Abstract
Translocations involving nucleoporin 98kD (NUP98) on chromosome 11p15 occur at relatively low frequency in acute myeloid leukemia (AML) but can be missed with routine karyotyping. In this study, high-resolution genome-wide copy number analyses revealed cryptic NUP98/NSD1 translocations in 3 of 92 cytogenetically normal (CN)–AML cases. To determine their exact frequency, we screened > 1000 well-characterized pediatric and adult AML cases using a NUP98/NSD1-specific RT-PCR. Twenty-three cases harbored the NUP98/NSD1 fusion, representing 16.1% of pediatric and 2.3% of adult CN-AML patients. NUP98/NSD1-positive AML cases had significantly higher white blood cell counts (median, 147 × 109/L), more frequent FAB-M4/M5 morphology (in 63%), and more CN-AML (in 78%), FLT3/internal tandem duplication (in 91%) and WT1 mutations (in 45%) than NUP98/NSD1-negative cases. NUP98/NSD1 was mutually exclusive with all recurrent type-II aberrations. Importantly, NUP98/NSD1 was an independent predictor for poor prognosis; 4-year event-free survival was < 10% for both pediatric and adult NUP98/NSD1-positive AML patients. NUP98/NSD1-positive AML showed a characteristic HOX-gene expression pattern, distinct from, for example, MLL-rearranged AML, and the fusion protein was aberrantly localized in nuclear aggregates, providing insight into the leukemogenic pathways of these AMLs. Taken together, NUP98/NSD1 identifies a previously unrecognized group of young AML patients, with distinct characteristics and dismal prognosis, for whom new treatment strategies are urgently needed.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by recurrent genetic aberrations. It is hypothesized that AML results from cooperative but functionally distinct (epi)genetic aberrations.1,2 Aberrations leading to uncontrolled proliferation and/or survival are classified as type-I and are often activating mutations in signal transduction molecules, for example, FLT3/internal tandem duplications (FLT3/ITDs). Type-II aberrations primarily block normal differentiation and include the AML-characteristic fusions, for example, promyelocytic leukemia–retinoic acid receptor α (PML/RARA), AML1/ETO, and CBFβ/MYH11. In the last decade, novel type-II aberrations were discovered, for example, mutations in NPM1 and CEBPA, and they are mainly found in patients with cytogenetically normal (CN)–AML.3,4 Despite this progress, there still is a significant group of AML cases in which the underlying genetic aberrations are unknown.
Type-II aberrations also include translocations of nucleoporin 98kD (NUP98) located on chromosome 11p15, although they account for < 1% of AML cases.5-8 Fusions of NUP98 to more than 20 different partner genes have been described previously,5 and they can be divided into homeobox genes (eg, HOXA9, -C11, and -D13) and nonhomeobox genes (DDX10, NSD1, and TOP1). NUP98-HOXA9, the most frequent fusion, has aberrant self-renewal capacity, blocks differentiation in transfected myeloid progenitors, and induces AML in mice.9,10
Genome-wide approaches proved to be powerful tools to dissect AML molecularly. High-resolution array-based comparative genome hybridization (A-CGH) and single-nucleotide polymorphism arrays (SNP-A) identified recurrent copy number aberrations and regions with loss of heterozygosity, although AML seemed to be relatively genomically stable compared with other malignancies.11-13 Mapping of genetic lesions will improve insight into the AML biology and may ultimately lead to development of new treatments.
In this study, we used high-resolution A-CGHs and SNP-As to identify novel genetic aberrations underlying CN-AML. The cytogenetically cryptic NUP98/NSD1 translocation was initially identified in 3 cases with these techniques. Subsequently, we performed a comprehensive study of > 1000 pediatric and adult AML cases, and we identified the NUP98/NSD1 translocation as a frequent cryptic event within pediatric CN-AML (16.1%) compared with adult CN-AML (2.3%). Moreover, NUP98/NSD1 seemed to be a novel independent predictor for dismal outcome. NUP98/NSD1-positive AML showed a distinct gene expression pattern, including high expression of HOXB cluster genes, providing insight into the leukemogenic pathways of these AMLs.
Methods
Study cohort
Two patient cohorts were included in this study, a pediatric cohort and an adult cohort. The pediatric cohort (n = 293; age, 0-18 years) consisted of children with available frozen bone marrow (BM) or peripheral blood samples taken at diagnosis that were provided by the Dutch Childhood Oncology Group (The Hague, The Netherlands; n = 141), the AML–Berliner-Frankfurt-Münster (BFM) Study Group (Germany and Czech Republic; n = 128), and the Saint-Louis Hospital (Paris, France; n = 24). In addition, of 3 NUP98/NSD1-positive cases, paired remission and relapse samples were available. The pediatric cohort was representative for pediatric AML, comparing patient characteristics with the AML-BFM98 series (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), although our cohort included more FAB-M4 and less FAB-M2 cases. Survival analysis was restricted to patients with de novo AML enrolled in Dutch Childhood Oncology Group and AML-BFM Study Group studies, excluding PML/RARA cases (n = 238). The median follow-up time of survivors of 4.2 years (range, 0.3-22.7 years), and overall probability of event-free survival (pEFS) and probability of overall survival (pOS) for the pediatric cohort were 44 ± 3% and 62 ± 3%, respectively.
The adult cohort consisted of AML patients (n = 808; age, 15-77 years, including 20 children aged 15-18 years) treated on consecutive Dutch-Belgian Cooperative Trial Group for Hematology Oncology protocols, with available frozen BM or peripheral blood taken at diagnosis.14 Survival analysis was restricted to patients with de novo AML excluding PML/RARA (n = 727). The median follow-up time of survivors was 4.2 years (range, 0.5-18.7 years), and overall pEFS and pOS for the adult cohort were 32 ± 2% and 39 ± 2%, respectively.
Morphologic classification and karyotyping were centrally reviewed by each study group. Molecular characterization included mutational analysis of NPM1, CEBPA, MLL, FLT3, KIT, N- and K-RAS; PTPN11 and WT1 for the pediatric cohort; and NPM1, FLT3, N-RAS, WT1, IDH1, and IDH2 for the adult cohort.14-20
Institutional review board approval had been obtained for these studies from Erasmus Medical Center. Informed consent was obtained from the patients in accordance with the Declaration of Helsinki.
Genome-wide copy number analysis
In 45 pediatric CN-AML cases, A-CGH was performed using the human genome CGH Microarray 105K (Agilent Technologies) according to the manufacturer's protocol.21 Microarray images were analyzed using feature extraction software (Version 8.1; Agilent Technologies), and data were subsequently analyzed with Genomic Workbench (Version 5.0.14; Agilent Technologies).
In 47 adult CN-AML samples, SNP-As (250K NspI DNA-mapping array; Affymetrix) were performed according to the manufacturer's protocol. Genotypes were calculated using BRLMM, copy numbers were assessed using dChipSNP, and data were subsequently visualized in SNPExpress.22
FISH
Split-signal FISH analysis of NUP98 was performed on thawed cytospin slides using the 44-kb overlapping bacterial artificial chromosome probes RP11-120E20 and RP11-348A20 (BACPAC Resources Center) as described previously.23 The NUP98/NSD1 translocation was confirmed using bacterial artificial chromosome probes RP11-348A20 and RP11-99N22.
Detection of the NUP98/NSD1 transcript
Presence of NUP98/NSD1 and the reciprocal NSD1-NUP98 were determined by RT-PCR. Primers and cycle conditions are presented in supplemental Table 2. Purified PCR products were directly sequenced on a PRISM 3100 genetic analyzer (Applied Biosystems) and analyzed using CLCWorkbench (Version 3.5.1; CLC Bio). NUP98/NSD1 was not detected in normal BM controls (n = 7).
NUP98/NSD1 transcript levels were measured in duplicate based on the intercalation of SYBR Green (Finnzymes) using RT-quantitative (q)PCR (TaqMan chemistry) on a PRISM 7900HT system (Applied Biosystems) and calculated relative to GAPDH expression. Amplification efficiency was nearly 100%, and the dissociation curves confirmed amplification of a single product for NUP98/NSD1 as well as GAPDH. The sensitivity of the RT-qPCR, as determined by serially diluting cDNA of a NUP98/NSD1 case (4432) in cDNA derived from an NUP98/NSD1-negative AML cell line (NB4; DSMZ), reached 10−3 to 10−4, with an input of only 20 ng of cDNA. Transcript levels were calculated relative to 4432 using the standard curve.
