Abstract
GVHD is still one of the major complications after allogeneic stem cell transplantation. Whereas murine data have clearly shown the beneficial effects of regulatory T cells (Tregs) on the prevention of GVHD, data from the human system are rare. Here, we present a comparative dynamic analysis of CD4+CD25hiCD127lo/− Tregs from patients with and without GVHD analyzing the whole genome profile over the first 6 months after stem cell transplantation, representing the most sensitive time window for tolerance induction. The Treg transcriptome showed a high stability. However, the comparison of Treg transcriptomes from patients with and without GVHD uncovered regulated gene transcripts highly relevant for Treg cell function. The confirmative protein analyses demonstrated a significantly higher expression of granzyme A, CXCR3, and CCR5 in Tregs of immune tolerant patients. These results point to a reduced suppressive function of Tregs from GVHD patients with diminished migration capacity to the target organs.
Introduction
Despite improvements in allogeneic stem cell transplantation (SCT), graft-versus-host disease GVHD is still one of the most serious complications with a high morbidity and mortality rate.1
Even though just accounting for 5% of the total CD4+ T-cell population,2 CD4+CD25hi T cells are critical regulators in the induction and maintenance of peripheral immune tolerance and have therefore been explored for their suppressive capability in experimental GVHD models. Removal of these regulatory T cells (Tregs) from the donor allograft accelerates GVHD, whereas the adaptive transfer of Tregs inhibits the allogeneic immune response.3-6
In recent years, surface molecules, transcription factors, and soluble molecules have been identified that are highly relevant for the induction of in vivo tolerance by Tregs.7 The hyaluronan receptor CD44 identifies a Treg cell population with highly suppressive capacity.8-10 Galectin-1 (LGALS1) is preferentially expressed in Tregs11 and has been described as a master regulator in cell homeostasis.12 Furthermore, the perforin/granzyme pathway is used by Tregs to control immune responses.13 The presence of receptors for inflammatory chemokines, such as CCR2, CXCR3, CCR4, CCR5, and CCR8, on regulatory T cell subsets14,15 suggests that Tregs are able to enter inflamed tissues and play an important role in experimental GVHD models.16-21
Recently, hematopoietic SCT qualified as first venue for donor Treg cell infusion.22,23 However, our knowledge on the complex immune regulatory role of Tregs in allogeneic SCT is incomplete, and it is still unknown why Tregs obviously fail to induce immune tolerance in patients developing a GVHD. We therefore performed a dynamic approach studying the transcriptome of Tregs in the most sensitive period of immune tolerance induction and compared patients with and without GVHD.
Methods
Patients' characteristics and healthy controls
The study was approved by the institutional ethics committee of Hannover Medical School. From March 2007 to December 2010, a total of 270 patients were transplanted at Hannover Medical School and 141 patients have given their written informed consent to participate in the study, in accordance with the Declaration of Helsinki. Blood samples of 80 mL were obtained every 4 weeks within the first 6 months. Inclusion criteria for further analysis were allogeneic peripheral blood stem cell transplantation, donor cell chimerism of mononucleated cells > 98%, no active infections (ie, cytomegalovirus), sufficient cell numbers of freshly FACS-sorted Tregs (> 30 000) with a purity > 95%. Strict inclusion criteria were defined to avoid bias because of the stem cell source (bone marrow vs peripheral blood stem cell transplantation) and different origin of Tregs (donor/host chimerism). Patients with active infections (ie, cytomegalovirus) have been excluded with respect to the possibility of Treg induction or activation by inflammatory cytokines produced in response to pathogen recognition by cells of the innate immune system. From 141 patients given their informed consent, only 98 patients fulfilled aforementioned inclusion criteria and have been analyzed in this study. Only in 10 of the 141 participating patients, the exclusion from further analysis was done because of low cell numbers. In the primary Treg transcriptome study, only patients with histologically confirmed, severe (grade III/IV) acute GVHD (aGVHD) before therapeutic steroid application were included. In the following studies, also patients with mild (grade 1/II) aGVHD were analyzed. Patients with limited and extensive chronic GVHD (cGVHD)were also included. aGVHD was staged according to modified Glucksberg criteria.24 Treg transcriptome studies (n = 26), miRNA profiling (n = 6), real-time RT-PCR experiments (n = 39), and protein analyses by flow cytometry (n = 27) were performed in independent patients at different time points. Equal amounts of RNA were pooled if this was necessary because of the low cell numbers. Healthy, age- and sex-matched persons were analyzed as controls (n = 33). All patients received a GVHD prophylaxis with cyclosporine and mycophenolate mofetil. In some patients a short course of methotrexate was additionally given as GVHD prophylaxis. Patient characteristics are given in Table 1.
Patient characteristics
| Characteristic/category . | Treg transcriptome study . | Confirmatory analyses at gene-expression and protein level . | |
|---|---|---|---|
| Primary . | Confirmatory . | ||
| No. of patients | 6 | 20 | 72 |
| Sex, female/male | 5/1 | 13/7 | 29/43 |
| Age, y | |||
| Median (range) | 52 (26-66) | 53 (19-66) | 50 (21-72) |
| Diagnosis | |||
| AML | 3 | 10 | 31 |
| ALL | 1 | 3 | 9 |
| MDS | 2 | 7 | 26 |
| NHL | — | — | 6 |
| Donor type | |||
| MRD | 4 | 10 | 29 |
| MUD | 2 | 10 | 43 |
| Donor match | |||
| Match/mismatch | 5/1 | 18/2 | 59/12 |
| Conditioning regimen | |||
| Flu/Cy | — | — | 4 |
| Flu/Mel | — | — | 2 |
| FLAMSA-like | 3 | 12 | 29 |
| ClaraC/TBI | 1 | 3 | 13 |
| Bu/Cy | 1 | 2 | 13 |
| TBI/Cy | 1 | 3 | 11 |
| No GVHD | 3 | 8 | 25 |
| GVHD | 3 | 12 | 47 |
| Acute | 1 | 4 | 19 |
| Acute/chronic | 2 | 3 | 22 |
| Chronic | — | 5 | 6 |
| GVHD grade | |||
| Acute | |||
| I | — | 2 | 9 |
| II | — | — | 4 |
| III | 1 | 1 | 2 |
| IV | 1 | 1 | 4 |
| Chronic | |||
| Limited | — | 4 | 4 |
| Extensive | — | 1 | 2 |
| Acute/chronic | |||
| I: limited | — | 2 | 7 |
| I: extensive | — | — | 3 |
| II: limited | — | — | 4 |
| II: extensive | — | 1 | 3 |
| III: limited | — | — | 1 |
| III: extensive | 1 | — | 2 |
| IV: limited | — | — | 1 |
| IV: extensive | — | — | 1 |
| Characteristic/category . | Treg transcriptome study . | Confirmatory analyses at gene-expression and protein level . | |
|---|---|---|---|
| Primary . | Confirmatory . | ||
| No. of patients | 6 | 20 | 72 |
| Sex, female/male | 5/1 | 13/7 | 29/43 |
| Age, y | |||
| Median (range) | 52 (26-66) | 53 (19-66) | 50 (21-72) |
| Diagnosis | |||
| AML | 3 | 10 | 31 |
| ALL | 1 | 3 | 9 |
| MDS | 2 | 7 | 26 |
| NHL | — | — | 6 |
| Donor type | |||
| MRD | 4 | 10 | 29 |
| MUD | 2 | 10 | 43 |
| Donor match | |||
| Match/mismatch | 5/1 | 18/2 | 59/12 |
| Conditioning regimen | |||
| Flu/Cy | — | — | 4 |
| Flu/Mel | — | — | 2 |
| FLAMSA-like | 3 | 12 | 29 |
| ClaraC/TBI | 1 | 3 | 13 |
| Bu/Cy | 1 | 2 | 13 |
| TBI/Cy | 1 | 3 | 11 |
| No GVHD | 3 | 8 | 25 |
| GVHD | 3 | 12 | 47 |
| Acute | 1 | 4 | 19 |
| Acute/chronic | 2 | 3 | 22 |
| Chronic | — | 5 | 6 |
| GVHD grade | |||
| Acute | |||
| I | — | 2 | 9 |
| II | — | — | 4 |
| III | 1 | 1 | 2 |
| IV | 1 | 1 | 4 |
| Chronic | |||
| Limited | — | 4 | 4 |
| Extensive | — | 1 | 2 |
| Acute/chronic | |||
| I: limited | — | 2 | 7 |
| I: extensive | — | — | 3 |
| II: limited | — | — | 4 |
| II: extensive | — | 1 | 3 |
| III: limited | — | — | 1 |
| III: extensive | 1 | — | 2 |
| IV: limited | — | — | 1 |
| IV: extensive | — | — | 1 |
GVHD grade was determined according to the Glucksberg criteria.
AML indicates acute myeloid leukemia; ALL, acute lymphatic leukemia; MDS, myeloid dysplastic syndrome; NHL, non-Hodgkin lymphoma; —, not applicable; MRD, matched related donor; MUD, matched unrelated donor; Flu, fludarabine; Cy, cyclophosphamide; Mel, melphalan; FLAMSA-like, fludarabine, cyrarabine, amsacrine44 ; ClaraC, clofarabine cytarabine; TBI, total body irradiation.; and GVHD, graft-versus-host disease.
Collection of peripheral blood samples and isolation of Tregs
Heparinized blood samples of 80 mL were obtained from patients every 30 days (± 5 days) after transplantation for 6 months (30, 60, 90, 120, 150, and 180 days). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation. Subsequently, T cells were stained with anti-human CD25 phycoerythrin (Miltenyi Biotec), anti-human CD14 allophycocyanin, anti-human CD4 fluorescein isothiocyanate, and anti-human CD127 Alexa-647 antibodies (BD Biosciences). CD4+CD25hi T cells were sorted immediately after isolation using FACSAria II (BD Biosciences) or MoFlow (Beckman Coulter). Cells were in addition negatively gated on CD14 allophycocyanin and reanalyzed for low expression of CD127 Alexa-647 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Purity of isolated CD4+CD25hiCD127lo/− Tregs was > 95%. After cell sorting, T cells were centrifuged and the pellet was resolved in RLT buffer (QIAGEN) according to the manufacturer's protocol. For additional flow cytometry studies, PBMCs were frozen at −70°C until analysis.
Affymetrix gene chip assays
Quality and integrity of the total RNA isolated from at least 30 000 sorted CD4+CD25high T cells were controlled by Agilent 2100 Bioanalyzer (Agilent Technologies). For the primary Treg transcriptome study, RNA of patients without GVHD (n = 3) and with histologically confirmed acute (III/IV)/cGVHD (n = 3) was pooled in equal ratios according to concentration and quality. Pooling was required because of limited Treg cell numbers in lymphocytopenic patients. For the confirmatory Treg transcriptome study, RNA of patients without GVHD (n = 8) and with acute (I-IV)/cGVHD (n = 12) was pooled. Biotin-labeled target synthesis was started from 5 μg of total RNA. The following reactions were done as previously described.25
Data analysis
All Affymetrix microarray data were RMA normalized. For further downstream analysis, Agilent GeneSpring GX 11 software (Agilent Technologies) was applied. In the primary Treg transcriptome study, signal intensities of 2 replicates were compared at different time points after hematopoietic SCT. Relative gene expression was determined by normalized intensity values. GeneSpring analysis was performed using the Treg transcriptome data of the following time points and clinical subgroups: no GVHD day 90, no GVHD day 150, GVHD day 90, aGVHD manifestation, GVHD day 150, and cGVHD manifestation (Figure 1A). Data from the primary Treg transcriptome study were confirmed by experiments analyzing additional 20 patients: without GVHD (n = 8 in 3 different pools) and with GVHD (n = 12 in 4 different pools) at 2 different time points (< 100 days and > 100 days). As demonstrated in Figure 1B, the following microarray analyses were done and resulting datasets comparatively analyzed: no GVHD < 100 days (array B), no GVHD > 100 days (arrays D and E), GVHD < 100 days (arrays H and I), and GVHD > 100 days (arrays L and M). In addition, the Treg transcriptome of healthy controls was analyzed to identify whether genes were differentially overexpressed or repressed in GVHD/immune tolerant patients: controls were age- and sex-matched to the primary transcriptome study resulting in 2 arrays (n = 3 independent controls/array). Student t test and 1-way ANOVA were used to identify significant expression changes. Cut-off was a transcript fold change of ≥ 2 or ≤ −2 in at least 1 comparison. The entire dataset was deposited in MIAME compliant format at the GEO database (http://ncbi.nlm.nih.gov/geo; accession number GSE 23332).
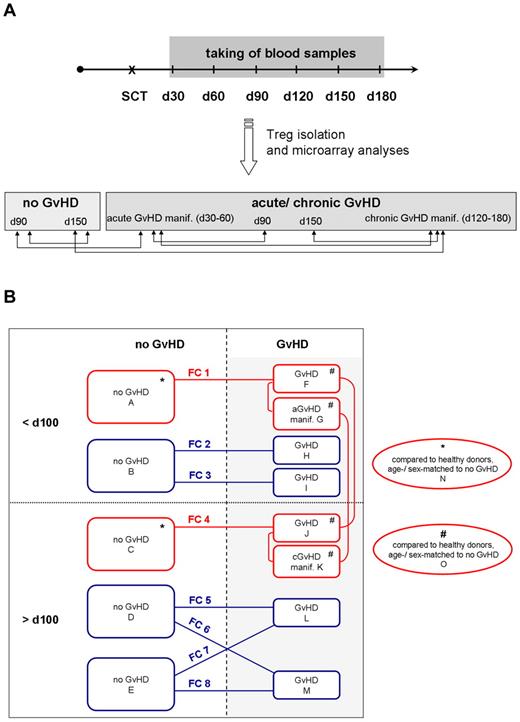
Design of Treg transcriptome studies. (A) Primary Treg transcriptome study. Blood samples were taken at regular intervals at 30, 60, 90, 120, 150, and 180 days after SCT, and CD4+CD25hiCD127lo/− Tregs were isolated by FACS. Samples were assigned to the no GVHD or aGVHD/cGVHD group according to clinical and histologic diagnoses and analyzed by microarray technology. Comparative analysis of Treg transcriptomes was performed as indicated by the arrows. (B) Primary and confirmatory Treg transcriptome studies. Microarrays of the primary Treg transcriptome study are red framed, analyzing patients in the no GVHD group at day 90 (array A) and day 150 (array C), in the GVHD group at day 90 (array F), with aGVHD manifestation (array G), at day 150 (array J), and with cGVHD manifestation (array K). Red lines demonstrate comparisons of the primary Treg transcriptome study, which also included a comparative analysis with healthy, age- and sex-matched donors (Table 2). Confirmatory microarray experiments are blue-framed, analyzing patients in the no GVHD group before 100 days (array B) and after 100 days (arrays D and E), in the GVHD group before 100 days (arrays H and I) and after 100 days (arrays L and M). In each array, CD4+CD25hiCD127lo/− Tregs with a purity more than 95% of n = 3 independent patients with exception of array D (n = 2) were analyzed. FC1-FC8 indicates the comparison of 2 arrays and resulting fold change values are summarized in Table 3.
Design of Treg transcriptome studies. (A) Primary Treg transcriptome study. Blood samples were taken at regular intervals at 30, 60, 90, 120, 150, and 180 days after SCT, and CD4+CD25hiCD127lo/− Tregs were isolated by FACS. Samples were assigned to the no GVHD or aGVHD/cGVHD group according to clinical and histologic diagnoses and analyzed by microarray technology. Comparative analysis of Treg transcriptomes was performed as indicated by the arrows. (B) Primary and confirmatory Treg transcriptome studies. Microarrays of the primary Treg transcriptome study are red framed, analyzing patients in the no GVHD group at day 90 (array A) and day 150 (array C), in the GVHD group at day 90 (array F), with aGVHD manifestation (array G), at day 150 (array J), and with cGVHD manifestation (array K). Red lines demonstrate comparisons of the primary Treg transcriptome study, which also included a comparative analysis with healthy, age- and sex-matched donors (Table 2). Confirmatory microarray experiments are blue-framed, analyzing patients in the no GVHD group before 100 days (array B) and after 100 days (arrays D and E), in the GVHD group before 100 days (arrays H and I) and after 100 days (arrays L and M). In each array, CD4+CD25hiCD127lo/− Tregs with a purity more than 95% of n = 3 independent patients with exception of array D (n = 2) were analyzed. FC1-FC8 indicates the comparison of 2 arrays and resulting fold change values are summarized in Table 3.
cDNA synthesis and quantitative RT-PCR
RNA isolation was performed as described previously26 from FACS-sorted Tregs. Isolated mRNA was reverse-transcribed using 200 U Superscript II (Invitrogen), oligo dT, and random hexamer primers (Invitrogen). The resulting cDNAs from the patients were pooled (n ≥ 3) because of limited availability of cell numbers and to obtain sufficient cDNA for subsequent real-time RT-PCR experiments. More than 5 candidate genes were analyzed with one cDNA pool: no GVHD patients (n = 12) in 4 cDNA pools, GVHD patients (n = 27) in 9 cDNA pools, and healthy controls (n = 9) in 3 cDNA pools. Primer sequences are presented in supplemental Table 1. Quantitative real-time RT-PCR was performed with the GeneAmp 7500 Sequence Detection System (ABI) using Brilliant SYBR Green QPCR Core Reagent Kit (Stratagene). Relative mRNA levels were determined using standard curves for each gene, and gene expression was performed in relation to the expression of the housekeeping genes RPS9 and GAPDH, respectively.
Treg cell phenotyping by flow cytometry
PBMCs (1 × 106 cells) were thawed, cultured overnight in standard cell culture media, and stained with monoclonal antibodies against CD4, CD25, Foxp3, CD127, CXCR3, CCR5, CCR1, GZMA, LAG3, and CD44. Phenotyping was done on a FACSCanto (BD Biosciences). The resulting data were further analyzed with FlowJo software (TreeStar) and calculated as percentage of CD4+CD25hiCD127lo/−Foxp3+ T cells.
Micro RNA profiling and data analysis
Total RNA from patients without (n = 3) and with GVHD (n = 3) before and after 100 days after SCT was extracted from freshly isolated CD4+CD25hiCD127lo/− Tregs using the miRNeasy Mini Kit (QIAGEN). In addition, cells from age- and sex-matched healthy controls (n = 3 for each group) were prepared accordingly. The resulting samples of equally pooled miRNA were analyzed by Febit Biomed applying the Geniom Biochip MPEA homo sapiens as described previously.27 Raw and normalized data are available at the GEO database (http://ncbi.nlm.nih.gov/geo; accession number GSE 26769). Data files containing logqmedian values for the different comparisons were analyzed using Genesis software (Institute for Genomics and Bioinformatics, Graz University of Technology, Austria). The logqmedian values were normalized and hierarchically clustered. Predicted targets of miRNAs were identified according to the mirBase database.
DNA demethylation analysis of the foxp3 gene locus in gut biopsies
Gut biopsies from patients clinically suspected to have developed intestinal GVHD were taken during endoscopic examinations and directly frozen with RNA later (Ambion) in liquid nitrogen. DNA was isolated from frozen intestinal biopsies (2 biopsies per patient) using the Qia-Amp DNA tissue kit (QIAGEN) according to the manufacturer's protocol. The analysis was performed in patients with cGVHD (n = 6; 4 females/2 males), aGVHD (n = 8; 2 females/6 males), and without any pathologic signs of GVHD (n = 4; 3 females/1 male). DNA demethylation analysis of foxp3 was carried out as previously described.28 In patients with female stem cell donors, the results were corrected with a factor of 2 because 1 of the 2 Treg specific demethylation region alleles is methylated as a result of X-inactivation. For this analysis, only patients with a donor cell chimerism > 98% were included.
Results
Stable Treg transcriptome in the early phase of allogeneic SCT
To investigate the dynamic changes of Treg transcriptomes during reconstitution of the immune system and in relation to the development of GVHD, comparative gene-expression profiling was performed in FACS-sorted CD4+CD25hiCD127lo/− Tregs from peripheral blood of transplanted patients without GVHD and at selected time points of aGVHD and cGVHD manifestation as well as at 3 and 5 months after SCT (day 90 and day 150, respectively). This primary Treg transcriptome study only included highly selected and well-defined patients with histologically confirmed severe aGVHD fulfilling predefined criteria24 to avoid bias. Microarray data revealed 46 076 transcripts with normalized intensity values of at least 20th percentile in one of the raw data samples. Because of the high dimensionality of the data with respect to the chronologic sequence and disease manifestation, principal component analysis was used to allow a more comprehensive overview. As shown in Figure 2, the principal component analysis revealed a strong correlation between the 6 Treg transcriptomes: the transcriptome data of patients without GVHD at day 90 and day 150 as well as for the GVHD group at day 90 and at the time of aGVHD highly correlated. In addition, the transcriptome at the time point of clinically apparent cGVHD showed a high correlation with the transcriptome of patients in the GVHD group at day 150 after SCT. These strong correlations point to a distinct molecular stability of Tregs in the chronologic sequence which in the early phase after SCT appears to be restricted to the respective group of patients with or without GVHD.
Transcriptome stability of regulatory T cells in SCT patients. Three-dimensional principal component analysis of the primary Treg transcriptome study and resulting 6 transcriptomes. Closely related transcriptomes are marked by loops. no GVHD d90 indicates Treg transcriptome analyzed on day 90 after SCT of patients with and without developing a GVHD; no GVHD d150, Treg transcriptome analyzed on day 150 after SCT of patients (without) developing a GVHD; aGVHD manif, Treg transcriptome of GVHD patients analyzed at the time point of clinical manifestation of the acute form of GVHD; and cGVHD manif, Treg transcriptome of GVHD patients analyzed at the time point of clinical manifestation of the chronic form of GVHD.
Transcriptome stability of regulatory T cells in SCT patients. Three-dimensional principal component analysis of the primary Treg transcriptome study and resulting 6 transcriptomes. Closely related transcriptomes are marked by loops. no GVHD d90 indicates Treg transcriptome analyzed on day 90 after SCT of patients with and without developing a GVHD; no GVHD d150, Treg transcriptome analyzed on day 150 after SCT of patients (without) developing a GVHD; aGVHD manif, Treg transcriptome of GVHD patients analyzed at the time point of clinical manifestation of the acute form of GVHD; and cGVHD manif, Treg transcriptome of GVHD patients analyzed at the time point of clinical manifestation of the chronic form of GVHD.
Expression profiling of circulating Tregs in patients with and without GVHD
Primary Treg transcriptome study.
Among the 46 076 gene transcripts, 3159 were expressed in the primary Treg transcriptome study with a ≥ 2 or ≤ −2 fold change for at least 1 of the predefined 6 comparisons (Figure 1A). Gene ontology analysis is shown in Figure 3A. Focusing on the molecular functions, the majority of gene transcripts (74.6%) encodes for binding-associated molecules. Furthermore, regulated gene transcripts belong to cellular (27.2%) and metabolic processes (17.1%) or to the immune system (8.2%). Further analyses were performed with emphasis on binding-associated molecules, representing the largest part in molecular function, and on immune responses because GVHD can be regarded as an allogeneic immune response. A Venn diagram analysis of the primary Treg transcriptomes was performed, which resulted in a set of 161 differentially expressed genes that clearly separated Tregs from patients with and without GVHD independent of the time point (Figure 3B). A selection of gene transcripts with the most prominent changes, including a comparative analysis of the Treg transcriptome from healthy controls, is shown in Table 2. Most strikingly, Tregs isolated from patients with severe aGVHD showed a down-regulation of molecules related to migration/homing of T cells to inflamed tissue and secondary lymphoid organs (ie, CCR5, CXCR3, CCR3, CXCR6, and CCR1). Interestingly, CCR5, CCR3, and CCR1 were significantly reduced at the transcriptional level in Tregs from GVHD patients early after SCT (< 100 days), whereas the gene transcripts of CXCR3 and CXCR6 were down-regulated before and after 100 days. The gene expression of LAG3, CD44, LGALS1, and granzyme A, which represent molecules associated with the suppressive function of Tregs, was reduced in the GVHD group early (< 100 days) and late (> 100 days) after SCT. Among others, cyclin-dependent kinase inhibitor 2B (CDKN2B, p15) and cyclin-dependent kinase 6 (CDK6) were up-regulated at the transcriptional level in patients developing a GVHD independent of the time. This is in contrast to immune tolerant patients in whom CD44, LGALS1, granzyme A, and migratory chemokine receptors CCR3, CCR1, and CXCR3 were up-regulated. The comparison with the Treg transcriptome of healthy controls uncovered that the suppressive molecules CD44, LGALS1, granzyme A, and migratory chemokine receptors CCR3, CCR1, and CXCR3 were up-regulated in Tregs of immune tolerant patients compared with nontransplanted, healthy controls. Most strikingly, CCR1 was > 10-fold higher expressed in immune tolerant patients than in nontransplanted healthy controls (Table 2). The differential analysis within the GVHD group (Table 3) demonstrated that the gene expression of CD44 and granzyme A was reduced to the same amount at different time points, including the time of GVHD manifestation. While being down-regulated at the time point of aGVHD compared with day 90 in the GVHD group, the expression levels of the other candidate genes were still higher (CCR1) or comparable (ie, LAG3, CCR5, CCR3, and CXCR3) to healthy donors. When looking at the immune tolerant group, it is obvious that migratory chemokine receptors were most prominently up-regulated in the early phase (CXCR6, CCR5, CCR3, and CXCR3).
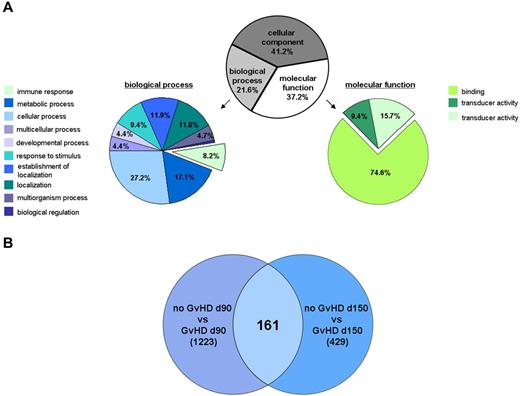
Primary Treg transcriptome study. (A) Gene ontology analysis of regulated gene transcripts in GVHD. Analysis was performed using the GeneSpring GX11 software on the 3159 transcripts with a fold change ≥ 2 or ≤ −2 for at least 1 of the predefined 6 comparisons (“Data analysis”) and a P value < .1. (B) Venn diagram analysis of the Treg transcriptomes based on the 3159 transcripts with a fold change ≥ 2 or ≤ −2 for at least 1 of the 6 comparisons (“Data analysis”). The differential analysis of the Treg transcriptomes from patients at day 90 and day 150, respectively, with and without developing a GVHD after SCT revealed, n = 1233 and n = 429, respectively, regulated gene transcripts. Independent of the analyzed time point, the differential Treg transcriptome analysis of patients with and without developing a GVHD identified n = 161 regulated gene transcripts with a fold change ≥ 2 or ≤ −2.
Primary Treg transcriptome study. (A) Gene ontology analysis of regulated gene transcripts in GVHD. Analysis was performed using the GeneSpring GX11 software on the 3159 transcripts with a fold change ≥ 2 or ≤ −2 for at least 1 of the predefined 6 comparisons (“Data analysis”) and a P value < .1. (B) Venn diagram analysis of the Treg transcriptomes based on the 3159 transcripts with a fold change ≥ 2 or ≤ −2 for at least 1 of the 6 comparisons (“Data analysis”). The differential analysis of the Treg transcriptomes from patients at day 90 and day 150, respectively, with and without developing a GVHD after SCT revealed, n = 1233 and n = 429, respectively, regulated gene transcripts. Independent of the analyzed time point, the differential Treg transcriptome analysis of patients with and without developing a GVHD identified n = 161 regulated gene transcripts with a fold change ≥ 2 or ≤ −2.
Primary Treg transcriptome study: selection of most prominently regulated gene transcripts in GVHD compared with no GVHD and healthy donors
| Gene symbol . | Gene title . | FC GVHD 90 days vs no GVHD 90 days . | FC GVHD 90 days vs HD . | FC no GVHD 90 days vs HD . | FC GVHD 150 days vs no GVHD 150 days . | FC GVHD 150 days vs HD . | FC no GVHD 150 days vs HD . | Probe set ID . | Unigene . |
|---|---|---|---|---|---|---|---|---|---|
| CDKN2B | Cyclin-dependent kinase inhibitor 2B | 2.8 | 4.5 | 2.0 | 3.2 | 6.5 | 2.5 | 236313_at | Hs.729 01 |
| CDK6 | Cyclin-dependent kinase 6 | 2.4 | 2.4 | 2.0 | 2.8 | 2.0 | NR | 224851_at | Hs.119882 |
| LAG3 | Lymphocyte-activation gene 3 | −2.3 | NR | NR | −2.1 | NR | NR | 206486_at | Hs.409523 |
| CD44 | CD44 molecule (Indian blood group) | −2.9 | NR | 2.0 | −4.4 | −1.6 | 2.6 | 209835_x_at | Hs.502328 |
| GZMA | Granzyme A | −3.4 | NR | 1.6 | −2.9 | NR | NR | 205488_at | Hs.907 08 |
| CXCR6 | Chemokine (C-X-C motif) receptor 6 | −3.8 | −3.3 | NR | −2.0 | NR | NR | 206974_at | Hs.345 26 |
| LGALS1 | Galectin-1 | −1.9 | NR | 1.7 | −2.8 | −1.8 | NR | 201105_at | Hs.445351 |
| CCR5 | Chemokine (C-C motif) receptor 5 | −4.4 | 1.5 | 1.5 | −1.9 | NR | −1.8 | 206991_s_at | Hs.536735 |
| CCR3 | Chemokine (C-C motif) receptor 3 | −5.7 | NR | 1.5 | −1.6 | 1.9 | −1.6 | 208304_at | Hs.506190 |
| CCR1 | Chemokine (C-C motif) receptor 1 | −6.4 | 3.8 | 11.0 | −1.5 | 1.9 | 1.6 | 205098_at | Hs.301921 |
| CXCR3 | Chemokine (C-X-C motif) receptor 3 | −6.3 | NR | 3.0 | −2.3 | 2.2 | 3.4 | 207681_at | Hs.198252 |
| Gene symbol . | Gene title . | FC GVHD 90 days vs no GVHD 90 days . | FC GVHD 90 days vs HD . | FC no GVHD 90 days vs HD . | FC GVHD 150 days vs no GVHD 150 days . | FC GVHD 150 days vs HD . | FC no GVHD 150 days vs HD . | Probe set ID . | Unigene . |
|---|---|---|---|---|---|---|---|---|---|
| CDKN2B | Cyclin-dependent kinase inhibitor 2B | 2.8 | 4.5 | 2.0 | 3.2 | 6.5 | 2.5 | 236313_at | Hs.729 01 |
| CDK6 | Cyclin-dependent kinase 6 | 2.4 | 2.4 | 2.0 | 2.8 | 2.0 | NR | 224851_at | Hs.119882 |
| LAG3 | Lymphocyte-activation gene 3 | −2.3 | NR | NR | −2.1 | NR | NR | 206486_at | Hs.409523 |
| CD44 | CD44 molecule (Indian blood group) | −2.9 | NR | 2.0 | −4.4 | −1.6 | 2.6 | 209835_x_at | Hs.502328 |
| GZMA | Granzyme A | −3.4 | NR | 1.6 | −2.9 | NR | NR | 205488_at | Hs.907 08 |
| CXCR6 | Chemokine (C-X-C motif) receptor 6 | −3.8 | −3.3 | NR | −2.0 | NR | NR | 206974_at | Hs.345 26 |
| LGALS1 | Galectin-1 | −1.9 | NR | 1.7 | −2.8 | −1.8 | NR | 201105_at | Hs.445351 |
| CCR5 | Chemokine (C-C motif) receptor 5 | −4.4 | 1.5 | 1.5 | −1.9 | NR | −1.8 | 206991_s_at | Hs.536735 |
| CCR3 | Chemokine (C-C motif) receptor 3 | −5.7 | NR | 1.5 | −1.6 | 1.9 | −1.6 | 208304_at | Hs.506190 |
| CCR1 | Chemokine (C-C motif) receptor 1 | −6.4 | 3.8 | 11.0 | −1.5 | 1.9 | 1.6 | 205098_at | Hs.301921 |
| CXCR3 | Chemokine (C-X-C motif) receptor 3 | −6.3 | NR | 3.0 | −2.3 | 2.2 | 3.4 | 207681_at | Hs.198252 |
Differential gene expression changes are presented as fold change (FC) comparing GVHD with immune tolerant (no GVHD) patients early (90 days) and late (150 days) after SCT and sex- and age-matched healthy donors (HD), respectively.
NR indicates not regulated.
Primary Treg transcriptome study: selection of most prominently regulated gene transcripts in the GVHD group with respect to different time points
| Gene symbol . | Gene title . | FC GVHD 90 days vs GVHD 150 days . | FC GVHD 90 days vs aGVHD manifestation . | FC GVHD 150 days vs cGVHD manifestation . | FC aGVHD vs cGVHD manifestation . | FC aGVHD manifestation vs HD . | FC cGVHD manifestation vs HD . | Probe set ID . | Unigene . |
|---|---|---|---|---|---|---|---|---|---|
| CDKN2B | Cyclin-dependent kinase inhibitor 2B | NR | 1.9 | NR | −3.9 | 2.9 | 9.8 | 236313_at | Hs.729 01 |
| CDK6 | Cyclin-dependent kinase 6 | NR | 1.5 | NR | NR | 1.5 | 1.5 | 224851_at | Hs.119882 |
| LAG3 | Lymphocyte-activation gene 3 | NR | −1.9 | NR | 1.9 | 1.6 | NR | 206486_at | Hs.409523 |
| CD44 | CD44 molecule (Indian blood group) | NR | NR | NR | NR | NR | NR | 209835_x_at | Hs.502328 |
| GZMA | Granzyme A | NR | NR | NR | NR | NR | NR | 205488_at | Hs.907 08 |
| CXCR6 | Chemokine (C-X-C motif) receptor 6 | −2.6 | NR | NR | NR | −2.2 | −1.6 | 206974_at | Hs.345 26 |
| LGALS1 | Galectin-1 | 2.0 | NR | −1.6 | NR | NR | NR | 201105_at | Hs.445351 |
| CCR5 | Chemokine (C-C motif) receptor 5 | −2.5 | −2.7 | NR | NR | 1.6 | NR | 206991_s_at | Hs.536735 |
| CCR3 | Chemokine (C-C motif) receptor 3 | −3.3 | −2.0 | NR | NR | NR | 1.8 | 208304_at | Hs.506190 |
| CCR1 | Chemokine (C-C motif) receptor 1 | NR | −2.2 | −1.8 | NR | 3.6 | 2.6 | 205098_at | Hs.301921 |
| CXCR3 | Chemokine (C-X-C motif) receptor 3 | −3.2 | −2.4 | NR | NR | NR | 1.9 | 207681_at | Hs.198252 |
| Gene symbol . | Gene title . | FC GVHD 90 days vs GVHD 150 days . | FC GVHD 90 days vs aGVHD manifestation . | FC GVHD 150 days vs cGVHD manifestation . | FC aGVHD vs cGVHD manifestation . | FC aGVHD manifestation vs HD . | FC cGVHD manifestation vs HD . | Probe set ID . | Unigene . |
|---|---|---|---|---|---|---|---|---|---|
| CDKN2B | Cyclin-dependent kinase inhibitor 2B | NR | 1.9 | NR | −3.9 | 2.9 | 9.8 | 236313_at | Hs.729 01 |
| CDK6 | Cyclin-dependent kinase 6 | NR | 1.5 | NR | NR | 1.5 | 1.5 | 224851_at | Hs.119882 |
| LAG3 | Lymphocyte-activation gene 3 | NR | −1.9 | NR | 1.9 | 1.6 | NR | 206486_at | Hs.409523 |
| CD44 | CD44 molecule (Indian blood group) | NR | NR | NR | NR | NR | NR | 209835_x_at | Hs.502328 |
| GZMA | Granzyme A | NR | NR | NR | NR | NR | NR | 205488_at | Hs.907 08 |
| CXCR6 | Chemokine (C-X-C motif) receptor 6 | −2.6 | NR | NR | NR | −2.2 | −1.6 | 206974_at | Hs.345 26 |
| LGALS1 | Galectin-1 | 2.0 | NR | −1.6 | NR | NR | NR | 201105_at | Hs.445351 |
| CCR5 | Chemokine (C-C motif) receptor 5 | −2.5 | −2.7 | NR | NR | 1.6 | NR | 206991_s_at | Hs.536735 |
| CCR3 | Chemokine (C-C motif) receptor 3 | −3.3 | −2.0 | NR | NR | NR | 1.8 | 208304_at | Hs.506190 |
| CCR1 | Chemokine (C-C motif) receptor 1 | NR | −2.2 | −1.8 | NR | 3.6 | 2.6 | 205098_at | Hs.301921 |
| CXCR3 | Chemokine (C-X-C motif) receptor 3 | −3.2 | −2.4 | NR | NR | NR | 1.9 | 207681_at | Hs.198252 |
Differential gene expression changes are presented as fold change (FC) comparing GVHD with immune tolerant (no GVHD) patients early (90 days) and late (150 days) after SCT and sex- and age-matched healthy donors (HD), respectively.
NR indicates not regulated.
Confirmatory Treg transcriptome study.
To validate the results of the primary Treg transcriptome study, additional patients with (n = 12) and without (n = 8) GVHD were analyzed by microarray technology. Because primary Treg transcriptome study showed a high correlation within the GVHD group between the early time point and aGVHD manifestation as well as between the late time point and cGVHD manifestation (Figure 2), this confirmatory study only looked at early (< 100 days) and late (> 100 days) time points in the first half-year after SCT. Patient characteristics are shown in Table 1. As summarized in Figure 1B, datasets were comparatively analyzed and displayed together with results of the primary study (Table 4): the down-regulation of the molecules granzyme A, CD44, LAG3, and LGALS1 could be confirmed in Tregs of at least one other group of GVHD patients compared with immune tolerant patients. The gene expression of LAG3 and LGALS1 was down-regulated late after SCT (> 100 days), whereas granzyme A and CD44 showed a reduced gene expression level in Tregs of GVHD patients independent of the time. The most prominent regulation was seen for granzyme A in Tregs of GVHD patients before 100 days with an up to 8-fold reduction of gene expression. The reduced gene expression of CCR1 could be confirmed early after SCT (< 100 days), whereas CCR5 and CXCR3 were most prominently regulated in the primary Treg transcriptome study, including patients with severe aGVHD.
Validation of primary Treg transcriptome study by confirmatory microarray experiments
| Gene symbol . | Gene title . | GVHD vs no GVHD < 100 days . | GVHD vs no GVHD > 100 days . | Probe set ID . | Unigene . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FC1 . | FC2 . | FC3 . | FC4 . | FC5 . | FC6 . | FC7 . | FC8 . | ||||
| CDKN2B | Cyclin-dependent kinase inhibitor 2B | 2.8 | 3.6 | 1.6 | 3.2 | NR | 1.7 | NR | 1.8 | 236313_at | Hs.729 01 |
| CDK6 | Cyclin-dependent kinase 6 | 2.4 | 2.3 | NR | 2.8 | 1.5 | NR | NR | NR | 224851_at | Hs.119882 |
| LAG3 | Lymphocyte-activation gene 3 | −2.3 | NR | NR | −2.1 | −1.9 | −1.7 | −1.8 | −1.6 | 206486_at | Hs.409523 |
| CD44 | CD44 molecule (Indian blood group) | −2.9 | NR | −1.6 | −4.4 | −3.4 | NR | −3.3 | NR | 204490_s_at | Hs.502328 |
| GZMA | Granzyme A | −3.4 | −2.7 | −8.0 | −2.9 | NR | −2.1 | NR | −1.9 | 205488_at | Hs.907 08 |
| CXCR6 | Chemokine (C-X-C motif) receptor 6 | −3.8 | 1.9 | 3.2 | −2.0 | NR | NR | −1.5 | NR | 206974_at | Hs.345 26 |
| LGALS1 | Galectin-1 | −1.9 | NR | NR | −2.8 | −1.8 | NR | −1.8 | NR | 201105_at | Hs.445351 |
| CCR5 | Chemokine (C-C motif) receptor 5 | −4.4 | NR | NR | −1.9 | NR | NR | NR | NR | 206991_s_at | Hs.536735 |
| CCR3 | Chemokine (C-C motif) receptor 3 | −5.7 | NR | 1.9 | −1.6 | 1.6 | 1.5 | NR | NR | 208304_at | Hs.506190 |
| CCR1 | Chemokine (C-C motif) receptor 1 | −6.4 | −5.8 | −19.6 | −1.5 | NR | 2.7 | NR | 1.7 | 205098_at | Hs.301921 |
| CXCR3 | Chemokine (C-X-C motif) receptor 3 | −6.3 | NR | NR | −2.3 | −1.5 | NR | −1.6 | NR | 207681_at | Hs.198252 |
| Gene symbol . | Gene title . | GVHD vs no GVHD < 100 days . | GVHD vs no GVHD > 100 days . | Probe set ID . | Unigene . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FC1 . | FC2 . | FC3 . | FC4 . | FC5 . | FC6 . | FC7 . | FC8 . | ||||
| CDKN2B | Cyclin-dependent kinase inhibitor 2B | 2.8 | 3.6 | 1.6 | 3.2 | NR | 1.7 | NR | 1.8 | 236313_at | Hs.729 01 |
| CDK6 | Cyclin-dependent kinase 6 | 2.4 | 2.3 | NR | 2.8 | 1.5 | NR | NR | NR | 224851_at | Hs.119882 |
| LAG3 | Lymphocyte-activation gene 3 | −2.3 | NR | NR | −2.1 | −1.9 | −1.7 | −1.8 | −1.6 | 206486_at | Hs.409523 |
| CD44 | CD44 molecule (Indian blood group) | −2.9 | NR | −1.6 | −4.4 | −3.4 | NR | −3.3 | NR | 204490_s_at | Hs.502328 |
| GZMA | Granzyme A | −3.4 | −2.7 | −8.0 | −2.9 | NR | −2.1 | NR | −1.9 | 205488_at | Hs.907 08 |
| CXCR6 | Chemokine (C-X-C motif) receptor 6 | −3.8 | 1.9 | 3.2 | −2.0 | NR | NR | −1.5 | NR | 206974_at | Hs.345 26 |
| LGALS1 | Galectin-1 | −1.9 | NR | NR | −2.8 | −1.8 | NR | −1.8 | NR | 201105_at | Hs.445351 |
| CCR5 | Chemokine (C-C motif) receptor 5 | −4.4 | NR | NR | −1.9 | NR | NR | NR | NR | 206991_s_at | Hs.536735 |
| CCR3 | Chemokine (C-C motif) receptor 3 | −5.7 | NR | 1.9 | −1.6 | 1.6 | 1.5 | NR | NR | 208304_at | Hs.506190 |
| CCR1 | Chemokine (C-C motif) receptor 1 | −6.4 | −5.8 | −19.6 | −1.5 | NR | 2.7 | NR | 1.7 | 205098_at | Hs.301921 |
| CXCR3 | Chemokine (C-X-C motif) receptor 3 | −6.3 | NR | NR | −2.3 | −1.5 | NR | −1.6 | NR | 207681_at | Hs.198252 |
Differential gene expression changes for regulated gene transcripts in the primary and confirmatory Treg transcriptome study are presented as fold change (FC) comparing GVHD patients with immune tolerant (no GVHD) patients early (< 100 days) and late (> 100 days) after SCT.
NR indicates not regulated.
Validation of microarray data by quantitative real-time RT-PCR
To validate the microarray data and to further analyze the statistical significance in a larger group of independent patients, we performed quantitative real-time RT-PCR in patients with (n = 27) and without (n = 12) GVHD early (< 100 days) and late (> 100 days) after SCT as well as in healthy controls (n = 9). We concentrated on differentially expressed candidates (LAG3, CD44, granzyme A, LGALS1, CXCR6, CCR5, CCR1, and CXCR3) showing a regulation of more than or equal to 2-fold or less than or equal to −2-fold in at least 2 predefined comparisons of the Treg transcriptome studies (Table 4). The regulation of gene expression could be confirmed in the majority of analyzed candidates. Results of statistically significantly regulated genes (P < .05) are shown in Figure 4. Early after SCT (< 100 days), Tregs of immune tolerant patients had significantly higher gene expression levels than GVHD patients for granzyme A, LAG3, LGALS1, CCR5, and CXCR3. Transcript levels for LAG3 remained significantly up-regulated (> 100 days) in patients without GVHD (P < .001). Real-time RT-PCR analysis for CD44 and CCR1 revealed only a significant up-regulation after 100 days in immune tolerant patients.
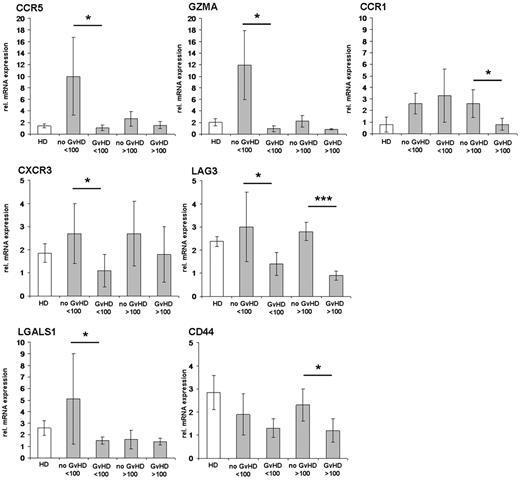
Validation of candidate genes identified in the Treg transcriptome studies by real-time RT-PCR. Differentially expressed candidate genes with a significant regulation for at least 2 predefined comparisons of the Treg transcriptome studies (Table 4) have been analyzed by real-time RT-PCR in an independent larger group of patients. Patients without GVHD and with GVHD manifestation before day 100 and after day 100 after SCT are summarized as “no GVHD” or “GVHD” < 100 days and > 100 days, respectively. Isolated RNAs from sorted CD4+CD25hiCD127lo/− T cells were transcribed, and cDNAs were pooled from n = 12 patients without GVHD in 4 cDNA pools, n = 27 patients with GVHD in 9 cDNA pools, and n = 9 healthy donors (HD) in 3 cDNA pools. The results of real-time RT-PCRs are presented as mean values of relative mRNA expression levels to the housekeeping genes RPS9 and GAPDH, respectively. *P < .05 (significant, 2-paired Student t test). ***P < .001 (highly significant, 2-paired Student t test).
Validation of candidate genes identified in the Treg transcriptome studies by real-time RT-PCR. Differentially expressed candidate genes with a significant regulation for at least 2 predefined comparisons of the Treg transcriptome studies (Table 4) have been analyzed by real-time RT-PCR in an independent larger group of patients. Patients without GVHD and with GVHD manifestation before day 100 and after day 100 after SCT are summarized as “no GVHD” or “GVHD” < 100 days and > 100 days, respectively. Isolated RNAs from sorted CD4+CD25hiCD127lo/− T cells were transcribed, and cDNAs were pooled from n = 12 patients without GVHD in 4 cDNA pools, n = 27 patients with GVHD in 9 cDNA pools, and n = 9 healthy donors (HD) in 3 cDNA pools. The results of real-time RT-PCRs are presented as mean values of relative mRNA expression levels to the housekeeping genes RPS9 and GAPDH, respectively. *P < .05 (significant, 2-paired Student t test). ***P < .001 (highly significant, 2-paired Student t test).
Micro-RNA profiling in Tregs of patients with and without GVHD
Micro-RNA expression was analyzed in Tregs of independent immune tolerant and GVHD patients early (< 100 days) and late (> 100 days) after SCT as well as of age- and sex-matched healthy controls. The following miRNAs, predictively targeting the identified candidate genes, were found to be differentially expressed with high significance: miR-671–5p (granzyme A, P < .01), miR-659 (LGALS1, P < .01), miR-18b* (CCR5, P < .05), miR-132* (CXCR3, P < .05), miR-501–3p (CCR1, P < .05), and miR-328 (CD44, P < .05; Figure 5).
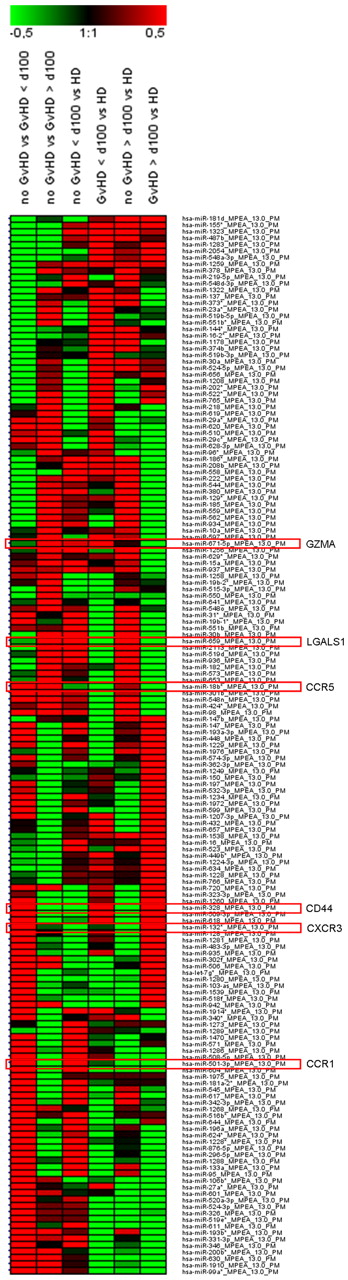
Regulated miRNAs in Tregs of SCT patients. Patients without GVHD and with GVHD manifestation before day 100 and after day 100 after SCT are summarized as “no GVHD” or “GVHD” > 100 days and > 100 days, respectively. CD4+CD25hiCD127lo/− Tregs were sorted from PBMCs of blood withdrawals with a purity > 95%. Isolated total RNA from n = 3 patients without GVHD and n = 3 patients with GVHD < 100 days and > 100 days were pooled, respectively. In addition, total RNAs were pooled from n = 3 healthy donors (HD). Results are presented as a heat map generated using Genesis software (Institute for Genomics and Bioinformatics, Graz University of Technology, Austria) and show 163 differentially regulated miRNAs with a log median fold change > 0.4 or < −0.5 in GVHD vs no GVHD < 100 days and/or GVHD vs no GVHD > 100 days. Red frames highlight miRNAs with predicted targets of gene transcripts encoding GZMA, LGALS1, CCR5, CD44, CXCR3, and CCR1.
Regulated miRNAs in Tregs of SCT patients. Patients without GVHD and with GVHD manifestation before day 100 and after day 100 after SCT are summarized as “no GVHD” or “GVHD” > 100 days and > 100 days, respectively. CD4+CD25hiCD127lo/− Tregs were sorted from PBMCs of blood withdrawals with a purity > 95%. Isolated total RNA from n = 3 patients without GVHD and n = 3 patients with GVHD < 100 days and > 100 days were pooled, respectively. In addition, total RNAs were pooled from n = 3 healthy donors (HD). Results are presented as a heat map generated using Genesis software (Institute for Genomics and Bioinformatics, Graz University of Technology, Austria) and show 163 differentially regulated miRNAs with a log median fold change > 0.4 or < −0.5 in GVHD vs no GVHD < 100 days and/or GVHD vs no GVHD > 100 days. Red frames highlight miRNAs with predicted targets of gene transcripts encoding GZMA, LGALS1, CCR5, CD44, CXCR3, and CCR1.
Differential protein expression of identified candidates in Tregs
To study differentially expressed candidate genes at the protein level, flow cytometric analyses were done in independent patients with (n = 17) and without (n = 10) GVHD before and after 100 days after SCT. We concentrated on the following differentially expressed molecules, which were identified both in the Treg transcriptome studies and confirmed by real-time RT-PCR: granzyme A, LAG3, CD44, CCR5, CCR1, and CXCR3. Results, including the FACS analysis of healthy, not transplanted controls (n = 9), are summarized in Figure 6. Tregs of immune tolerant patients showed a significantly (P < .05) higher expression of granzyme A (3.4% vs 1.9%), CCR5 (16.5% vs 5.5%), and CXCR3 (18.3% vs 4.2%) early after SCT (< 100 days) compared with GVHD patients, calculated as the mean percentage of CD4+CD25hiFoxp3+CD127lo/− Tregs. Moreover, the density of surface expression presented as mean fluorescence intensities of these molecules is also increased (Figure 6B). The candidate gene LAG3 (4% vs 2.9%, < 100 days and 6.4% vs 4.7%, > 100 days) was not significantly regulated at the protein level but tended to have a higher expression in Tregs of immune tolerant patients (Figure 6A). In contrast to the gene expression analysis, flow cytometric analysis of CCR1 revealed a higher expression in GVHD patients after 100 days (3% vs 0.8%), and no significant expression differences were found before 100 days. Although nearly all CD4+CD25hiFoxp3+CD127lo/− Tregs expressed CD44, histograms did not show significant differences in mean fluorescence intensities (data not shown).
Differential protein expression of identified candidates in Tregs. PBMCs were isolated at indicated time points early (< 100 days) and late (> 100 days) after SCT from patients without GVHD (n = 10), with aGVHD/cGVHD (n = 17), and healthy donors (n = 9). (A) Expression values were calculated as a percentage of CD4+CD25hiCD127lo/−Foxp3+ Tregs, and their mean values are presented as bar graphs. (B) Representative overlays from histogram plots are shown for candidates with significant (*P < .05) expression differences. Dashed black lines indicate patients without GVHD; and solid gray lines, patients with aGVHD/cGVHD. Gray shaded areas represent the respective isotype controls. Bold black (gray) numbers indicate the mean fluorescence intensities of CD4+CD25hiCD127lo/−Foxp3+ Tregs in patients without (with) GVHD.
Differential protein expression of identified candidates in Tregs. PBMCs were isolated at indicated time points early (< 100 days) and late (> 100 days) after SCT from patients without GVHD (n = 10), with aGVHD/cGVHD (n = 17), and healthy donors (n = 9). (A) Expression values were calculated as a percentage of CD4+CD25hiCD127lo/−Foxp3+ Tregs, and their mean values are presented as bar graphs. (B) Representative overlays from histogram plots are shown for candidates with significant (*P < .05) expression differences. Dashed black lines indicate patients without GVHD; and solid gray lines, patients with aGVHD/cGVHD. Gray shaded areas represent the respective isotype controls. Bold black (gray) numbers indicate the mean fluorescence intensities of CD4+CD25hiCD127lo/−Foxp3+ Tregs in patients without (with) GVHD.
Reduction of nTregs in the intestinal mucosa of GVHD patients
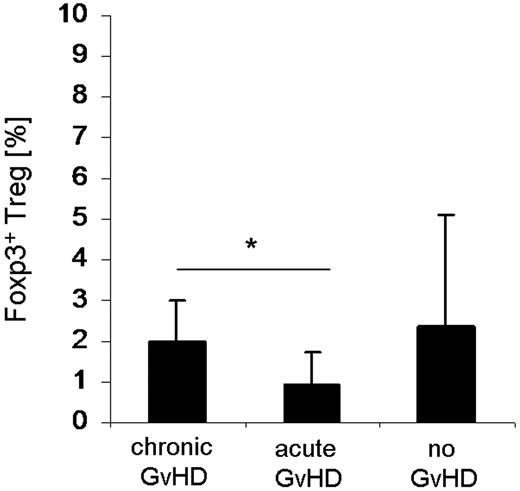
As the significantly reduced surface expression of CCR5 and CXCR3 in circulating Tregs of GVHD patients may lead to a diminished migration capacity of Tregs into the target organs of GVHD, we analyzed intestinal biopsies taken from patients with clinically suspected intestinal GVHD for Treg-specific DNA demethylation within the foxp3 gene locus. This method allows the quantification of infiltrating Tregs stably expressing Foxp3. The percentage of Foxp3+ Tregs was significantly (P < .05) reduced in the intestinal mucosa of patients with aGVHD (n = 8; 2 females/6 males) that was accompanied by an even higher grade of infiltration by T lymphocytes (data not shown) in contrast to those with cGVHD (n = 6; 4 females/2 males; Figure 7). Patients without histologic signs for GVHD (n = 4; 3 females/1 male) tended to have a higher percentage of mucosa-infiltrating Tregs, whereas the infiltration grade with T lymphocytes was lower.
Quantification of Tregs infiltrating the intestinal mucosa from SCT patients. Gut biopsies were endoscopically taken from transplanted patients with clinically suspected intestinal GVHD. DNA was isolated from 2 gut biopsies each taken from patients with cGVHD (n = 6; 4 females/2 males), aGVHD (n = 8; 2 females/6 males), and without any histologic signs of GVHD (n = 4; 3 females/1 male). Percentage of FOXP3 Treg specific demethylation region demethylation levels was determined by quantitative PCR (“DNA demethylation analysis of the foxp3 gene locus in gut biopsie”). *P < .05 (Student t test).
Quantification of Tregs infiltrating the intestinal mucosa from SCT patients. Gut biopsies were endoscopically taken from transplanted patients with clinically suspected intestinal GVHD. DNA was isolated from 2 gut biopsies each taken from patients with cGVHD (n = 6; 4 females/2 males), aGVHD (n = 8; 2 females/6 males), and without any histologic signs of GVHD (n = 4; 3 females/1 male). Percentage of FOXP3 Treg specific demethylation region demethylation levels was determined by quantitative PCR (“DNA demethylation analysis of the foxp3 gene locus in gut biopsie”). *P < .05 (Student t test).
Discussion
In aGVHD, the immune system insufficiently controls alloreactive effector T cells, which are expanded and recruited at the site of inflammation. As the data from the human system implicate that reduced Treg cell numbers in the periphery29-31 and target organs32 contribute to the development of GVHD, insufficient reconstitution of Tregs after SCT might be one aspect of uncontrolled expansion of alloreactive donor T-cell clones. In contrast to these former studies concentrating on numerical Treg deficiency in GVHD, our study focused on the identification of phenotypic and possible functional differences by a global Treg transcriptome analysis. Normally working Tregs play a substantial role for the homeostasis of the immune system after allogeneic SCT with respect to their migration capacity to secondary lymphoid organs and sites of inflammation, clonal expansion, and suppressor function. The involvement of chemokines and their receptors in the recruitment of effector T cells to target organs during GVHD has been reported in a number of experimental models.33-36 CCR5 seems to be essential for the recruitment of Tregs to GVHD target tissues as prerequisite for their suppressive function on alloreactive T cells, as Tregs from CCR5−/− mice do not accumulate in the target organs and fail to suppress GVHD.35 Furthermore, CXCR3-transfected Tregs have been shown to migrate more efficiently to GVHD target organs, such as liver, lung, and intestine, thus ameliorating GVHD.37
Our Treg transcriptome studies confirm these data in humans, revealing a substantially higher gene expression of several chemokine receptors (CCR5, CCR6, CXCR3, CXCR6, CCR3, and CCR1) in immune tolerant patients not developing a GVHD. Additional experiments confirmed the significantly higher gene expression of CCR5, CXCR3, and CCR1 in a larger cohort of transplanted patients. Moreover, miRNA profiling identified the differential expression of miR-18b*, miR-132*, and miR-501–3p, which target predictively CCR5, CXCR3, and CCR1, respectively, in Tregs from independent patients with and without GVHD. Subsequent protein analyses demonstrated a significantly higher expression of CCR5 and CXCR3 at the cell surface of circulating Tregs from immune tolerant patients compared with GVHD patients early after SCT. The comparison with Tregs of healthy donors revealed that Tregs of immune tolerant patients expressed more CCR5 and CXCR3 and GVHD patients significantly less. Tregs of immune tolerant patients with higher surface expression of CCR5 and CXCR3 might therefore properly migrate to secondary lymphoid organs and inflammatory sites capable of controlling potentially alloreactive T-cell clones in GVHD target organs efficiently in contrast to the Tregs of GVHD patients obviously failing to control alloreactivity sufficiently. This concept is strengthened by the quantification of Tregs infiltrating the intestinal mucosa as possible target organ of GVHD. Even though the gut biopsies of patients with aGVHD displayed a higher grade of infiltration by mature T cells, the percentage of infiltrating Tregs was reduced compared with gut biopsies of transplanted patients without GVHD. Whereas in a former study32 a contamination with Foxp3-expressing activated T cells could not be excluded, the demethylation status of the foxp3 gene locus in intestinal biopsies as found in our study provides evidence that we are dealing with terminally differentiated regulatory T cells with a stable phenotype.38 Our finding that the CXCR3 mRNA levels of Tregs from patients with severe GVHD were higher at day 90 and day 150 than at the time points of aGVHD/cGVHD manifestation might indicate that Tregs of GVHD patients highly expressing migratory chemokine receptors, such as CCR5 and CXCR3, might have been recruited and trapped in secondary lymphoid organs and therefore cannot be detected in the peripheral blood and GVHD target organs. The Treg transcriptome studies revealed significant differences in cell cycle regulation and proliferation of patients with and without GVHD. Expression levels for CDK6 and cyclin-dependent kinase inhibitor 2B (CDKN2B), inhibitor of CDK4, were up-regulated in GVHD. Considerable overexpression of CDK6 was also detected by others analyzing mononucleated cell samples from cGVHD patients,39 indicating that cell cycle progression from G1 to S phase may be a critical step in the pathogenesis of cGVHD. CyclinD-CDK4 and CyclinD-CDK6 complexes are associated with cell cycle reentry from G0 phase. These regulated candidates might contribute to the poor quantitative recovery of Tregs during the post-transplant immune reconstitution observed in GVHD patients.30,31 Thus, Tregs of GVHD patients might fail to effectively suppress alloreactive effector T cells because of numerical deficiency and possible defects in clonal expansion after antigen-specific activation.
Beyond the regulation of genes related to migration/homing and cell cycle progression/proliferation, our findings suggest a loss of function in Tregs from GVHD patients. Confirmative real-time RT-PCR experiments in a larger group of independent patients with GVHD showed a significantly lower gene expression of CD44, LGALS1, LAG3, and granzyme A than in immune tolerant patients. Each of these molecules is associated with the suppressive function of Tregs.7,10 These findings were again supported by miRNA profiling. Notably, subsequent protein analyses could delineate a significantly lower expression of granzyme A in Tregs from GVHD patients than from immune tolerant patients early after SCT. Granzyme A is predominantly expressed in activated Tregs and induces cytolysis in a perforin-dependent, FAS-FASL-independent manner.13 Very recently, Cai et al showed that granzyme B was not required for Treg cell-mediated suppression of GVHD.40 Granzyme A was reported to be more potent than granzyme B13 and mediated cytolysis also to mature dendritic cells expressing endogenous inhibitors to granzyme B.41 Therefore, Tregs from immune tolerant patients might control alloreactive immune responses more effectively because of improved cytolytic capacities mediated by granzyme A. A significantly higher gene expression level of LGALS1 could be detected in Tregs of immune tolerant patients. Further studies will have to analyze the protein expression of LGALS1 in transplanted patients. Interestingly, LGALS1 promotes growth arrest and apoptosis of activated T cells,42 and its recombinant application has shown beneficial effects in a murine GVHD model.43 Whereas the gene expression of LAG3 and CD44 has been significantly regulated, this could not be confirmed at the protein level.
In the present study, all patients received an immunosuppressive prophylactic regimen with cyclosporine, mycophenolate mofetil (with or without methotrexate), and blood was taken from the patients in the primary Treg transcriptome study with severe aGVHD before steroid application. However, at later time points, patients with and without GVHD received different immunosuppressive medications, and pharmacologic effects on Tregs have to be considered. Interestingly, Treg transcriptomes in the primary study showed a high correlation in the principal component analysis despite differences in immunosuppressive therapy.
In conclusion, we propose that Tregs of GVHD patients may not sufficiently migrate to the inflammatory sites and may not effectively control clonal expansion of alloreactive T cells because of impaired suppressive/cytolytic properties. Further investigations need to clarify whether these potentially defective Tregs are transplanted with the graft or whether they emerge during reconstitution in the allogeneic microenvironment. The answer to these questions might influence current clinical studies on adoptive Treg cell transfer and might help to optimize cellular strategies for immune intervention by specific manipulation of Tregs for the prophylaxis and treatment of life-threatening GVHD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the clinical coworkers from the stem cell transplantation unit and the endoscopy team from Hannover Medical School for patient caring and their support in probe sampling, Matthias Ballmeier and his coworkers for FACS-based cell sorting, and Petra Hagendorff for technical assistance in performing microarrays.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB738).
Authorship
Contribution: S.N.U. designed, performed, and analyzed experiments and wrote the manuscript; S.V. performed and analyzed experiments; R.G. was involved in microarray experiments; J.G. performed experiments; U.B. analyzed gut biopsies; S.B. and M.S. supported probe sampling from patients; D.B. was involved in microarray experiments; A.G. designed the study and carefully revised the manuscript; and A.F. designed the study, analyzed results of experiments, and wrote the manuscript.
Conflict-of-interest disclosure: U.B. is an employee of Epiontis GmbH, a company whose technique was applied in this work. The remaining authors declare no competing financial interests.
Correspondence: Anke Franzke, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany; e-mail: franzke.anke@mh-hannover.de.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal