO:0000502, a large multiple subunit complex that catalyzes protein degradation. This complex consists of the barrel-shaped proteasome core complex and 1 or 2 associated proteins or complexes that act in regulating entry into or exit from the core. | 5 | 0.2174 | 1.55E-10 |
| Gene set name . | No. genes in gene set, K . | Description . | No. genes in overlap, k . | k/K . | P . |
|---|---|---|---|---|---|
| KEGG_PROTEASOME | 48 | Proteasome | 11 | 0.2292 | 6.52E-14 |
| BIOCARTA_PROTEASOME_PATHWAY | 19 | Proteasome complex | 6 | 0.3158 | 1.59E-13 |
| WONG_PROTEASOME_GENE_MODULE | 46 | Genes that comprise the proteasome gene module | 7 | 0.1522 | 3.77E-13 |
| PROTEASOME_COMPLEX | 23 | Genes annotated by the GO term GO:0000502, a large multiple subunit complex that catalyzes protein degradation. This complex consists of the barrel-shaped proteasome core complex and 1 or 2 associated proteins or complexes that act in regulating entry into or exit from the core. | 5 | 0.2174 | 1.55E-10 |
Shown are the top 4 gene sets.
Overlaps were computed with gene sets C1, C3, C5, CPG, CP:Biocarta, and CP:KEGG.
Alterations in proteasome gene expression and favorable and unfavorable genes. (A) Bar plots of alterations in proteasome (PSM) gene expression within 48 hours of test-dosing with Bz in TT3 versus dexamethasone (Dex) in the control arm of TT2, thalidomide (Thal) in the experimental arm of TT2, lenalidomide (Len) in a phase 2 trial and melphalan (Mel) in TT4 (applied 48 hours after Bz test-dosing). Within each plot, the heights of the bars indicate the mean expression changes within 48 hours of drug administration; the vertical line on each bar represents a 95% confidence interval, indicating significant (or nonsignificant) changes when not crossing (or crossing) the x-axis. These bar plots show that the PSM genes were uniquely altered significantly by Bz but not by the other agents. (B) Total number of favorable/unfavorable genes and the genes overlapping in the GEP70 and GEP80 models.
Alterations in proteasome gene expression and favorable and unfavorable genes. (A) Bar plots of alterations in proteasome (PSM) gene expression within 48 hours of test-dosing with Bz in TT3 versus dexamethasone (Dex) in the control arm of TT2, thalidomide (Thal) in the experimental arm of TT2, lenalidomide (Len) in a phase 2 trial and melphalan (Mel) in TT4 (applied 48 hours after Bz test-dosing). Within each plot, the heights of the bars indicate the mean expression changes within 48 hours of drug administration; the vertical line on each bar represents a 95% confidence interval, indicating significant (or nonsignificant) changes when not crossing (or crossing) the x-axis. These bar plots show that the PSM genes were uniquely altered significantly by Bz but not by the other agents. (B) Total number of favorable/unfavorable genes and the genes overlapping in the GEP70 and GEP80 models.
Correlations of GEP80 Model with FISH
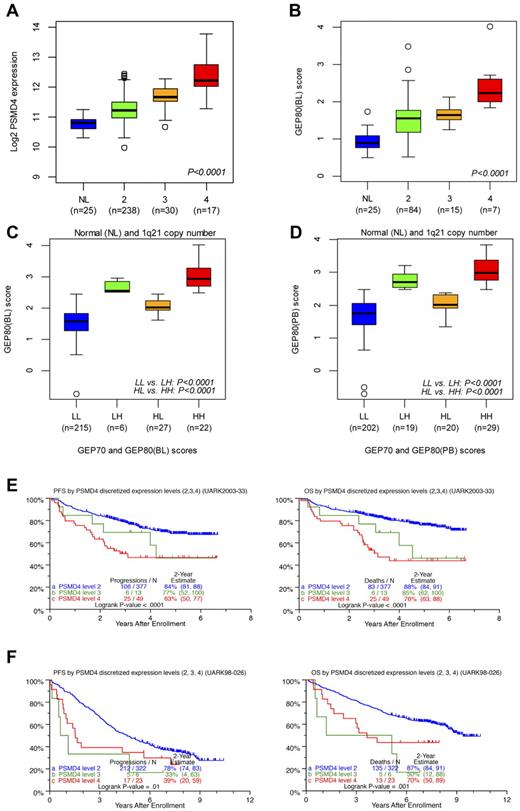
Correlating the GEP80 model with FISH-derived 1q21 copy number showed that PSMD4 expression levels increased significantly with FISH-derived 1q21 copy number (Figure 6A), as did GEP80(BL)–defined risk scores (Figure 6B). The latter finding also persisted in the context of combined GEP70 and GEP80 risk designations (low/low, low/high, high/low, and high/high), whether applied to samples at baseline (Figure 6C) or after Bz (Figure 6D). We then identified optimal cut points in PSMD4 expression levels, such that patients with 4 copies were designated as having high PSMD4 expression, patients with 3 copies medium expression, and those with 2 copies low expression. The cross-validated error rate was estimated to be 16%. In TT3, both PFS and OS progressively shortened with an increase in copy number–linked PSMD4 expression (Figure 6E). In the case of TT2, PFS was equally inferior in patients with 3 and 4 copy numbers compared with the 2-copy number group; a more graded effect, as in TT3, was noted for OS (Figure 6F).
Relationships of PSMD4 expression levels, chromosome 1q21 copy number, and GEP80 risk scores. (A) Box plot of baseline PSMD4 expression level in normal donors (NLs) and in patients with multiple myeloma with 1q21 (CKS1B) DNA copy numbers of 2, 3, and ≥4 from interphase FISH experiment. The plot shows that PSMD4 expression levels were highly correlated with 1q21 (CKS1B) copy number (P < .0001). (B) Box plot of the GEP80(BL) score in normal donors (NLs) and in patients with multiple myeloma with 1q21 copy numbers of 2, 3, and ≥ 4, showing increased risk score with higher copy number. (C) Box plot of GEP80(BL) score in risk groups defined by GEP70 and GEP80(BL) models: low 70-gene risk/low 80-gene risk (LL), low 70-gene risk/high 80-gene risk (LH), high 70-gene risk/low 80-gene risk (HL), and high 70-gene risk/high 80-gene risk (HH). (D) Box plot of GEP80(PB) score in risk groups defined by GEP70 and GEP80(PB) models: low 70-gene risk/low 80-gene risk (LL), low 70-gene risk/high 80-gene risk (LH), high 70-gene risk/low 80-gene risk (HL), and high 70-gene risk/high 80-gene risk (HH). (E) PFS (left) and OS (right) in TT3 (UARK2003-33) according to the PSMD4 expression levels (low, medium, and high) corresponding to 1q21 (CKS1B) copy number (2, 3, and 4). Note the graded effect of increasing PSMD4 levels on shortening PFS and OS in the case of TT3. (F) PFS (left) and OS (right) in TT2 (UARK98-026) according to the PSMD4 expression levels (low, medium, and high) corresponding to 1q21 (CKS1B) copy number (2, 3, and 4). Only patients with low-tertile PSMD4 expression levels fare better.
Relationships of PSMD4 expression levels, chromosome 1q21 copy number, and GEP80 risk scores. (A) Box plot of baseline PSMD4 expression level in normal donors (NLs) and in patients with multiple myeloma with 1q21 (CKS1B) DNA copy numbers of 2, 3, and ≥4 from interphase FISH experiment. The plot shows that PSMD4 expression levels were highly correlated with 1q21 (CKS1B) copy number (P < .0001). (B) Box plot of the GEP80(BL) score in normal donors (NLs) and in patients with multiple myeloma with 1q21 copy numbers of 2, 3, and ≥ 4, showing increased risk score with higher copy number. (C) Box plot of GEP80(BL) score in risk groups defined by GEP70 and GEP80(BL) models: low 70-gene risk/low 80-gene risk (LL), low 70-gene risk/high 80-gene risk (LH), high 70-gene risk/low 80-gene risk (HL), and high 70-gene risk/high 80-gene risk (HH). (D) Box plot of GEP80(PB) score in risk groups defined by GEP70 and GEP80(PB) models: low 70-gene risk/low 80-gene risk (LL), low 70-gene risk/high 80-gene risk (LH), high 70-gene risk/low 80-gene risk (HL), and high 70-gene risk/high 80-gene risk (HH). (E) PFS (left) and OS (right) in TT3 (UARK2003-33) according to the PSMD4 expression levels (low, medium, and high) corresponding to 1q21 (CKS1B) copy number (2, 3, and 4). Note the graded effect of increasing PSMD4 levels on shortening PFS and OS in the case of TT3. (F) PFS (left) and OS (right) in TT2 (UARK98-026) according to the PSMD4 expression levels (low, medium, and high) corresponding to 1q21 (CKS1B) copy number (2, 3, and 4). Only patients with low-tertile PSMD4 expression levels fare better.
Analysis of baseline variables linked to PFS and OS
When applied to both TT3 protocols, low albumin and hemoglobin concentrations; high levels of β2-microglobulin, LDH, C-reactive protein, and creatinine; and the presence of cytogenetic abnormalities conferred inferior PFS and OS on univariate analysis (Table 4). High-risk designations by GEP70 or GEP80 models (whether applied to samples at baseline or after Bz), GEP-derived delTP53, and high Proliferation Index represented highly adverse features for PFS and OS. FISH 1q21 copy number–discretized PSMD4 levels imparted inferior PFS and OS, whereas the FISH-derived CKS1B 1q21 copy number had borderline deleterious effects on PFS and significantly affected OS. In the multivariate stepwise Cox regression analysis, GEP80(BL)–defined high-risk status was the only surviving adverse genetic variable, along with high LDH and low albumin, that predicted inferior OS and PFS.
Univariate and multivariate Cox regression analyses of PFS and OS for combined training (TT3A, UARK2003-33) and test sets (TT3B, UARK 2006-66)
| Variable . | n/N (%) . | PFS . | OS . | ||
|---|---|---|---|---|---|
| HR (95% CI) . | P* . | HR (95% CI) . | P* . | ||
| Univariate Cox regression analysis† | |||||
| Age ≥ 65 y | 71/270 (26) | 1.30 (0.81-2.11) | .281 | 1.02 (0.57-1.80) | .957 |
| Albumin level < 3.5 g/dL | 89/269 (33) | 2.03 (1.29-3.20) | .002 | 1.93 (1.16-3.23) | .012 |
| B2M level ≥ 3.5 mg/L | 133/267 (50) | 2.01 (1.27-3.19) | .003 | 1.96 (1.16-3.30) | .011 |
| ISS Stage III | 68/267 (25) | 2.10 (1.32-3.33) | .002 | 2.09 (1.24-3.52) | .006 |
| Creatinine level ≥ 2.0 mg/dL | 22/269 (8) | 2.07 (1.09-3.93) | .025 | 2.08 (1.02-4.22) | .043 |
| CRP level ≥ 8 mg/L | 98/269 (36) | 2.05 (1.31-3.20) | .002 | 1.93 (1.17-3.20) | .010 |
| Hb level < 10 g/dL | 79/269 (29) | 2.19 (1.39-3.44) | < .001 | 2.33 (1.40-3.88) | .001 |
| LDH level ≥ 190 U/L | 70/269 (26) | 3.99 (2.55-6.24) | < .001 | 4.13 (2.49-6.84) | < .001 |
| Cytogenetic abnormalities | 104/267 (39) | 2.36 (1.51-3.70) | < .001 | 3.43 (2.03-5.80) | < .001 |
| Baseline 70-gene high risk | 49/270 (18) | 4.22 (2.65-6.71) | < .001 | 5.00 (3.00-8.35) | < .001 |
| GEP CD-1 subgroup | 16/270 (6) | 1.00 (0.36-2.74) | .999 | 1.33 (0.48-3.69) | .579 |
| GEP CD-2 subgroup | 32/270 (12) | 0.53 (0.21-1.31) | .168 | 0.40 (0.13-1.29) | .126 |
| GEP HY subgroup | 74/270 (27) | 0.97 (0.59-1.61) | .921 | 0.98 (0.55-1.73) | .934 |
| GEP LB subgroup | 28/270 (10) | 0.39 (0.14-1.08) | .069 | 0.39 (0.12-1.24) | .110 |
| GEP MF subgroup | 18/270 (7) | 2.31 (1.19-4.49) | .014 | 2.18 (1.03-4.58) | .040 |
| GEP MS subgroup | 33/270 (12) | 1.17 (0.60-2.27) | .647 | 0.95 (0.43-2.10) | .907 |
| GEP PR subgroup | 34/270 (13) | 2.53 (1.47-4.34) | < .001 | 2.92 (1.62-5.25) | < .001 |
| GEP proliferation index ≥ 10 | 34/270 (13) | 3.67 (2.25-5.97) | < .001 | 3.91 (2.29-6.69) | < .001 |
| GEP TP53 deletion | 29/270 (11) | 2.71 (1.54-4.76) | < .001 | 3.40 (1.87-6.19) | < .001 |
| Baseline 80-gene high risk | 28/270 (10) | 6.60 (3.91-11.15) | < .001 | 8.76 (4.99-15.38) | < .001 |
| Post-Bz 80-gene high risk | 48/270 (18) | 5.05 (3.20-7.97) | < .001 | 6.15 (3.70-10.23) | < .001 |
| FISH 1q21 copy number (2,3,4) | 107 | 1.67 (0.96-2.88) | .068 | 1.92 (1.10-3.36) | .022 |
| PSMD4 discretized levels (2,3,4) | 269 | 1.67 (1.28-2.18) | < .001 | 1.95 (1.48-2.58) | < .001 |
| Multivariate Cox regression analysis‡ | |||||
| Albumin level < 3.5 g/dL | 89/269 (33) | 1.90 (1.20-2.99) | .006 | 1.73 (1.03-2.90) | .038 |
| Baseline 80-gene high risk | 28/269 (10) | 3.77 (2.14-6.62) | < .001 | 5.25 (2.83-9.75) | < .001 |
| LDH level ≥ 190 U/L | 70/269 (26) | 2.94 (1.81-4.76) | < .001 | 2.63 (1.51-4.60) | < .001 |
| Variable . | n/N (%) . | PFS . | OS . | ||
|---|---|---|---|---|---|
| HR (95% CI) . | P* . | HR (95% CI) . | P* . | ||
| Univariate Cox regression analysis† | |||||
| Age ≥ 65 y | 71/270 (26) | 1.30 (0.81-2.11) | .281 | 1.02 (0.57-1.80) | .957 |
| Albumin level < 3.5 g/dL | 89/269 (33) | 2.03 (1.29-3.20) | .002 | 1.93 (1.16-3.23) | .012 |
| B2M level ≥ 3.5 mg/L | 133/267 (50) | 2.01 (1.27-3.19) | .003 | 1.96 (1.16-3.30) | .011 |
| ISS Stage III | 68/267 (25) | 2.10 (1.32-3.33) | .002 | 2.09 (1.24-3.52) | .006 |
| Creatinine level ≥ 2.0 mg/dL | 22/269 (8) | 2.07 (1.09-3.93) | .025 | 2.08 (1.02-4.22) | .043 |
| CRP level ≥ 8 mg/L | 98/269 (36) | 2.05 (1.31-3.20) | .002 | 1.93 (1.17-3.20) | .010 |
| Hb level < 10 g/dL | 79/269 (29) | 2.19 (1.39-3.44) | < .001 | 2.33 (1.40-3.88) | .001 |
| LDH level ≥ 190 U/L | 70/269 (26) | 3.99 (2.55-6.24) | < .001 | 4.13 (2.49-6.84) | < .001 |
| Cytogenetic abnormalities | 104/267 (39) | 2.36 (1.51-3.70) | < .001 | 3.43 (2.03-5.80) | < .001 |
| Baseline 70-gene high risk | 49/270 (18) | 4.22 (2.65-6.71) | < .001 | 5.00 (3.00-8.35) | < .001 |
| GEP CD-1 subgroup | 16/270 (6) | 1.00 (0.36-2.74) | .999 | 1.33 (0.48-3.69) | .579 |
| GEP CD-2 subgroup | 32/270 (12) | 0.53 (0.21-1.31) | .168 | 0.40 (0.13-1.29) | .126 |
| GEP HY subgroup | 74/270 (27) | 0.97 (0.59-1.61) | .921 | 0.98 (0.55-1.73) | .934 |
| GEP LB subgroup | 28/270 (10) | 0.39 (0.14-1.08) | .069 | 0.39 (0.12-1.24) | .110 |
| GEP MF subgroup | 18/270 (7) | 2.31 (1.19-4.49) | .014 | 2.18 (1.03-4.58) | .040 |
| GEP MS subgroup | 33/270 (12) | 1.17 (0.60-2.27) | .647 | 0.95 (0.43-2.10) | .907 |
| GEP PR subgroup | 34/270 (13) | 2.53 (1.47-4.34) | < .001 | 2.92 (1.62-5.25) | < .001 |
| GEP proliferation index ≥ 10 | 34/270 (13) | 3.67 (2.25-5.97) | < .001 | 3.91 (2.29-6.69) | < .001 |
| GEP TP53 deletion | 29/270 (11) | 2.71 (1.54-4.76) | < .001 | 3.40 (1.87-6.19) | < .001 |
| Baseline 80-gene high risk | 28/270 (10) | 6.60 (3.91-11.15) | < .001 | 8.76 (4.99-15.38) | < .001 |
| Post-Bz 80-gene high risk | 48/270 (18) | 5.05 (3.20-7.97) | < .001 | 6.15 (3.70-10.23) | < .001 |
| FISH 1q21 copy number (2,3,4) | 107 | 1.67 (0.96-2.88) | .068 | 1.92 (1.10-3.36) | .022 |
| PSMD4 discretized levels (2,3,4) | 269 | 1.67 (1.28-2.18) | < .001 | 1.95 (1.48-2.58) | < .001 |
| Multivariate Cox regression analysis‡ | |||||
| Albumin level < 3.5 g/dL | 89/269 (33) | 1.90 (1.20-2.99) | .006 | 1.73 (1.03-2.90) | .038 |
| Baseline 80-gene high risk | 28/269 (10) | 3.77 (2.14-6.62) | < .001 | 5.25 (2.83-9.75) | < .001 |
| LDH level ≥ 190 U/L | 70/269 (26) | 2.94 (1.81-4.76) | < .001 | 2.63 (1.51-4.60) | < .001 |
95% CI indicates 95% confidence interval; B2M, β2-microglobulin; ISS, International Staging System; CRP, C-reactive protein; and Hb, hemoglobin.
Determined with Wald chi-square test in Cox regression.
All univariate P values were reported regardless of significance. Variables with univariate P < .10 were considered for selection in the multivariate analysis.
Multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the 0.05 level.
Discussion
Pharmacogenomic investigations after test-dose applications of Bz have shown that high expression levels of proteasome genes are linked to inferior prognosis in both of our Bz-based TT3 protocols. The GEP80 model also discriminated outcomes whether applied to samples at baseline or after Bz. These findings, observed in TT3A, were validated in TT3B and also applied to both arms of TT2 (control and thalidomide). With the use of sample before Bz and at baseline in TT3, the GEP80 model provided segregation of high-risk subsets of 9% and 3%, respectively, in the GEP70 low-risk group and low-risk subsets of 41% and 55%, respectively, in the GEP70 high-risk setting. In TT2, the GEP80 model failed to discern a low-risk subset among the patients with GEP70 high-risk disease, and the GEP80 model discriminated only one patient with high-risk status among patients identified to have GEP70 low-risk disease.
To predict the risk category of a patient with newly diagnosed disease who will be treated with a Bz-based regimen, one can apply the GEP80 model to the patient's log2-transformed gene expression profiles that have been preprocessed and normalized with the Affymetrix MAS5.0 algorithm. The risk score is then calculated as the mean log2 expression of those unfavorable probe sets minus the mean log2 expression of the favorable probe sets listed in Table 2. The patient should be classified as having high-risk disease if the score is ≥ 2.48; otherwise, the patient should be considered to have low-risk disease.
PSMD4 was only 1 of 3 genes common to both GEP70 and GEP80 models. PSMD4 and other proteasome genes were uniquely up-regulated by Bz but not by dexamethasone, immunomodulatory agents, and melphalan. Correlative FISH analysis of CKS1B copy number showed that PSMD4 expression levels were highly sensitive to copy number. This enabled FISH-guided distinction of 3 PSMD4 expression levels (high, medium, low) that had prognostic implications in both TT3 and TT2, mainly by segregating a superior subset in case of low-tertile expression levels (Figure 6E-F). Prognostically, GEP80(BL)–defined high-risk status was the sole genetic parameter that survived multivariate PFS and OS models, along with low albumin and high LDH levels, although all the other standard and both cytogenetic and molecular genetic variables also contributed when examined alone.
We have previously reported that both the copy number and the percentage of cells with amp1q21 invariably increase comparing samples obtained at diagnosis and at relapse.24 Because genes residing on chromosome 1q21 contribute critically to the high-risk designation in the GEP70 model, an increase in risk score from baseline to relapse portends poor survival after relapse.25 These data suggest that clonal evolution in myeloma can be traced to a copy number–dependent increased expression of genes within the 1q21 amplicon. We found a significant up-regulation in the expression of proteasome genes after a 48-hour Bz test dose (Figure 5A; supplemental Tables 2.1 and 2.2). These changes, which ranged from 1.1- to 1.2-fold, could reflect the active up-regulation of these genes in the CD138-selected cells in response to suppression of the proteasome. However, an alternative explanation, and one that we support, is that these changes reflect the differential sensitivity of PCs to Bz-induced cell death within 48 hours of Bz dosing.
CD138 selection does not discriminate between normal and malignant PCs, nor does it distinguish clonal myeloma cells with 2, 3, or ≥ 4 copies of 1q21.24 We have reported that myeloma cell samples often exhibit an intraclonal heterogeneity with respect to amp1q21 and that a higher copy number at diagnosis portends a higher risk of death.24 More importantly, we have noted that the proportion of cells with amp1q21 always increases at relapse, suggesting that cells with more copies of 1q21 have a resistant phenotype.
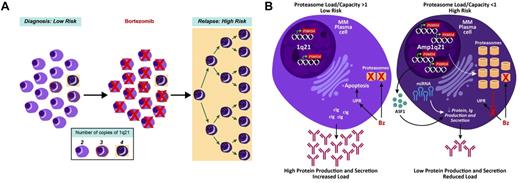
Consistent with this finding on serial samples at diagnosis and relapse, we have also noted that the GEP70 score inevitably increases at relapse and that a shift from low to high risk imparts a significantly shorter survival after relapse.25 Recent murine studies showed that Bz treatment reduces normal PCs in the spleen and BM by > 95%.26 In the current study, we demonstrated that PSMD4 expression is copy number dependent, with normal PCs from healthy donors having the lowest levels and > 90% of clonal cells from myeloma patients exhibiting > 2 copies of 1q21. Taken together, we propose that the minor yet significant up-regulation of PSMD4 and other proteasome genes 48 hours after Bz dosing is not because of a drug-induced up-regulation of the gene but rather to the preferential killing of normal PCs and myeloma cells with 2 copies of 1q21 (Figure 7A).
Proposed model of high-risk multiple myeloma development in reference to 1q21/PSMD4 copy number. (A) At time of first presentation, low-risk multiple myeloma has cells with variable 1q21/PSMD4 copy number (2, 3, or 4), whereby higher copy numbers are associated with Bz resistance. (B) A major mechanism of Bz's action could be to upset the protein load to protein capacity (PL:PC) balance in the myeloma cells. This imbalance leads to unfolded protein response (UPR)–induced apoptosis. The 4-copy 1q21/PSMD4 myeloma cells have an expanded proteasome and global increase in miRNAs, which causes increased protein capacity and decreased protein production. At relapse, the 4-copy myeloma cells are able to evade the UPR.
Proposed model of high-risk multiple myeloma development in reference to 1q21/PSMD4 copy number. (A) At time of first presentation, low-risk multiple myeloma has cells with variable 1q21/PSMD4 copy number (2, 3, or 4), whereby higher copy numbers are associated with Bz resistance. (B) A major mechanism of Bz's action could be to upset the protein load to protein capacity (PL:PC) balance in the myeloma cells. This imbalance leads to unfolded protein response (UPR)–induced apoptosis. The 4-copy 1q21/PSMD4 myeloma cells have an expanded proteasome and global increase in miRNAs, which causes increased protein capacity and decreased protein production. At relapse, the 4-copy myeloma cells are able to evade the UPR.
We have been able to show that high-risk myeloma in the Bz era is associated with high expression levels of proteasome genes. If proteasome activity can be completely extinguished by Bz, how might high-risk disease be related to Bz resistance? Given the rapid recycling of the proteasome,27 high levels of mRNA may allow for the rapid rescue of proteasome activity, so that higher doses or shorter intervals between doses of Bz may be required for patients with high-risk disease to benefit. Alternatively, protein translation inhibitors may prevent the creation of said proteasome subunits. In addition, the extra-proteasomal effects of PSMD4 may counter the activity of Bz.28 Thus, as a polyubiquitin receptor, PSMD4 could bind to and interfere with the function of tumor suppressor genes, causing them to be destined for degradation; however, in the absence of 1q21 gains, Bz would be able to counteract these effects.
Another possibility is that this protein can act as a chaperone-like molecule during the Bz-induced unfolded protein response (UPR) and prevent the subsequent UPR-induced apoptosis of myeloma cells.29 Given the secretory nature of PCs, another intriguing possibility is that PSMD4 fragments can suppress protein secretion. PSMD4 can undergo proteolytic cleavage, and an internal fragment, identified as antisecretory factor 1, has been shown to be a potent inhibitor of mucus secretion in the gut.30 It is conceivable that antisecretory factor 1 exerts its antisecretory function at the transcriptional level and, therefore, could possibly suppress protein production by myeloma cells, thus reducing the load on the endoplasmic reticulum and proteasome. There is a growing body of literature to indicate that both differentiation and death of PCs is regulated by the load and capacity of the proteasome and UPR.31
Our study has clearly shown that myeloma cells with high proteasome mRNA activity, as evidenced by high PSMD4 expression, portend an inferior prognosis. Because protein production is often reduced or even lost in end-stage disease, we may face, in high-risk myeloma, a scenario of very high proteasome activity and, hence, high protein-degradation capacity but low protein load, resulting in reduced UPR-induced apoptosis by Bz. This mechanism of Bz resistance can potentially be overcome by targeting the UPR with other novel agents in combination with proteasome inhibitors, such as inhibitors of heat-shock protein 9032 and histone deacetylase.33 We have also reported that there is global up-regulation of miRNAs in high-risk myeloma, which in turn is associated with decreased protein translation,34 so that strategies targeting miRNAs in high-risk multiple myeloma may restore Bz sensitivity by increasing protein translation and, consequently, the UPR (Figure 7B).
In summary, investigations of the short-term effects of Bz in TT3 have shown insights into novel mechanisms of resistance to this agent. As we have embarked on GEP-defined risk-based assignments of therapy in TT4 and TT5,35 we are also performing pharmacogenomic analyses after test-dosing of melphalan (10 mg/m2) toward identifying the basis for synergistic interaction of this genotoxic drug with novel agents.36
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors thank the M. J. Murdock Charitable Trust for providing support for a genomics server at Cancer Research And Biostatistics (CRAB), Seattle, where part of the statistical analysis for this research was conducted.
This work was supported by the National Cancer Institute (grant CA 55 813).
National Institutes of Health
Authorship
Contribution: J.D.S. and B.B. conceptualized the work; B.B., S.U., C.J.H., J.D.S., and J.E. wrote the paper; B.B., F.v.R., S.U., S.W., B.N., Y.A., and E.A. contributed patients and performed clinical research; O.S. and R.W. performed gene expression profiling; E.T., P.Q., and I.H. supervised and discussed gene expression profiling analyses; and J.C., P.Q., Q.Z., and Y.Z. performed statistical analyses.
Conflict-of-interest disclosure: J.D.S., B.B., P.Q., and Y.Z. have filed patents related to technology that is described in the present study. J.D.S. is a founder and has an ownership stake in Signal Genetics LLC, a biotechnology company that has licensed said technology from the University of Arkansas for purposes of commercial development purposes; holds patents, or has submitted patent applications, on the use of GEP in cancer medicine; receives royalties related to patent licenses from Genzyme Novartis and Signal Genetics; has received research funding from Celgene, Millennium, and Novartis; has advised Celgene, Genzyme, Millennium, and Novartis; and has received speaking honoraria from Celgene, ArrayBioPharma, Centocor Ortho Biotech, Genzyme, Millennium, and Novartis. B.B. has received research funding from Celgene and Novartis, is a consultant to Celgene and Genzyme, has received speaking honoraria from Celgene and Millennium, and is a co-inventor on patents and patent applications related to use of GEP in cancer medicine. The remaining authors declare no competing financial interests.
Correspondence: John D. Shaughnessy Jr, 4301 W Markham, Little Rock, AR 72205; e-mail: shaughnessyjohndjr@uams.edu.