Immunofluorescence
Cellular localization of the NUP98/NSD1 fusion protein was examined using immunofluorescence. Thawed cytospin slides with leukemic cells were fixed and permeabilized with ice-cold 96% methanol. After washing with PBS, slides were incubated with a primary antibody (sc-101546 [α-N-terminal NUP98] or sc-32479 [α-C-terminal NSD1]; Santa Cruz Biotechnology). This was followed by incubation with a fluorescently labeled secondary antibody (DyLight 488 goat anti–rat IgG or DyLight 549 rabbit anti–goat, respectively; Jackson ImmunoResearch Laboratories). Cells were visualized with a Zeiss LSM700-microscope 63×/1.4 NA oil objective, scanned at 2048 × 2048 pixels in 3 channels (8-bit resolution), and the resulting images were acquired and processed with ZEN 2009 light edition software (Carl Zeiss).
Gene expression profiling
Gene expression profiling data (HGU133 Plus 2.0 microarray; Affymetrix) were available of 274 pediatric cases.24 Original data are available in the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo; accession GSE17855). Checking RNA quality, microarray processing, data acquisition, and data normalization have been previously described previously.24 Differentially expressed genes were calculated using t test-based statistics (Bioconductor package LIMMA; http://www.bioconductor.org/) in the statistical data analysis environment R (Version 2.7.0; http://www.r-project.org/). The P values were corrected for multiple testing according to the false discovery rate (FDR)–procedure of Hochberg and Benjaminini (Bioconductor package Multtest).25 Supervised clustering and principal component analyses were performed using GeneMath XT 1.6.1 software (Applied Maths). Unsupervised clustering analysis was performed and visualized as described previously.16,23
Expression of microRNA-196b and -10a
MicroRNA (miR)–10a and -196b expression was determined in a selection of the pediatric cohort (n = 90 and n = 84, respectively), reflecting the different genetic subgroups in pediatric AML. miR-expression levels were measured in duplicate on a PRISM-7900HT system using a stem-loop based RT-qPCR according to the manufacturer's protocol (Applied Biosystems). The threshold was manually set at 0.15, and the comparative cycle threshold method was used to calculate the miR-expression levels relative to the endogenous miR-control RNU24.
Statistical analysis
Statistical analyses were performed with SPSS 17.0 (SPSS) and SAS/STAT 9.2 (SAS Institute). Categorical variables were compared using the χ2 or Fisher exact test, and continuous variables using Mann-Whitney U test. To assess outcome, the following parameters were used: complete remission (CR, defined as < 5% blasts in the bone marrow, with regeneration of trilineage hematopoiesis plus absence of leukemic cells in the cerebrospinal fluid or elsewhere), cumulative incidence of relapse or nonresponse (pCIR, defined as time between diagnosis and relapse; resistant disease was included as an event on day 0), pEFS (defined as time between diagnosis and first event, including failure to achieve remission, relapse, death by any cause, or second malignancy), and pOS (defined as time between diagnosis and death). pEFS and pOS were estimated by the Kaplan-Meier method and compared using the log-rank test. CIR curves were constructed by the method of Kalbfleisch and Prentice and were compared using Gray test.26 The independency of prognostic factors was examined by multivariate Cox regression analysis. All tests were 2-tailed, and P values < .05 were considered statistically significant.
Results
Genome-wide copy number profiling identified NUP98 aberrations
Using high-resolution A-CGH, we detected an 11p15-aberration in 2 of 45 pediatric CN-AML cases, involving the NUP98 gene. One case harbored a 0.4-Mb duplication involving the 5′ part of NUP98 and the other case a 0.1-Mb deletion of the 3′ end (Figure 1A). In both deletions, the genomic breakpoint was located between the probes in NUP98 exons 11 and 13. Because most translocation breakpoints of NUP98 cluster in introns 11-12 and 12-13,5 we suspected an unbalanced NUP98 translocation, and this translocation was confirmed by split-signal FISH in both cases (Figure 1B). To identify the partner genes, we selected candidate genes resulting in cryptic translocations with NUP98, including the nuclear receptor-binding SET domain protein 1 (NSD1) gene located on chromosome 5q35, 4 Mb from the telomere. RT-PCR revealed the NUP98/NSD1 transcript in both cases, and sequence analysis showed an identical in-frame fusion of NUP98 exon 12 and NSD1 exon 6 (Figure 1C-E). FISH analysis confirmed these fusions (Figure 1F).
Discovery of NUP98/NSD1 fusion in 2 CN-AML cases. (A) Array-CGH profiles of chromosome 11p15 showing the ratio of tumor DNA/control DNA (blue tracing) versus the inverted experiment (red tracing). The profile of patient 3495 (top panel) shows a part of the 0.4-Mb duplication involving the 5′ part of NUP98 (indicated by the blue vertical arrow), and the profile of patient 4716 shows the 0.1-Mb deletion involving the 3′ part of NUP98 (indicated by the red vertical bar). The horizontal arrows indicate the bar representing the NUP98 gene. (B) Split-signal FISH analysis of NUP98 for patients 3495 and 4716 using a partly overlapping green and red probe located in NUP98 (cen indicates probe situated more centromeric; tel indicates probe situated more telomeric. (C) RT-PCR analysis using NUP98- and NSD1-specific primers, and GAPDH primers as internal control, reveals a specific NUP98/NSD1 fusion transcript in patients 3495 and 4716. The reciprocal NSD1-NUP98 transcript was not detected in both patients (pos indicates positive control, [patient 5007]; neg indicates negative control [normal bone marrow]; ntc indicates non-template control). (D) Sequence analysis confirmed an identical in-frame fusion between NUP98 exon 12 and NSD1 exon 6 in both patients. (E) The NUP98/NSD1 fusion protein will harbor the GLFG-repeats of NUP98, and among others the PHD fingers and SET domain of NSD1. (F) Dual-color FISH analysis using a green probe for NUP98 and a red probe for NSD1 confirmed the fusion of NUP98 and NSD1 at the chromosomal level.
Discovery of NUP98/NSD1 fusion in 2 CN-AML cases. (A) Array-CGH profiles of chromosome 11p15 showing the ratio of tumor DNA/control DNA (blue tracing) versus the inverted experiment (red tracing). The profile of patient 3495 (top panel) shows a part of the 0.4-Mb duplication involving the 5′ part of NUP98 (indicated by the blue vertical arrow), and the profile of patient 4716 shows the 0.1-Mb deletion involving the 3′ part of NUP98 (indicated by the red vertical bar). The horizontal arrows indicate the bar representing the NUP98 gene. (B) Split-signal FISH analysis of NUP98 for patients 3495 and 4716 using a partly overlapping green and red probe located in NUP98 (cen indicates probe situated more centromeric; tel indicates probe situated more telomeric. (C) RT-PCR analysis using NUP98- and NSD1-specific primers, and GAPDH primers as internal control, reveals a specific NUP98/NSD1 fusion transcript in patients 3495 and 4716. The reciprocal NSD1-NUP98 transcript was not detected in both patients (pos indicates positive control, [patient 5007]; neg indicates negative control [normal bone marrow]; ntc indicates non-template control). (D) Sequence analysis confirmed an identical in-frame fusion between NUP98 exon 12 and NSD1 exon 6 in both patients. (E) The NUP98/NSD1 fusion protein will harbor the GLFG-repeats of NUP98, and among others the PHD fingers and SET domain of NSD1. (F) Dual-color FISH analysis using a green probe for NUP98 and a red probe for NSD1 confirmed the fusion of NUP98 and NSD1 at the chromosomal level.
In the adult cohort, high-resolution SNP-A was performed in 47 CN-AML cases. An 11p15-aberration including NUP98 was detected in 1 case (supplemental Figure 1). The identical NUP98/NSD1 fusion, as found in both pediatric cases, was detected in this case.
NUP98/NSD1 and other NUP98 fusions
To determine the frequency of NUP98/NSD1, we screened 1101 AML cases using RT-PCR. The NUP98/NSD1 transcript was detected in 23 cases (2.1%; Table 1), all carrying the identical in-frame fusion of NUP98 exon 12 and NSD1 exon 6. The reciprocal NSD1/NUP98 transcript was detected in 13 of the 23 cases (57%) only. Three of 10 NUP98/NSD1-positive cases, analyzed by genomic profiling, demonstrated numerical changes of the NUP98 or NSD1 gene adjacent to the breakpoint, indicative of an unbalanced translocation.
Individual characteristics of the NUP98/NSD1-positive (n = 23) and the other NUP98-translocated (n = 3) AML cases
| ID no. . | NUP98 parter gene . | Cohort . | Age, y . | Sex . | WBC, × 109/L . | FAB . | Cytogenetic aberration(s) . | Aberrant 5q35 or 11p15 on A-CGH or SNP-A . | Reciprocal product . | Mutations . | Therapy protocol . | CR . | Relapse (relapse-free survival in mo) . | Dead (OS in mo) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0506 | NSD1 | ped | 10 | M | 332.0 | M4 | Normal | No | Yes | FLT3/ITD + WT1 | MRC12 | + | − (17.2) | + (7.2)* |
| 3397 | NSD1 | ped | 15 | M | 154.4 | M5 | +8 | NA | Yes | FLT3/ITD | MRC12 | + | + (4.4) | + (5.6) |
| 3495 | NSD1 | ped | 14 | M | 57.1 | NA | Normal | dupl 11p15 | No | FLT3/ITD | MRC12 | + | + (6.3) | − (6.3)† |
| 4380 | NSD1 | ped | 4 | M | 7.4 | M4 | Normal | No | No | N-RAS + WT1 | BFM04 | + | + (7.3) | − (64.8)* |
| 4417 | NSD1 | ped | 17 | M | 189.0 | M2 | Normal | No | Yes | FLT3/ITD + WT1 | BFM04 | + | + (5.3) | + (30.2)* |
| 4432 | NSD1 | ped | 6 | M | 324.0 | M2 | Normal | No | Yes | FLT3/ITD + WT1 | BFM04 | + | + (4.9) | + (8.0) |
| 4716 | NSD1 | ped | 4 | M | 377.6 | M1 | Normal | del 11p15 | No | FLT3/ITD | MRC12 | + | + (7.9) | + (9.4) |
| 4730 | NSD1 | ped | 9 | M | 121.3 | M4 | del 9q | NA | Yes | FLT3/ITD | MRC12 | + | + (8.7) | − (14.4)* |
| 4733 | NSD1 | ped | 2 | M | 267.3 | M4 | Normal | No | Yes | FLT3/ITD | MRC12 | + | + (6.9) | + (9.9) |
| 5007 | NSD1 | ped | 14 | F | NA | NA | del 9q | NA | Yes | FLT3/ITD + WT1 | LAME | NA | NA | NA |
| 5144 | NSD1 | ped | 16 | F | 187.0 | M1 | Normal | NA | Yes | FLT3/ITD + WT1 | BFM98 | + | + (4.6) | + (8.2) |
| 5166 | NSD1 | ped | 13 | M | 226.0 | M1/2 | Normal | NA | No | FLT3/ITD | BFM04 | + | − | − (32.0)* |
| 6328 | NSD1 | ped | 2 | M | 153.0 | M4 | Normal | No | Yes | FLT3/ITD | MRC15 | + | + (7.2) | − (31.8)* |
| 2176 | NSD1 | adult | 41 | M | 263.4 | M4 | Normal | NA | Yes | FLT3/ITD | HO04 | − | + (1.1) | |
| 2280 | NSD1 | adult | 49 | F | 6.1 | M2 | Normal | del 11p15 | No | WT1 | HO29 | − | + (2.0) | |
| 2305 | NSD1 | adult | 27 | M | 58.0 | M5 | Normal | No | No | FLT3/ITD + WT1 | HO29 | + | + (11.5) | − (92.3)* |
| 4333 | NSD1 | adult | 31 | M | 177.0 | M5 | Normal | NA | No | FLT3/ITD | HO42 | − | + (1.5) | |
| 6463 | NSD1 | adult | 63 | F | 78.0 | M5 | inv(5)(q1?2q3?4) | NA | No | FLT3/ITD | HO43 | − | + (9.2) | |
| 6714 | NSD1 | adult | 47 | F | 140.0 | M5 | Normal | NA | Yes | FLT3/ITD‡ | HO04 | + | − (19.9) | + (19.9)* |
| 6884 | NSD1 | adult | 19 | F | 126.4 | M5 | Marker§ | NA | No | FLT3/ITD‡ | HO04A | + | + (4.6) | + (8.5) |
| 7191 | NSD1 | adult | 45 | F | 131.0 | M5 | Normal | NA | No | FLT3/ITD‡ | HO42 | − | + (10.0) | |
| 11678 | NSD1 | adult | 54 | F | 49.2 | M4 | Normal | NA | Yes | FLT3/ITD | HO42A | − | + (3.8) | |
| 13983 | NSD1 | adult | 36 | M | 97.2 | M4 | Normal | NA | Yes | FLT3/ITD + WT1 | HO42A | + | + (3.6) | + (8.8) |
| 0297 | JARID1A | ped | 1 | M | 8.4 | M7 | Complex | NA | No | None | MRC12 | + | − | − (128.4) |
| 4096 | TOP1 | ped | 12 | F | 214.0 | M4 | t(11;20)(p15;q1?2) | NA | Yes | CEBPAs + WT1 | MRC12 | + | + (9.8) | + (19.0) |
| 6974 | DDX10 | adult | 32 | M | 4.9 | M6 | +8,inv(11)(p15;q22) | NA | NA | N-RAS‡ | HO29 | + | + (13.9) | + (22.3) |
| ID no. . | NUP98 parter gene . | Cohort . | Age, y . | Sex . | WBC, × 109/L . | FAB . | Cytogenetic aberration(s) . | Aberrant 5q35 or 11p15 on A-CGH or SNP-A . | Reciprocal product . | Mutations . | Therapy protocol . | CR . | Relapse (relapse-free survival in mo) . | Dead (OS in mo) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0506 | NSD1 | ped | 10 | M | 332.0 | M4 | Normal | No | Yes | FLT3/ITD + WT1 | MRC12 | + | − (17.2) | + (7.2)* |
| 3397 | NSD1 | ped | 15 | M | 154.4 | M5 | +8 | NA | Yes | FLT3/ITD | MRC12 | + | + (4.4) | + (5.6) |
| 3495 | NSD1 | ped | 14 | M | 57.1 | NA | Normal | dupl 11p15 | No | FLT3/ITD | MRC12 | + | + (6.3) | − (6.3)† |
| 4380 | NSD1 | ped | 4 | M | 7.4 | M4 | Normal | No | No | N-RAS + WT1 | BFM04 | + | + (7.3) | − (64.8)* |
| 4417 | NSD1 | ped | 17 | M | 189.0 | M2 | Normal | No | Yes | FLT3/ITD + WT1 | BFM04 | + | + (5.3) | + (30.2)* |
| 4432 | NSD1 | ped | 6 | M | 324.0 | M2 | Normal | No | Yes | FLT3/ITD + WT1 | BFM04 | + | + (4.9) | + (8.0) |
| 4716 | NSD1 | ped | 4 | M | 377.6 | M1 | Normal | del 11p15 | No | FLT3/ITD | MRC12 | + | + (7.9) | + (9.4) |
| 4730 | NSD1 | ped | 9 | M | 121.3 | M4 | del 9q | NA | Yes | FLT3/ITD | MRC12 | + | + (8.7) | − (14.4)* |
| 4733 | NSD1 | ped | 2 | M | 267.3 | M4 | Normal | No | Yes | FLT3/ITD | MRC12 | + | + (6.9) | + (9.9) |
| 5007 | NSD1 | ped | 14 | F | NA | NA | del 9q | NA | Yes | FLT3/ITD + WT1 | LAME | NA | NA | NA |
| 5144 | NSD1 | ped | 16 | F | 187.0 | M1 | Normal | NA | Yes | FLT3/ITD + WT1 | BFM98 | + | + (4.6) | + (8.2) |
| 5166 | NSD1 | ped | 13 | M | 226.0 | M1/2 | Normal | NA | No | FLT3/ITD | BFM04 | + | − | − (32.0)* |
| 6328 | NSD1 | ped | 2 | M | 153.0 | M4 | Normal | No | Yes | FLT3/ITD | MRC15 | + | + (7.2) | − (31.8)* |
| 2176 | NSD1 | adult | 41 | M | 263.4 | M4 | Normal | NA | Yes | FLT3/ITD | HO04 | − | + (1.1) | |
| 2280 | NSD1 | adult | 49 | F | 6.1 | M2 | Normal | del 11p15 | No | WT1 | HO29 | − | + (2.0) | |
| 2305 | NSD1 | adult | 27 | M | 58.0 | M5 | Normal | No | No | FLT3/ITD + WT1 | HO29 | + | + (11.5) | − (92.3)* |
| 4333 | NSD1 | adult | 31 | M | 177.0 | M5 | Normal | NA | No | FLT3/ITD | HO42 | − | + (1.5) | |
| 6463 | NSD1 | adult | 63 | F | 78.0 | M5 | inv(5)(q1?2q3?4) | NA | No | FLT3/ITD | HO43 | − | + (9.2) | |
| 6714 | NSD1 | adult | 47 | F | 140.0 | M5 | Normal | NA | Yes | FLT3/ITD‡ | HO04 | + | − (19.9) | + (19.9)* |
| 6884 | NSD1 | adult | 19 | F | 126.4 | M5 | Marker§ | NA | No | FLT3/ITD‡ | HO04A | + | + (4.6) | + (8.5) |
| 7191 | NSD1 | adult | 45 | F | 131.0 | M5 | Normal | NA | No | FLT3/ITD‡ | HO42 | − | + (10.0) | |
| 11678 | NSD1 | adult | 54 | F | 49.2 | M4 | Normal | NA | Yes | FLT3/ITD | HO42A | − | + (3.8) | |
| 13983 | NSD1 | adult | 36 | M | 97.2 | M4 | Normal | NA | Yes | FLT3/ITD + WT1 | HO42A | + | + (3.6) | + (8.8) |
| 0297 | JARID1A | ped | 1 | M | 8.4 | M7 | Complex | NA | No | None | MRC12 | + | − | − (128.4) |
| 4096 | TOP1 | ped | 12 | F | 214.0 | M4 | t(11;20)(p15;q1?2) | NA | Yes | CEBPAs + WT1 | MRC12 | + | + (9.8) | + (19.0) |
| 6974 | DDX10 | adult | 32 | M | 4.9 | M6 | +8,inv(11)(p15;q22) | NA | NA | N-RAS‡ | HO29 | + | + (13.9) | + (22.3) |
ped indicates pediatric; adult, adult; F, female; M, male; NA, not available; WBC, white blood cell count; FAB, French-American-British subtype; and CEBPAs, CEBPA single mutation.
These patients received an allo-SCT (0506, 5166, and 6714 in CR1 and 4380, 4417, 4730, 6328, and 2305 after relapse).
No follow-up available after relapse.
These samples were not tested for WT1 mutations due to lack of material.
Karyotype: 47,XX,+21?/46,XX.
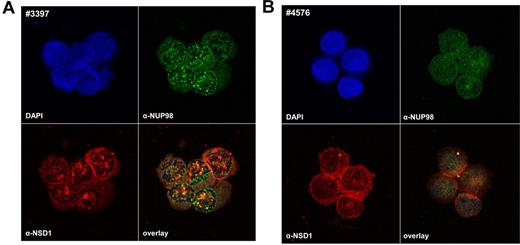
We observed an aberrant cellular pattern when staining NUP98 and NSD1 in NUP98/NSD1-positive leukemic cells (Figure 2). NUP98/NSD1-positive samples showed nuclear speckles, in contrast to NUP98/NSD1-negative samples that displayed a fine distribution of NUP98 and NSD1 in the nucleus and cytoplasm. This indicates that the NUP98/NSD1 fusion protein forms nuclear aggregates.
Aberrant nuclear-staining pattern of NUP98/NSD1-positive samples. Immunofluorescence of leukemic cells with antibodies directed against the N-terminus of NUP98 (green) and the C-terminus of NSD1 (red) is shown for a NUP98/NSD1-positive (A) and a NUP98/NSD1-negative patient sample (B). Patient 3397 shows a pattern of nuclear speckles for NUP98 staining as well as NSD1 staining, indicating accumulation of the NUP98/NSD1 fusion protein in aggregates in the nucleus, in contrast to patient 4576. DAPI indicates 4,6-diamidino-2-phenylindole.
Aberrant nuclear-staining pattern of NUP98/NSD1-positive samples. Immunofluorescence of leukemic cells with antibodies directed against the N-terminus of NUP98 (green) and the C-terminus of NSD1 (red) is shown for a NUP98/NSD1-positive (A) and a NUP98/NSD1-negative patient sample (B). Patient 3397 shows a pattern of nuclear speckles for NUP98 staining as well as NSD1 staining, indicating accumulation of the NUP98/NSD1 fusion protein in aggregates in the nucleus, in contrast to patient 4576. DAPI indicates 4,6-diamidino-2-phenylindole.
We found 4 additional AML cases carrying cytogenetically visible 11p15-aberrations, suggesting involvement of NUP98. One pediatric case carried the NUP98/JARID1A fusion that we described previously.27 Another pediatric case had a t(11;20)(p15;q1?2), and FISH analysis and RT-PCR confirmed the in-frame NUP98/TOP1 and reciprocal TOP1/NUP98 fusion (supplemental Figure 2). One adult case had an inv(11)(p15q22), and RT-PCR showed presence of the in-frame NUP98/DDX10 (supplemental Figure 2). The second adult case had an inv(11)(p15q13) besides a t(15;17)(q22;q21), but NUP98 involvement could not be investigated because of lack of material. In addition, 9 selected acute lymphoblastic leukemia samples did not harbor NUP98/NSD1 (see supplemental Methods).
Age dependency of NUP98/NSD1
Thirteen of 313 (4.2%) NUP98/NSD1-positive cases were found in children (0-18 years) and 10/788 (1.3%) in adults (> 18 years; P = .003). When analyses were limited to CN-AML, we observed an even higher frequency of NUP98/NSD1 in children (16.1%) compared with adults (2.3%; P < .001; Figure 3A-B). Of note, NUP98/NSD1 cases were not observed in children < 2 years of age.
NUP98/NSD1 is a frequent event in pediatric CN-AML. (A) Pie charts showing percentage of the different type-II aberrations found in pediatric CN-AML (n = 62; left pie chart) and in adult CN-AML (n = 344; right pie chart). The mutational analysis of CEBPA and MLL was not available for the adult cohort. (B) Histogram representing the percentage of the NUP98/NSD1 positive cases within the different age categories in CN-AML shows a decreasing frequency of NUP98/NSD1 with age.
NUP98/NSD1 is a frequent event in pediatric CN-AML. (A) Pie charts showing percentage of the different type-II aberrations found in pediatric CN-AML (n = 62; left pie chart) and in adult CN-AML (n = 344; right pie chart). The mutational analysis of CEBPA and MLL was not available for the adult cohort. (B) Histogram representing the percentage of the NUP98/NSD1 positive cases within the different age categories in CN-AML shows a decreasing frequency of NUP98/NSD1 with age.
Characteristics of NUP98/NSD1 cases
All but 1 of the NUP98/NSD1 cases presented with de novo AML (patient 11678 presented with therapy-related AML; Table 1). NUP98/NSD1 cases had a higher white blood cell count compared with other cases (median, 147 × 109/l vs 26 × 109/l; P < .001) and were associated with FAB-M4/M5 morphology (65.2% vs 39.2%; P = .01; Table 2). However, when analyzing the adult and pediatric cohort separately (supplemental Table 3A-B), the association with M4/M5 morphology was only observed in the adult cohort.
Clinical and molecular characteristics of NUP98/NSD1-positive versus -negative cases
| . | NUP98/NSD1-positive cases . | NUP98/NSD1-negative cases . | P . |
|---|---|---|---|
| Total, N (%) | 23 (2.1) | 1078 (97.9) | |
| Pediatric cohort | 13 (4.4) | 280 (95.6) | .001 |
| Adult cohort | 10 (1.2) | 798 (98.8) | |
| Age, y | .004 | ||
| Median | 16.8 | 40.0 | |
| Range | 2.3-63.0 | 0.0-77.0 | |
| Sex, % | 0.21 | ||
| Female | 34.8 | 48.0 | |
| Male | 65.2 | 52.0 | |
| WBC, × 109/L (N = 1071) | < .001 | ||
| Median | 146.5 | 26.2 | |
| Range | 6.1-377.6 | 0.3-510.0 | |
| FAB type, N (%) | .26 | ||
| M0 | 54 (5.0) | ||
| M1 | 3 (13.0) | 193 (17.9) | .13* |
| M2 | 3 (13.0) | 260 (24.1) | |
| M3 | 42 (3.9) | ||
| M4 | 8 (34.8) | 208 (19.3) | .01† |
| M5 | 7 (30.4) | 215 (19.9) | |
| M6 | 21 (1.9) | ||
| M7 | 11 (1.0) | ||
| RAEB/RAEB-t | 49 (4.5) | ||
| Other | 2 (0.2) | ||
| Missing | 2 (8.7) | 23 (2.1) | |
| Karyotype, N (%) | .004 | ||
| t(8;21) | 74 (6.9) | ||
| inv(16) | 91 (8.4) | ||
| t(15;17) | 39 (3.6) | ||
| 11q23 | 87 (8.1) | ||
| CN-AML | 18 (78.3) | 388 (36.0) | |
| Other | 5 (21.7) | 355 (32.9) | |
| Missing | 44 (4.1) | ||
| Mutations, N (%) | |||
| NPM1 (n = 1084) | 263 (24.8) | .006 | |
| CEBPA (n = 268)‡ | 15 (5.9) | > .99 | |
| MLL-PTD (n = 244)‡ | 6 (2.6) | > .99 | |
| FLT3-ITD (n = 1089) | 21 (91.3) | 234 (22.0) | < .001 |
| FLT3-TKD (n = 1072) | 88 (8.4) | .25 | |
| N-RAS (n = 1069) | 1 (4.3) | 146 (14.0) | .35 |
| KIT (n = 280)‡ | 20 (7.5) | .61 | |
| PTPN11 (n = 280)‡ | 4 (1.5) | > .99 | |
| WT1 (n = 680) | 9 (45.0) | 46 (7.0) | < .001 |
| IDH1 (n = 808)§ | 50 (6.3) | > .99 | |
| IDH2 (n = 808)§ | 91 (11.4) | 0.61 | |
| . | NUP98/NSD1-positive cases . | NUP98/NSD1-negative cases . | P . |
|---|---|---|---|
| Total, N (%) | 23 (2.1) | 1078 (97.9) | |
| Pediatric cohort | 13 (4.4) | 280 (95.6) | .001 |
| Adult cohort | 10 (1.2) | 798 (98.8) | |
| Age, y | .004 | ||
| Median | 16.8 | 40.0 | |
| Range | 2.3-63.0 | 0.0-77.0 | |
| Sex, % | 0.21 | ||
| Female | 34.8 | 48.0 | |
| Male | 65.2 | 52.0 | |
| WBC, × 109/L (N = 1071) | < .001 | ||
| Median | 146.5 | 26.2 | |
| Range | 6.1-377.6 | 0.3-510.0 | |
| FAB type, N (%) | .26 | ||
| M0 | 54 (5.0) | ||
| M1 | 3 (13.0) | 193 (17.9) | .13* |
| M2 | 3 (13.0) | 260 (24.1) | |
| M3 | 42 (3.9) | ||
| M4 | 8 (34.8) | 208 (19.3) | .01† |
| M5 | 7 (30.4) | 215 (19.9) | |
| M6 | 21 (1.9) | ||
| M7 | 11 (1.0) | ||
| RAEB/RAEB-t | 49 (4.5) | ||
| Other | 2 (0.2) | ||
| Missing | 2 (8.7) | 23 (2.1) | |
| Karyotype, N (%) | .004 | ||
| t(8;21) | 74 (6.9) | ||
| inv(16) | 91 (8.4) | ||
| t(15;17) | 39 (3.6) | ||
| 11q23 | 87 (8.1) | ||
| CN-AML | 18 (78.3) | 388 (36.0) | |
| Other | 5 (21.7) | 355 (32.9) | |
| Missing | 44 (4.1) | ||
| Mutations, N (%) | |||
| NPM1 (n = 1084) | 263 (24.8) | .006 | |
| CEBPA (n = 268)‡ | 15 (5.9) | > .99 | |
| MLL-PTD (n = 244)‡ | 6 (2.6) | > .99 | |
| FLT3-ITD (n = 1089) | 21 (91.3) | 234 (22.0) | < .001 |
| FLT3-TKD (n = 1072) | 88 (8.4) | .25 | |
| N-RAS (n = 1069) | 1 (4.3) | 146 (14.0) | .35 |
| KIT (n = 280)‡ | 20 (7.5) | .61 | |
| PTPN11 (n = 280)‡ | 4 (1.5) | > .99 | |
| WT1 (n = 680) | 9 (45.0) | 46 (7.0) | < .001 |
| IDH1 (n = 808)§ | 50 (6.3) | > .99 | |
| IDH2 (n = 808)§ | 91 (11.4) | 0.61 | |
WBC indicates white blood count.
P value represents FAB M1/M2 versus other FAB type.
P value represents FAB M4/M5 versus other FAB type.
Only screened in pediatric cohort.
Only screened in adult cohort.
Only 5 of the 23 NUP98/NSD1 cases showed cytogenetic aberrations (Table 1). NUP98/NSD1 was mutually exclusive with all type-II aberrations, that is, AML-characteristic fusion genes, NPM1 mutations, CEBPA double mutations, and MLL-PTD (Table 2). In contrast, NUP98/NSD1 was frequently associated with type-I aberrations: 91% of NUP98/NSD1 cases had an FLT3/ITD versus 22% of NUP98/NSD1-negative cases (P < .001). WT1 mutations were present in 9/20 (45%) NUP98/NSD1 cases versus 46/660 (7%) of NUP98/NSD1-negative cases (P < .001). One case harbored an N-RAS mutation (Tables 1–2).
Prognostic relevance of NUP98/NSD1
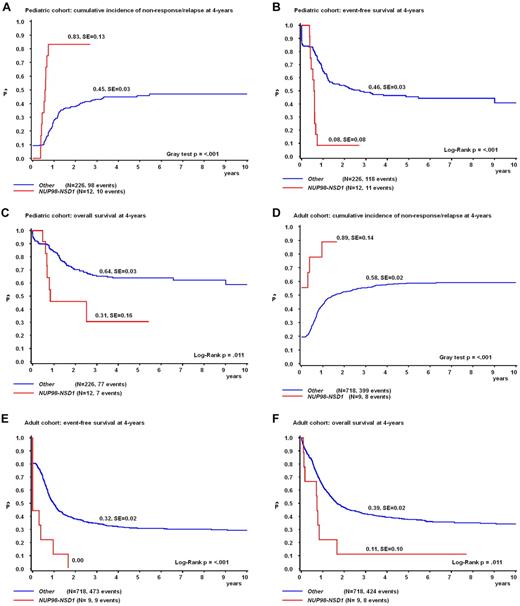
The prognostic impact was analyzed separately in the pediatric and adult cohort. The CR rate of NUP98/NSD1-positive cases in the pediatric cohort was 100%, but 10 of the 12 cases relapsed early. The pCIR at 4 years was 83 ± 13% for NUP98/NSD1-positive versus 45 ± 3% for negative cases (P < .001; Figure 4A). NUP98/NSD1 cases had a poor pEFS and pOS (4-years pEFS, 8 ± 8% vs 46 ± 3%, P < .001; and 4-years pOS, 31 ± 16% vs 64 ± 3%, P = .011 for NUP98/NSD1-positive vs negative cases, respectively; Figure 4B-C). In the adult cohort, 6 of the 10 NUP98/NSD1-positive patients never reached CR, and these cases had a significantly higher pCIR, and worse pEFS and pOS than NUP98/NSD1-negative cases (4-years CIR, 89 ± 14% vs 58 ± 2%, P < .001; 4-years pEFS, 0% vs 32 ± 2%, P < .001; and 4-years pOS, 11 ± 10% vs 39 ± 2%, P = .011, respectively; Figure 4D-F).
NUP98/NSD1 confers a poor outcome in pediatric and adult AML. Survival curves of the pediatric cohort (n = 238; A-C) and adult cohort (n = 727; D-F) depicting the cumulative incidence of nonresponse/relapse (CIR) according to the Kalbfleisch and Prentice method (A,D) and Kaplan-Meier estimates of pEFS (B,E) and pOS (C,F).
NUP98/NSD1 confers a poor outcome in pediatric and adult AML. Survival curves of the pediatric cohort (n = 238; A-C) and adult cohort (n = 727; D-F) depicting the cumulative incidence of nonresponse/relapse (CIR) according to the Kalbfleisch and Prentice method (A,D) and Kaplan-Meier estimates of pEFS (B,E) and pOS (C,F).
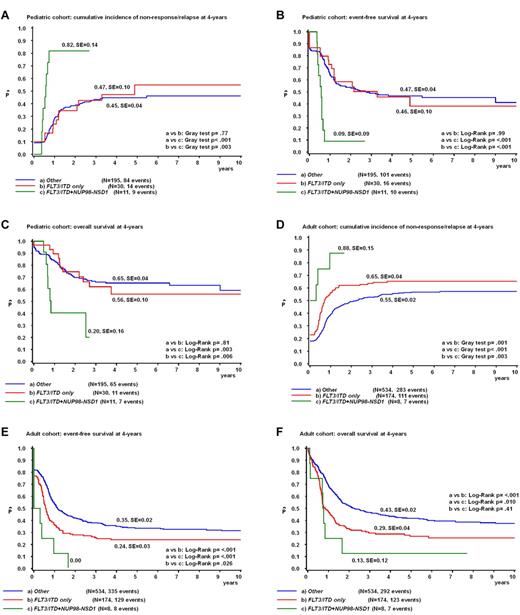
As all but 2 NUP98/NSD1-positive cases carried simultaneously an FLT3/ITD, we then analyzed the prognostic relevance of NUP98/NSD1 versus other patients carrying an FLT3/ITD. Among pediatric FLT3/ITD-positive (FLT3/ITD+) cases (n = 41) as well as adult FLT3/ITD+ cases (n = 182), NUP98/NSD1 cases had a worse outcome than the other FLT3/ITD+ cases (pediatric cohort: 4-years pCIR, 82 ± 14% vs 47 ± 10%, P = .003; 4-years pEFS, 9 ± 9% vs 46 ± 10%, P < .001; and 4-years pOS, 20 ± 16% vs 56 ± 10%, P = .006; and adult cohort: 4-years pCIR, 88 ± 15% vs 65 ± 4%, P = .003; 4-years pEFS, 0% vs 24 ± 3%, P = .026; and 4-years pOS, 13 ± 12% vs 29 ± 4%, P = .41; Figure 5A-F). When excluding cases with NPM1 mutations from the FLT3/ITD+ group, NUP98/NSD1 conferred a worse outcome than FLT3/ITD+/NPM1 wild-type cases in the pediatric cohort. The outcome of NUP98/NSD1-positive cases and FLT3/ITD+/NPM1 wild-type cases was both very poor in the adult cohort (supplemental Figure 3A-F).
NUP98/NSD1 clearly identifies a poor prognostic subgroup within pediatric and adult FLT3/ITD-positive AML. Survival curves of the pediatric cohort (A-C) and adult cohort (D-F) depicting the CIR according to the Kalbfleisch and Prentice method (A,D) and Kaplan-Meier estimates of the pEFS (B,E) and pOS (C,F) according to the NUP98/NSD1 and FLT3/ITD status.
NUP98/NSD1 clearly identifies a poor prognostic subgroup within pediatric and adult FLT3/ITD-positive AML. Survival curves of the pediatric cohort (A-C) and adult cohort (D-F) depicting the CIR according to the Kalbfleisch and Prentice method (A,D) and Kaplan-Meier estimates of the pEFS (B,E) and pOS (C,F) according to the NUP98/NSD1 and FLT3/ITD status.
When all analyses were restricted to CN-AML cases, the impact of NUP98/NSD1 among the diverse subtypes of the FLT3/ITD+ group was less clear, which may be influenced by the small numbers (supplemental Figure 4A-F).
In a multivariate Cox regression model including the variables adult versus pediatric cohort, favorable risk cytogenetics, age (> 60 years), and FLT3/ITD, NUP98/NSD1 was an independent poor prognostic factor for the probability of relapse-free survival (hazard ratio [HR], 2.6; P < .001), for pEFS (HR, 2.5; P < .001), and for pOS (HR, 1.7; P = .049; supplemental Table 4).
Regarding the other 3 NUP98-translocated cases, numbers were too small to perform outcome analyses (Table 1).
Minimal residual disease levels in NUP98/NSD1-positive patients
Of 3 NUP98/NSD1-positive patients, BM samples at morphologic remission and relapse were available. In all remission samples, we detected the NUP98/NSD1 transcript at high levels (10−1-10−3; supplemental Figure 5). In all relapse samples, NUP98/NSD1 was present at similar levels compared with diagnosis, indicating clonal stability.
Gene expression profiling
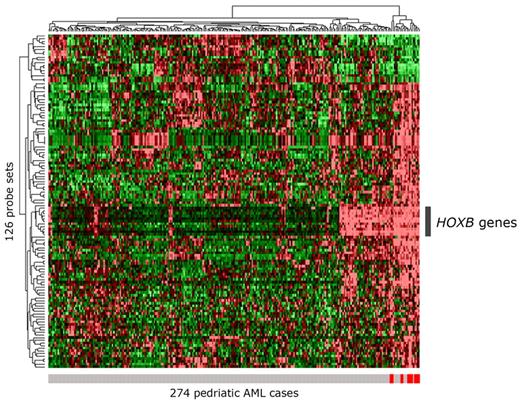
Supervised analysis of gene expression levels of 13 NUP98/NSD1 versus 261 other pediatric de novo AML cases resulted in 126 discriminative probe sets (FDR-adjusted P value < .05; supplemental Table 5A). Hierarchical clustering on these probe sets did not group the NUP98/NSD1 cases exclusively together (Figure 6). A partially similar profile was found in 46 other cases, based on highly expressed HOXB cluster genes. Fifteen of these cases carried an NPM1 mutation, known to be associated with high HOXB expression.28 Unsupervised clustering analysis showed identical results (supplemental Figure 6).
Clustering of 274 pediatric AML cases based on the 126 most discriminative probe sets for NUP98/NSD1. Hierarchical clustering of 274 pediatric AML cases based on the 126 most discriminative probe sets (FDR-adjusted P value < .05) for NUP98/NSD1 AML is presented in a heat map. The 13 NUP98/NSD1 cases are indicated with a red vertical bar below the heat map; the other AML cases are indicated with a gray vertical bar. In the heat map, cells represent relative log 2 expression values and have been color-coded on a scale ranging from bright green (−2) to bright red (+2), with black indicating no change relative to the median. Besides clustering of the NUP98/NSD1 cases, a group of other AML cases show a partly similar gene expression profile mainly based on the highly expressed HOXB probe sets, indicated by the gray bar at the right of the heat map.
Clustering of 274 pediatric AML cases based on the 126 most discriminative probe sets for NUP98/NSD1. Hierarchical clustering of 274 pediatric AML cases based on the 126 most discriminative probe sets (FDR-adjusted P value < .05) for NUP98/NSD1 AML is presented in a heat map. The 13 NUP98/NSD1 cases are indicated with a red vertical bar below the heat map; the other AML cases are indicated with a gray vertical bar. In the heat map, cells represent relative log 2 expression values and have been color-coded on a scale ranging from bright green (−2) to bright red (+2), with black indicating no change relative to the median. Besides clustering of the NUP98/NSD1 cases, a group of other AML cases show a partly similar gene expression profile mainly based on the highly expressed HOXB probe sets, indicated by the gray bar at the right of the heat map.
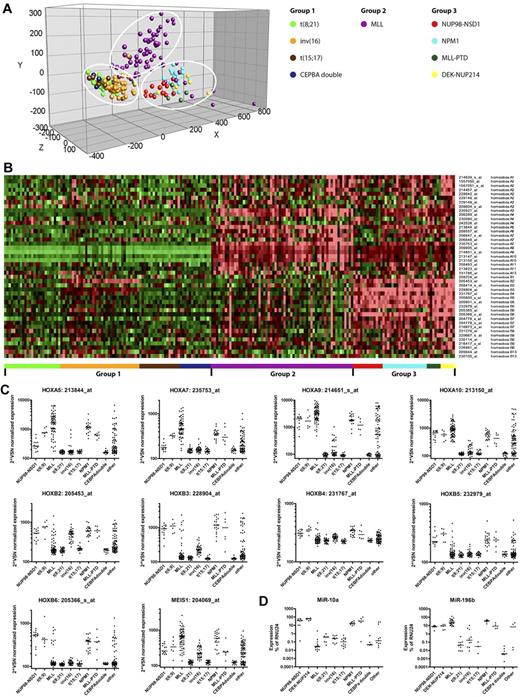
Triggered by this HOXB signature, we next investigated the expression pattern of all HOX cluster genes among AML groups characterized by specific type-II aberrations. Principal component analysis on all HOXA and -B probe sets showed distinct clustering of these AML cases in 3 different groups (Figure 7A-B). Group 1 is characterized by low or absent expression of HOXA and -B genes and included cases with AML1-ETO, CBFβ-MYH11, PML-RARA, and CEBPA double-mutant AML. Group 2 is characterized by solely high HOXA expression and represented the majority of MLL-rearranged cases. Group 3 is characterized by both high HOXA and -B expression and included cases with NPM1 mutations, cases with DEK/NUP214, and the NUP98/NSD1 cases. NUP98/NSD1 cases were characterized by high HOXA9, -A10, -B2, -B3, -B4, -B5, and -B6 expression (Figure 7C). No expression of HOXC and HOXD genes was observed. MEIS1, a well-known HOX-cofactor gene, was also expressed in NUP98/NSD1-positive cases. NUP98/NSD1-positive cases could be separated from the other “group 3 members” by lower expression of HOXA5 and -A7. Because miR-196b and -10a are transcriptionally coregulated with the HOXA and -B locus, respectively, we investigated their expression in NUP98/NSD1-AML. NUP98/NSD1 cases indeed showed high expression of miR-196b and miR-10a (Figure 7D). Combined high expression of miR-196b and miR-10a was reported previously for NPM1-mutated AML,29 and we indeed also observed high expression for NPM1-mutated cases as well as for the other “group 3” members.
Distinct expression pattern of the HOXA and -B cluster genes in pediatric AML with NUP98/NSD1. (A) Principal component analysis of pediatric AML subgroups characterized by specific type-II aberrations (n = 210) was carried out based on all HOXA and –B–annotating probe sets present on the HGU133 Plus 2.0 microarray (Affymetrix). Each color-coded circle represents an individual AML case. Three distinct groups are observed in the principal component analysis, which are indicated by the white circles. (B) Heat map showing the expression of all HOXA and B-annotating probe sets in which the cells represent relative log 2 expression values and are color-coded on a scale ranging from bright green (−2) to bright red (+2), with black indicating no change relative to the median. The pediatric AML cases are grouped together based on their specific type-II aberration as indicated by the color-coded bars below the heat map, and the probe sets are alphabetically ordered. Three groups with distinct expression patterns are observed, that is, group 1, characterized by low or absent expression of HOXA and -B genes; group 2, characterized by solely high expression of HOXA genes (mainly HOXA5-A10); and group 3, characterized by expression of both HOXA and -B genes (mainly HOXA5-A10 and HOXB2-B6). The latter included the NUP98/NSD1 cases. (C) Variance stabilization and normalization (VSN)–normalization-normalized expression levels of HOX-annotating probe sets A5, A7, A9,and A10 and B2, B3, B4, B5, and B6 and a probe set annotating MEIS1 were depicted per graph for all pediatric AML cases (n = 274) grouped by their specific type-II aberration. NUP98/NSD1 cases were characterized by high HOXA9, -A10, -B2, -B3, -B4, -B5, -B6, and MEIS1 expression. (D) Relative expression levels of miRNA-10a and -196b by stem-loop qRT-PCR in pediatric AML cases (n = 90 and n = 84, respectively), representing the different genetic subgroups in pediatric AML. Both miRNA-10a and -196b are highly expressed in the group 3-members, whereas only miRNA-196b, but not miRNA-10a is highly expressed in the majority of the MLL-rearranged cases, correlating exactly with HOXA and -B expression, respectively.
Distinct expression pattern of the HOXA and -B cluster genes in pediatric AML with NUP98/NSD1. (A) Principal component analysis of pediatric AML subgroups characterized by specific type-II aberrations (n = 210) was carried out based on all HOXA and –B–annotating probe sets present on the HGU133 Plus 2.0 microarray (Affymetrix). Each color-coded circle represents an individual AML case. Three distinct groups are observed in the principal component analysis, which are indicated by the white circles. (B) Heat map showing the expression of all HOXA and B-annotating probe sets in which the cells represent relative log 2 expression values and are color-coded on a scale ranging from bright green (−2) to bright red (+2), with black indicating no change relative to the median. The pediatric AML cases are grouped together based on their specific type-II aberration as indicated by the color-coded bars below the heat map, and the probe sets are alphabetically ordered. Three groups with distinct expression patterns are observed, that is, group 1, characterized by low or absent expression of HOXA and -B genes; group 2, characterized by solely high expression of HOXA genes (mainly HOXA5-A10); and group 3, characterized by expression of both HOXA and -B genes (mainly HOXA5-A10 and HOXB2-B6). The latter included the NUP98/NSD1 cases. (C) Variance stabilization and normalization (VSN)–normalization-normalized expression levels of HOX-annotating probe sets A5, A7, A9,and A10 and B2, B3, B4, B5, and B6 and a probe set annotating MEIS1 were depicted per graph for all pediatric AML cases (n = 274) grouped by their specific type-II aberration. NUP98/NSD1 cases were characterized by high HOXA9, -A10, -B2, -B3, -B4, -B5, -B6, and MEIS1 expression. (D) Relative expression levels of miRNA-10a and -196b by stem-loop qRT-PCR in pediatric AML cases (n = 90 and n = 84, respectively), representing the different genetic subgroups in pediatric AML. Both miRNA-10a and -196b are highly expressed in the group 3-members, whereas only miRNA-196b, but not miRNA-10a is highly expressed in the majority of the MLL-rearranged cases, correlating exactly with HOXA and -B expression, respectively.
Further exploration of the most discriminative probe sets for NUP98/NSD1 showed up-regulation of 108 and down-regulation of 18 probe sets. Besides HOXB genes, the up-regulated sets included other cancer-associated transcription factors, such as VENTX, NKX2-3, UTF1, and NFIX, and 2 annotated NRG4, encoding a ligand for epidermal growth factor receptors. Among the down-regulated probe sets were 2 annotating STK24 that induces apoptotic pathways. When restricting analyses to CN-AML (n = 54), 9 probe sets (FDR-adjusted P value < .05) were discriminative for NUP98/NSD1 (n = 10; supplemental Table 5B) and included VENTX, UTF1, and NRG4. Hierarchical clustering clearly separated NUP98/NSD1 cases as a distinct group within CN-AML (supplemental Figure 7A). NUP98/NSD1 cases also clustered together based on discriminative probe sets between NUP98/NSD1 versus other FLT3/ITD-positive cases, excluding that their expression profile is solely driven by FLT3/ITD (supplemental Figure 7B).
Discussion
In this study, we provided evidence that NUP98/NSD1 is a recurrent translocation characterizing a novel clinically relevant group of AML patients. The fusion gene was shown to result from cryptic translocations not visible by conventional karyotyping. The previously reported NUP98/NSD1 cases also were not observed in the karyogram;5,13,30-35 hence, additional molecular techniques are required to identify these patients at diagnosis. Given the detrimental prognosis, we suggest that NUP98/NSD1 analysis should be added to the panel of molecular diagnostics in AML.
NUP98/NSD1 represented a frequent event in pediatric CN-AML (16.1%), comparable with the frequency of NPM1 mutations and CEBPA double mutations in this group (Figure 3A). In adult CN-AML, NUP98/NSD1 was less frequent (2.3%). This age-dependent frequency resembles core-binding factor AML that also occurs more frequently in children. Interestingly, both NUP98-HOXA9 and DEK/NUP214 are also typically associated with a younger age.36,37
NUP98/NSD1 was mutually exclusive with other type-II aberrations. Wang et al38 demonstrated that NUP98/NSD1 inhibited cellular differentiation, establishing NUP98/NSD1 as a type-II aberration. FLT3/ITD was present in the majority of NUP98/NSD1 cases, suggestive of a novel nonrandom association between type-I and -II aberrations. Furthermore, 45% of NUP98/NSD1 cases also harbored a WT1 mutation, although the exact role of WT1 mutations in leukemogenesis is unresolved. Recent evidence suggests that other NUP98 fusions also are frequently associated with WT1 mutations,36,39 and we showed previously that 33% of DEK/NUP214 cases also harbored WT1 mutations.17 This makes it conceivable that WT1 mutations have an additive function in NUP98- and NUP214-rearranged leukemogenesis.
NUP98/NSD1 was identified as an independent factor for dismal clinical outcome. Despite the current intensive treatment regimens, AML cases with NUP98/NSD1 were either refractory to induction chemotherapy or relapsed within 1 year of diagnosis. Four-year pEFS rates were < 10% for both pediatric and adult cases. The largest study on NUP98-rearrangements to date (n = 11) also reported poor outcome for NUP98-HOXA9 AML cases.36 Within the unfavorable FLT3/ITD+ AML subgroup, cases with NUP98/NSD1 did worse than patients carrying FLT3/ITD without NUP98/NSD1. We further subdivided FLT3/ITD+ AML cases according to the presence of an NPM1 mutation. After excluding NPM1-mutated cases, NUP98/NSD1-positive cases did equally poor as FLT3/ITD+/NPM1 wild-type cases in adult AML but still significantly worse in pediatric AML. Of note, within CN-AML, the impact of NUP98/NSD1 among the diverse subtypes of the FLT3/ITD+ group was not clear, which might be limited by the small numbers. Three investigated NUP98/NSD1 cases showed high minimal residual disease levels, correlating with the early relapses in these patients. Novel therapeutic strategies are urgently needed for this therapy resistant patient group.
Knowledge of the underlying biology of NUP98/NSD1 is important because it may identify novel therapeutic targets. Our expression profiles showed up-regulation of oncogenic transcription factors such as VENTX,40 and of NRG4, encoding an epidermal growth factor receptor ligand involved in proliferation41 and down-regulation of the proapoptotic gene STK24.42 Moreover, NUP98/NSD1-positive AML showed a distinct HOXA and -B expression signature and concomitant high miR-196b and miR-10a expression. This HOX activation pattern was distinct from MLL-rearranged cases that were characterized by HOXA activation only. NUP98/NSD1 cases resembled the HOX expression pattern of AML with NPM1 mutations DEK/NUP214 and MLL-PTD; however, they could be discriminated by lower HOXA5 and -A7 expression. NUP98-homeobox fusions bind DNA through the homeodomain of the fusion partner. Recruitment of CREBBP/p300 by the GLFG-repeats of NUP98 results in histone acetylation and subsequent transcriptional activation of target genes.43 NSD1 does not possess a homeodomain, but Wang et al38 reported that PHD fingers I to IV of NSD1 allowed binding to the HOXA7 and -A9 promoter. Binding capacity to promoters in the HOXB cluster was not reported. We observed high HOXA9 and -A10 expression in NUP98/NSD1 patient samples but did not observe high HOXA5 and -A7 expression, as seen in NUP98/NSD1-transfected progenitors by Wang et al38 As transforming capacities of some HOXA and -B genes are established,44 it would be of interest to investigate the mechanism and additive effect of high HOXB expression in AML with NUP98/NSD1. Importantly, Wang et al38 linked H3K36-methyltransferase activity of the SET domain of NSD1 to HOXA activation in NUP98/NSD1-transfected progenitors. Therefore, novel therapeutic options might arise from epigenetic studies, because NUP98/NSD1 showed to exert its leukemogenic function through 2 histone-modifying activities, that is, H3K36-methyltransferase activity and histone acetylation activity of the recruited CREBBP/p300-complex.38 The latter is probably present in all NUP98 fusions, because the GLFG-repeats, preserved in all NUP98 translocations, recruit the CREBBP/p300-complex. Therefore, specific histone acetyltransferase inhibitors might be potentially effective in NUP98-rearranged AML.45
Recently, Takeda et al46 suggested a novel mechanism by which NUP98 fusions dysregulate transcription. They showed that NUP98/HOXA9 and NUP98/DDX10 inhibited CRM1-dependent nuclear export, resulting in nuclear entrapment of transcriptional regulators, and thereby enhanced transcription of their downstream targets. We showed that NUP98/NSD1 aberrantly localized in nuclear aggregates, suggesting that this mechanism also may apply for NUP98/NSD1.
In conclusion, the cryptic NUP98/NSD1 translocation defines a previously unrecognized group of young AML patients with dismal outcome. Routine screening for NUP98/NSD1 at diagnosis will be essential for proper identification and stratification of these patients.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs J. P. P. Meijerink and R. W. Stam for providing material from pediatric acute lymphoblastic leukemia cases with high HOX-expression.
I.H.I.M.H. has been supported by a grant from the Pediatric Oncology Foundation Rotterdam (KOCR) foundation, Rotterdam, The Netherlands. M.P. has been supported by a grant (CM08/00027) from Instituto de Salud Carlos III (ISCIII), Spanish Ministry of Health, Spain.
National Institutes of Health
Authorship
Contribution: I.H.I.M.H., M.M.v.d.H.-E., R.P., and C.M.Z. designed the study; E.S., G.-J.J.L.K., J.T., A.B., D.R., U.C., and P.J.M.V. made this research possible by collecting patient samples and clinical data; I.H.I.M.H., S.T.C.J.M.A.-P., M.P., S.A., J.E.K., and J.F.G. performed the laboratory research; I.H.I.M.H., M.M.v.d.H.-E., H.B.B., P.J.M.V., and C.M.Z. analyzed and interpreted the data; M.Z. performed statistical analysis; I.H.I.M.H., M.M.v.d.H.-E., and C.M.Z. wrote the manuscript; and all authors critically reviewed the manuscript and gave their final approval.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: C. Michel Zwaan, Department of Pediatric Oncology/Hematology, Erasmus MC-Sophia Children's Hospital, Dr Molewaterplein 60, 3015 GJ Rotterdam, The Netherlands; e-mail: c.m.zwaan@erasmusmc.nl.

![Figure 1. Discovery of NUP98/NSD1 fusion in 2 CN-AML cases. (A) Array-CGH profiles of chromosome 11p15 showing the ratio of tumor DNA/control DNA (blue tracing) versus the inverted experiment (red tracing). The profile of patient 3495 (top panel) shows a part of the 0.4-Mb duplication involving the 5′ part of NUP98 (indicated by the blue vertical arrow), and the profile of patient 4716 shows the 0.1-Mb deletion involving the 3′ part of NUP98 (indicated by the red vertical bar). The horizontal arrows indicate the bar representing the NUP98 gene. (B) Split-signal FISH analysis of NUP98 for patients 3495 and 4716 using a partly overlapping green and red probe located in NUP98 (cen indicates probe situated more centromeric; tel indicates probe situated more telomeric. (C) RT-PCR analysis using NUP98- and NSD1-specific primers, and GAPDH primers as internal control, reveals a specific NUP98/NSD1 fusion transcript in patients 3495 and 4716. The reciprocal NSD1-NUP98 transcript was not detected in both patients (pos indicates positive control, [patient 5007]; neg indicates negative control [normal bone marrow]; ntc indicates non-template control). (D) Sequence analysis confirmed an identical in-frame fusion between NUP98 exon 12 and NSD1 exon 6 in both patients. (E) The NUP98/NSD1 fusion protein will harbor the GLFG-repeats of NUP98, and among others the PHD fingers and SET domain of NSD1. (F) Dual-color FISH analysis using a green probe for NUP98 and a red probe for NSD1 confirmed the fusion of NUP98 and NSD1 at the chromosomal level.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/13/10.1182_blood-2011-04-346643/4/m_zh89991178620001.jpeg?Expires=1769096134&Signature=jx2FhT6hX5ji5~M59711j8js9~c2YdF1Ln21AKQTXUlGNwSAPGbRGbw4yw091ONwDKlwLaT6CVQt0lHgubN9vm1gc9-buiheBZGYqYBzFO7OX1hBK4blF2xph7nOdIKTby3c4l~-0j87stPjejraphCWtxDGkksde4wk8AjvBXDMcM7ImLc5X6h~1haoHyTxql~SAEf~jhqVOl8RMtHvOODbF0Q6qlTswekspICDClXNhpw3bzFWxiEP44~u5496bTje~R43Fyy2ISXq-c0eN0tFs9eKlVR4bGvEaxEVE78WTQjziN8s3KmoYzO3HZ947TeH5-ogbezWezGDoispnQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal