Abstract
The efficient engraftment in immune-deficient mice achieved with both acute lymphoblastic leukemia (ALL) cell lines and primary samples has facilitated identification of the antileukemia activity of a wide variety of agents. Despite widespread usage, however, little is known about the early ALL localization and engraftment kinetics in this model, limiting experimental read-outs primarily to survival and endpoint analysis at high disease burden. In this study, we report that bioluminescent imaging can be reproducibly achieved with primary human ALL samples. This approach provides a noninvasive, longitudinal measure of leukemia burden and localization that enhances the sensitivity of treatment response detection and provides greater insight into the mechanism of action of antileukemia agents. In addition, this study reveals significant cell line– and species-related differences in leukemia migration, especially early in expansion, which may confound observations between various leukemia models. Overall, this study demonstrates that the use of bioluminescent primary ALL allows the detection and quantitation of treatment effects at earlier, previously unquantifiable disease burdens and thus provides the means to standardize and expedite the evaluation of anti-ALL activity in preclinical xenograft studies.
Introduction
Murine leukemia models have been useful and versatile tools for identifying and targeting underlying disease mechanisms.1 However, preclinical studies involving human samples are more immediately applicable when investigating new drugs or therapies. Initial studies with human leukemia in immune-deficient mice, such as the NOD/SCID (NOD.CB17-Prkdcscid/J), were a great step forward and remain valuable for rapid iterations in the study of novel therapies at the preclinical level.2,3 More recently, use of severely immune-deficient NOD/SCID/γc−/− (NSG; NOD.Cg-Prkdcscid/IL2rgtm1Wjl/SzJ and NOG; NOD.Cg-Prkdcscid/Il2rgtm1Sug/Jic) mice, which lack functional B, T, and NK cells, has allowed for more consistent and reproducible engraftment of normal human cells and primary leukemias.4-6
Primary acute lymphoblastic leukemia (ALL) xenografts in immune-deficient mice accurately represent human disease and gene expression and may have predictive value for clinical variables, such as drug resistance and time to relapse.7-11 Although this model is a powerful and widely used tool for the ongoing development of new drugs and biologic therapeutics for ALL,12-19 the kinetics of every primary patient sample are different, some engrafting in 4 weeks and others taking as long as one year, when measured by the appearance of human ALL blasts in peripheral blood.5,10,20 The endpoints of these studies are typically limited to survival analysis and time to grossly detectable disease. Monitoring the peripheral blood compartment for blasts is the primary method of disease detection, leaving the early engraftment kinetics and localization, therefore, largely unknown.21-24 Furthermore, the time from appearance of peripheral blasts to death is also highly variable in our experience, taking from as little as a few days to as long as several months. Animals may show no outward signs of engraftment, and samples are occasionally lost when mice die before displaying evidence of disease. Noninvasive measures, including magnetic resonance imaging, ultrasound, and positron emission tomography/computed tomography (PET/CT) scans rely on specific organ changes as a surrogate marker of disease. For individual samples, this may correlate well to disease development, such as with leukemia, which reliably produces splenic enlargement; but as the library of primary patient samples grows, it is clear that not all human leukemias behave the same when engrafted in a mouse. The early migration and anatomic niche population of each leukemia may be different, and current standard endpoint analyses (organ size, peripheral blood) do not give a complete picture of disease expansion or treatment response.
The application of bioluminescent reporters has enabled the noninvasive, longitudinal monitoring of hematologic malignant cells for in vivo studies of cellular migration and expansion, as well evaluation of responses to cytotoxic therapy.25-30 Luciferase reporters provide great sensitivity because of the extremely low background luminescence in wild-type animals and represent a broadly applicable approach to noninvasive disease monitoring.31-33 Firefly luciferase is particularly versatile as a bioluminescent reporter, as a significant portion of its emission spectrum is > 600 nm, wavelengths that achieve good tissue penetrance.34 Although these reporters require introduction of a novel substrate to achieve luminescence, the 2 most common substrates for bioluminescent reporters, luciferin and colenterazine, are well tolerated by animals. The kinetics after intraperitoneal administration of D-luciferin are reliable and reproducible with peak emission times exceeding several minutes.34
Lentiviral transduction has been shown to be an effective method for gene transfer into primary human ALL blasts.35-37 In this study, we use lentivirus-mediated gene transfer of luciferase to generate a panel of bioluminescent primary ALL samples and perform a systematic study of early engraftment kinetics using multiple ALL cell lines and primary samples. By enabling quantification of early disease burdens and identification of specific organ involvement, the ability to generate bioluminescent primary ALL samples reproducibly will provide the means for standardization and more accurate comparison of preclinical of ALL treatment responses.
Methods
Cells and patient samples
The human BCP ALL lines Nalm-6 and RS4-11 were obtained from ATCC, and 380 was kindly provided by Dr Dario Campana (St Jude). Mouse BCP ALL cell lines 289 and t309 were previously derived from primary leukemias developing in Eμ-Ret fusion protein transgenic mice on a BALB/cJ background.38-40 Mouse cell lines were cultured in RPMI medium with 20% FBS and 3% IL-7 supernatant, as previously described.40 Human lines were cultured in RPMI medium with 10% FBS and 1% penicillin/streptomycin. Human ALL samples were obtained as deidentified samples from the Leukemia Bank at the University of Pennsylvania and the ALL Cell Bank maintained by the Children's Oncology Group. The samples were collected for both cell banks under the Children's Hospital of Philadelphia Institutional Review Board–approved protocols and distributed under standard material transfer agreements. ALL-1 was obtained from peripheral blood of an adult ALL patient; ALL-2 and ALL-3 were obtained from bone marrow of pediatric ALL patients at diagnosis.
Mouse xenograft and allograft studies
Briefly, 6- to 10-week-old NOD.Cg-Prkdcscid/IL2rgtm1Wjl/SzJ (NSG) and NOD.CB17-Prkdcscid/J (NS) mice were obtained from The Jackson Laboratory or bred in-house and maintained under specific pathogen-free conditions. Animals were injected via tail vein with 106 viable murine or human leukemia cells in 0.2 mL sterile PBS unless otherwise indicated. Peripheral blood was obtained by retro-orbital bleeding, and the presence of ALL blasts was determined by flow cytometry. Human CD19 and CD45 and mouse B220 and BP-1 expression was detected by staining with fluorescently conjugated monoclonal antibodies (BD Biosciences). All experiments were conducted under the supervision of the facility's Institutional Animal Care and Use Committee according to an Institutional Animal Care and Use Committee–approved protocol.
Lentiviral transduction of primary leukemia samples
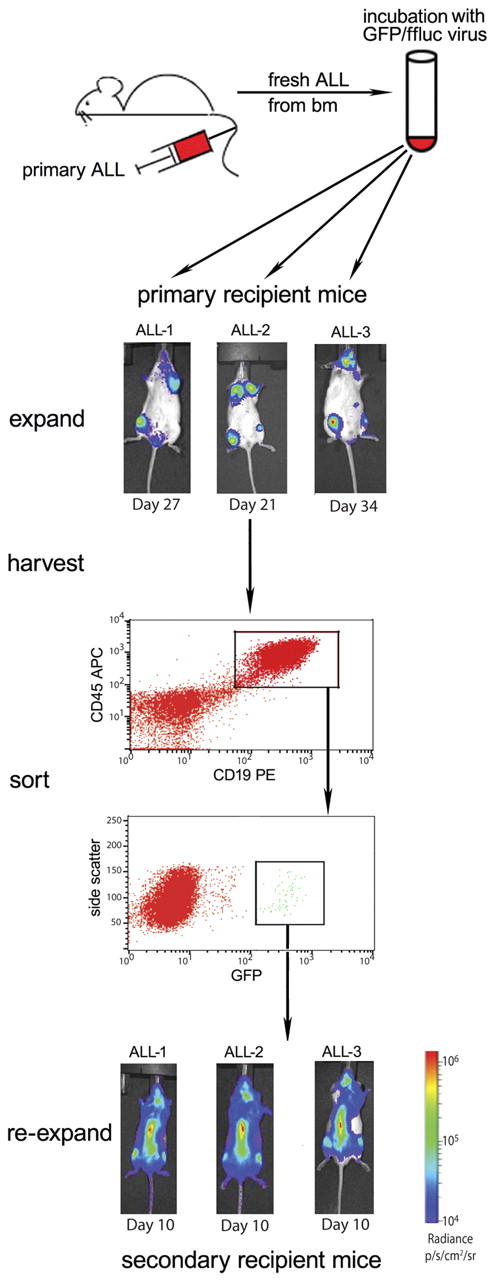
The self-inactivating lentiviral construct encoding firefly luciferase (ffluc) and green fluorescent protein (GFP), using the EF-1α promoter and a 2A ribosomal skip sequence, used in these experiments is described elsewhere.41,42 Primary human BCP ALL samples were expanded in NSG mice. Human ALL cells (2.5-5 × 104), isolated from the spleens of moribund recipient NSG mice, were incubated directly ex vivo in RPMI 1640 media supplemented with 10% FBS and 1% penicillin/streptomycin containing the lentivirus at a multiplicity of infection of 50:1 for 16 hours in polystyrene 5-mL tubes. Viable, unsorted cells were washed in excess volumes of sterile PBS 4 times and injected via tail vein into a single NSG mouse for expansion. The recipient mice were evaluated weekly by bioluminescence imaging for the presence of luciferase activity. Moribund recipient mice were killed, and human CD45, CD19, and GFP triple-positive cells were purified from spleen and bone marrow by flow cytometric sorting. Purified cells were then injected via tail vein into secondary recipient mice for further expansion and purification. This protocol is summarized in Figure 1. The secondary expansion cells were then used in the treatment assays described. All human cell lines and samples were subjected to genomic fingerprinting via a combination of short tandem repeat (STR) and single nucleotide polymorphism (SNP) analysis to verify the identity of the cells before and after luciferase transduction and in vivo expansion.43,44
Our strategy to generate stable ffluc+ primary ALL samples. Bulk transduction of primary patient ALL, freshly obtained from NSG mouse bone marrow, via lentivirus results in low-level GFP/ffluc expression, which can be purified by flow sorting. Three primary patient ALL samples (ALL-1, ALL-2, and ALL-3) were harvested from moribund NSG mice, transduced overnight, and viable cells injected into primary recipients via tail vein. All 3 samples engrafted with a small GFP+ population, which was then purified using the gating strategy shown (sample ALL-1 sorting is shown). This enrichment results in stable, > 99% bioluminescent primary ALL with resultant early detection and evenly disseminated disease in secondary (shown in bottom panels) and later recipient mice.
Our strategy to generate stable ffluc+ primary ALL samples. Bulk transduction of primary patient ALL, freshly obtained from NSG mouse bone marrow, via lentivirus results in low-level GFP/ffluc expression, which can be purified by flow sorting. Three primary patient ALL samples (ALL-1, ALL-2, and ALL-3) were harvested from moribund NSG mice, transduced overnight, and viable cells injected into primary recipients via tail vein. All 3 samples engrafted with a small GFP+ population, which was then purified using the gating strategy shown (sample ALL-1 sorting is shown). This enrichment results in stable, > 99% bioluminescent primary ALL with resultant early detection and evenly disseminated disease in secondary (shown in bottom panels) and later recipient mice.
Bioluminescent imaging
Anesthetized mice were imaged using a Xenogen Spectrum system and Living Image Version 4.0 software. Mice were given an intraperitoneal injection of 150 mg/kg D-luciferin (Caliper Life Sciences). Previous titration indicated time to peak of photon emission to be 5 minutes, with peak emission lasting for 6-10 minutes. Each animal was imaged alone (for photon quantitation) or in groups of up to 5 mice (for display purposes) in the anterior-posterior prone position at the same relative time point after luciferin injection (6 minutes). Data were collected until the mid-range of the linear scale was reached (600-60 000 counts) or maximal exposure settings reached (f stop 1, large binning, and 120 seconds), and then converted to photons/second/cm2/steradian (p/s/cm2/sr) to normalize each image for exposure time, f stop, binning and animal size. This approach avoids collection of saturated images. Mice without luciferase-containing cells were imaged at maximal settings, and a mean value of 3.6 × 105 p/s/cm2/sr was obtained. Mice with luciferase-expressing leukemia reliably became moribund when photon flux approached 2 × 1011 p/s/cm2/sr. For anatomic localization, a pseudocolor map representing light intensity was superimposed over the grayscale body-surface reference image. For confirmation of organ involvement, moribund animals were imaged and then killed and organs were removed, including vertebral body, skull base (calvarium), brain, liver, spleen, thymus, sternum, kidney with adrenal, gonads, and femur. Organs were then imaged immediately for luciferase signal (within the 10-minute peak emission) followed by homogenization or perfusion to isolate cells for flow cytometry. Diffuse luminescence tomography (DLIT) was performed and analyzed as per the manufacturer's instructions.45,46 This algorithm reconstructs the depth of light sources based on the known properties of the emitted light from the luciferase, the index of attenuation of the tissues involved, and an iterative algorithm based on spectral filters acquired in sequence.
Treatment response monitoring
Primary ALL cells from the second passage mice were purified to > 99% ffluc/GFP+ and injected via tail vein into NSG or NOD/SCID mice as indicated (106 cells/mouse). Mice with equivalent levels of bioluminescent disease at day 4 were randomized to either vehicle control or experimental groups. NSG mice were treated by oral gavage with 5 mg/kg sirolimus (SIR) in 0.5 mL 5% dextrose once daily for 24 days where indicated. NOD/SCID mice were treated with immunostimulatory DNA containing unmethylated CpG dinucleotides (CpG ODN), 100 μg in 0.2 mL sterile PBS intraperitoneally every 4 days for 3 doses where indicated.
Statistical considerations
Analysis was performed in Prism 4 (Graphpad Software). In vitro data represent means of duplicates, and comparisons of means were made via Mann-Whitney test. For comparison among multiple groups, Kruskal-Wallis analysis was performed with Dunn Multiple Comparison tests to compare individual groups. Survival curves were compared using the log-rank test with a Bonferroni correction for comparing multiple datasets.
Results
Primary patient ALL can be made bioluminescent with a self-inactivating lentiviral vector encoding firefly luciferase and GFP
The engraftment of primary ALL in immune-deficient mice has been shown to be a selective event.8,47,48 To optimize the transduction of ALL cells with xenografting potential, we transduced primary ALL cells harvested after an initial passage through NSG mice. We have applied the methodology depicted in Figures 1 to 3 separate primary BCP ALL samples (ALL-1, ALL-2, and ALL-3). Over several weeks, a distinct bioluminescent population of each transduced, unsorted sample emerged, although it remained a fraction of the total body disease as indicated by peripheral blood monitoring once tumor appeared. Animals were selected for harvest when the peripheral disease burden (human blasts detected by flow cytometry) approached moribund levels, and human GFP+ ALL cells were isolated from bone marrow and spleen. A small percentage of harvested cells were GFPhigh (0.5%-5%), and 5 × 104 viable GFP+ cells were sorted and reinjected into a second set of NSG mice, this time at > 99% purity for GFP/ffluc expressing cells. These animals developed detectable bioluminescent disease more quickly than the first-passage mice, giving a true bioluminescent picture of the early development of disease in a xenograft model of primary patient leukemia (Figure 1). When moribund, the second passage mice of ALL-1 were killed, and the spleen and marrow harvested and combined. Human leukemia was 85% GFPhigh, and this population was divided and reinjected, either unsorted at 85% purity or after flow sorting to > 99% GFPhigh. The third passage mice were followed for natural history, and the populations from spleen and marrow at terminal disease burdens later showed stable GFPhigh expression (85% or 99%, respectively). This indicates that the transduced population is stable through repeated passages and that the appearance of peripheral blasts and time to death are unchanged from the nontransduced population. In addition, genomic fingerprinting via a combination of STR and SNP analyses revealed no changes in the cells after transduction, or after 3 passages in mice (data not shown).
Bioluminescence can be used to infer organ localization of ALL cells
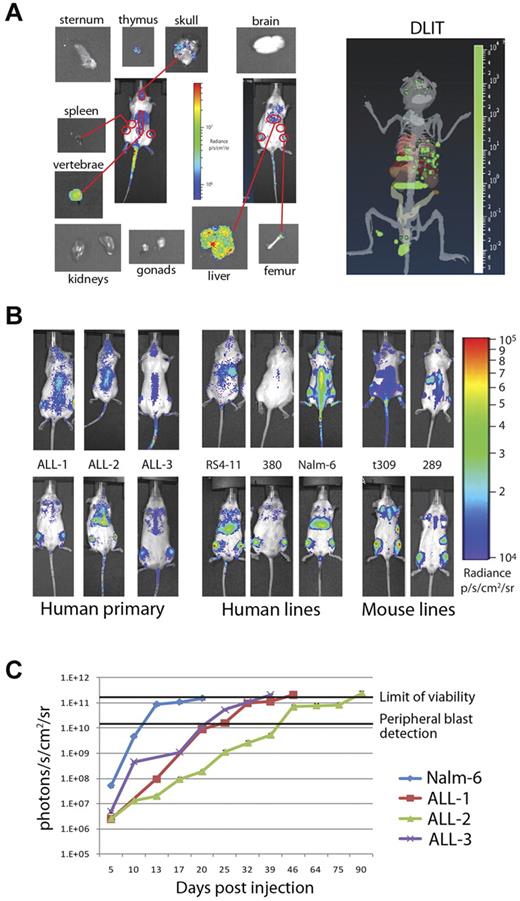
Two-dimensional pseudocolor intensity maps can approximate organ localization but may be dependent on mouse orientation (prone, supine, and decubitus) and sensitivity of the charge-coupled device camera. We combined information from the 2-dimensional intensity maps of mice engrafted with ffluc+ primary ALL with 3-dimensional reconstruction using the DLIT technology. We were able to isolate areas that corresponded to involved organs on the 2-dimensional intensity maps in combination with DLIT. A representative map generated using primary ALL-1 is shown in Figure 2A. To confirm organ involvement and avoid false-positive assumptions, we imaged isolated organs from killed mice (Figure 2A). For each of the ALL samples, the primary sites of early engraftment were the bone marrow and liver, with minor variations seen in other organs (supplemental Figure 1; see the Supplemental Materials link at the top of the article).
Bioluminescence can be used to distinguish patterns and timing of organ involvement for both human and mouse leukemia. (A) Prone, supine, and organ images of an NSG mouse injected with 1 × 106 ffluc+ primary ALL-1 cells. Outlined 2D regions of interest correspond to organ involvement validated at day 3 of engraftment by isolated imaging. DLIT image of mouse internal anatomy, with skin and gut removed for clarity, reconstructed from images is shown (right panel). Green voxels represent point sources of light within putative target organs, verified by ex vivo imaging and flow cytometry. (B) Day 3 intensity maps of NSG mice injected with 1 × 106 ffluc+ leukemia cells as indicated. These representative mice show the differing patterns of organ involvement at this early time point. Human ALL cells establish within the liver and bone marrow (primary ALL, RS4-11, and Nalm-6) or bone marrow only (380), whereas both mouse cell lines studied rapidly home to the spleen and bone marrow (t309 and 289). (C) Time course of leukemia engraftment reveals consistent bioluminescent correlation with appearance of peripheral blasts and maximal moribund disease burden. Nalm-6 (20-22 days), primary ALL-1 (40-50 days), primary ALL-2 (80-100 days), and primary ALL-3 (35-40 days) all become moribund at 2 × 1011 photons/s/cm2. Appearance of peripheral blasts (> 1%) happens consistently at systemic bioluminescent tumor burdens of > 1 × 1010 photons/s/cm2. In ALL-2, the stable bioluminescent burden from 46 to 75 days corresponds with stable but high peripheral blast burden (30%-50%) before a progression to end stage between 80 and 100 days. Results are presented as mean ± SE (n = 5 for primary ALL samples, n = 10 for Nalm-6).
Bioluminescence can be used to distinguish patterns and timing of organ involvement for both human and mouse leukemia. (A) Prone, supine, and organ images of an NSG mouse injected with 1 × 106 ffluc+ primary ALL-1 cells. Outlined 2D regions of interest correspond to organ involvement validated at day 3 of engraftment by isolated imaging. DLIT image of mouse internal anatomy, with skin and gut removed for clarity, reconstructed from images is shown (right panel). Green voxels represent point sources of light within putative target organs, verified by ex vivo imaging and flow cytometry. (B) Day 3 intensity maps of NSG mice injected with 1 × 106 ffluc+ leukemia cells as indicated. These representative mice show the differing patterns of organ involvement at this early time point. Human ALL cells establish within the liver and bone marrow (primary ALL, RS4-11, and Nalm-6) or bone marrow only (380), whereas both mouse cell lines studied rapidly home to the spleen and bone marrow (t309 and 289). (C) Time course of leukemia engraftment reveals consistent bioluminescent correlation with appearance of peripheral blasts and maximal moribund disease burden. Nalm-6 (20-22 days), primary ALL-1 (40-50 days), primary ALL-2 (80-100 days), and primary ALL-3 (35-40 days) all become moribund at 2 × 1011 photons/s/cm2. Appearance of peripheral blasts (> 1%) happens consistently at systemic bioluminescent tumor burdens of > 1 × 1010 photons/s/cm2. In ALL-2, the stable bioluminescent burden from 46 to 75 days corresponds with stable but high peripheral blast burden (30%-50%) before a progression to end stage between 80 and 100 days. Results are presented as mean ± SE (n = 5 for primary ALL samples, n = 10 for Nalm-6).
Having validated the identification of involved organs, we then compared the 3 primary ALL samples to 5 similarly labeled BCP ALL lines (3 human, 2 mouse) for early engraftment kinetics and localization over the first 7 days. The cells are not detectable in the peripheral blood in the first 7 days and are only detectable at minimal levels in femoral bone marrow (0.1%-1% of total cells) by flow cytometry on day 7 (data not shown). Although all human-derived ALL showed preferential early migration to the bone marrow and liver, we observed differences depending both on species of origin as well between cell lines (Figure 2B). The human cell lines (RS4-11, 380, and Nalm-6) exhibited more variability than primary ALL samples. Nalm-6 and RS4-11 rapidly populated the liver and bone marrow but were not detectable in the spleen, whereas 380 rapidly populated the bone marrow but spared the liver and spleen in these early time points. In contrast, the 2 mouse BCP ALL lines (t309 and 289) homed to the bone marrow and spleen, but not liver, within the first 72 hours. Over several weeks, all samples eventually populated all target organs, including liver, bone marrow, and spleen, but the preference and timing of each varied considerably.
Despite variation between ALL samples and cell lines in the rates of engraftment in different target organs, we observed a clear reproducibility in the impact of systemic disease burden on the appearance of blasts in peripheral blood and terminal morbidity (Figure 2C). Detection of > 1% ALL blasts in peripheral blood by flow cytometry did not occur consistently until bioluminescent leukemia burdens of > 1 × 1010 p/s/cm2/sr were reached. Notably, although the level of maximal peripheral disease varies among samples, a whole-body photon flux of 1 to 2 × 1011 p/s/cm2/sr is consistently close to terminal disease burden in all samples. As mice without luciferase-containing cells yielded a mean value of 3.6 × 105 p/s/cm2/sr, a detection range of 6 orders of magnitude is achieved with this approach, greatly expanding the window for treatment response evaluation.
Bioluminescent primary patient leukemia samples enable earlier and more sensitive disease response monitoring
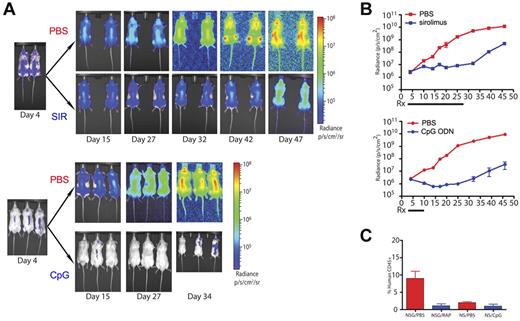
To evaluate the ability to detect treatment response provided by the use of bioluminescent primary ALL and to validate that the transduction process did not significantly alter the overall outcomes, we used 2 treatment models with which we have considerable experience. First, we measured the in vivo response of bioluminescent primary ALL to treatment with sirolimus, an mTOR inhibitor that we have previously demonstrated significantly prolongs survival of mice with xenografted leukemia.13,40 Whereas untreated animals had continual progression of their disease, the bioluminescent burden of disease in sirolimus-treated animals stabilized at a low level (Figures 3A-B). Over time, this represented a significantly reduced leukemia burden (P < .0001), delayed appearance of peripheral blasts, and resulted in extension of overall survival. Once therapy is discontinued, leukemia burden immediately begins to rise, as indicated by bioluminescence levels, even though peripheral blasts are still not detected. Time to appearance of peripheral blasts and moribund disease was not altered from the parent, nontransduced sample (data not shown). Second, in the NOD/SCID model, we applied our published protocol for CpG ODN-induced anti-ALL immune activity.14 In this model, NOD/SCID mice treated with CpG ODN demonstrated a rapid significant reduction in disease burden (P = .0001), whereas control animals had unchecked, progressive disease (Figure 3A-B). Again, after the cessation of treatment, ongoing imaging of the treated mice reveals a steady increase in disease burden.
Bioluminescence enables the visualization and quantification of therapeutic responses of primary ALL significantly earlier than other measurable parameters. (A) Intensity maps of total body disease developing over time in NSG mice treated with sirolimus (SIR) for 24 days (top panels) or in NOD/SCID mice treated with CpG ODN (CpG) over 8 days (bottom panels). Untreated (PBS) controls for both strains are shown. Results obtained with patient sample ALL-1 are shown. (B) Graphs with quantification of the bioluminescent ALL responses over time shown in panel A to treatment with sirolimus (top panel) or CpG ODN (bottom panel). (C) Flow cytometric evaluation of peripheral blood ALL burden at day 45 in the control, sirolimus-treated, and CpG ODN-treated mice shown in panels A and B. Overall, this experiment is not designed to compare the treatment strategies but rather to demonstrate the additional detail of ALL treatment response revealed by bioluminescent imaging.
Bioluminescence enables the visualization and quantification of therapeutic responses of primary ALL significantly earlier than other measurable parameters. (A) Intensity maps of total body disease developing over time in NSG mice treated with sirolimus (SIR) for 24 days (top panels) or in NOD/SCID mice treated with CpG ODN (CpG) over 8 days (bottom panels). Untreated (PBS) controls for both strains are shown. Results obtained with patient sample ALL-1 are shown. (B) Graphs with quantification of the bioluminescent ALL responses over time shown in panel A to treatment with sirolimus (top panel) or CpG ODN (bottom panel). (C) Flow cytometric evaluation of peripheral blood ALL burden at day 45 in the control, sirolimus-treated, and CpG ODN-treated mice shown in panels A and B. Overall, this experiment is not designed to compare the treatment strategies but rather to demonstrate the additional detail of ALL treatment response revealed by bioluminescent imaging.
In both experimental settings, significant treatment effects were clearly apparent weeks before they are detectable by nonbioluminescence methods (Figure 3C). The contrast between the early disease stabilization achieved with sirolimus and the rapid reduction of disease burden by CpG ODN treatment is clearly revealed by the use of bioluminescent primary ALL blasts. In addition, the occurrence of spontaneous regression of established bioluminescent disease in one untreated NOD/SCID mouse was observed (data not shown). Although it has long been observed that some NOD/SCID animals will “fail to engraft” human samples, one assumption has been that the cells died initially. We observed that, in some cases, the disease can engraft but is then spontaneously cleared after 7 days rather than failing to engraft at all; as we observed this only in NOD/SCID and never in NSG mice, the presence of NK cells may be responsible for this disease clearance.
Discussion
In this study, we report that bioluminescent disease monitoring, an approach used widely in other cancer models,49 is achievable with primary human ALL samples. Lentivirus constructs in which Firefly luciferase is coexpressed with GFP provide a sortable marker to isolate cells expressing the bioluminescent reporter. Using this strategy, we demonstrate the reproducible generation of primary ALL samples containing > 99%, stable ffluc-expressing cells. Although there is concern that the in vitro modification of primary leukemia cells could impair engraftment potential,50 the luciferase tagging protocol used here does not appear to significantly alter leukemia propagation; we achieve reliable and stable secondary and tertiary passage rates comparable with those of nontransduced cells from the same original primary ALL sample. In addition, ffluc+ leukemia samples result in disease that is genetically and phenotypically identical to the nontransduced leukemia samples based on STR and SNP analysis, disease kinetics, and response to therapy in our established models.
Our comparison of ALL responses after administration of 2 well-described treatments reveals the ability of the bioluminescence approach to provide more mechanistic and quantitative detail than current evaluation protocols. The use of luciferase-expressing ALL cells clearly and rapidly distinguishes between the apparent global disease-stabilizing effects of sirolimus and the cytotoxic activity of CpG ODN that underlie their superficially comparable inhibition of ALL progression by survival analysis. In addition, the information gained by learning about early anatomic niche localization may also open new avenues of study into the importance of organ specific homing molecules or drug penetration into sites of leukemic harbor. As certain therapies, whether pharmacologic or immunologic, may or may not treat various anatomic niches (eg, liver, CNS, gonads), understanding if or when those areas are infiltrated by the leukemia and whether disease in those areas responds to treatment would contribute significantly to results from preclinical studies.
The noninvasive, longitudinal monitoring of primary ALL xenografts we describe will expedite the investigation and discovery of novel therapies by allowing early detection at minimal disease levels, earlier response measurements, and identification of sites of leukemic relapse or resistance. By enabling (1) the identification of mice with similar organ involvement and equal disease burdens and (2) the precise quantification of pretreatment and post-treatment disease levels and (3) the selection of more appropriate endpoints, bioluminescent imaging will provide a method for standardization of treatment response monitoring in primary ALL xenograft models. This approach may prevent mischaracterization of therapies as ineffective based on current endpoint analysis (overall survival and peripheral blood blast counts). Finally, and not insignificantly, the ability to eliminate nonengrafted mice before the onset of experimental treatments, combined with the more accurate quantification of responses, will reduce the number of animals required to achieve statistical significance in preclinical testing.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Institutes of Health (K12CA076931, D.M.B.; and R01CA102646 and R01CA116660), the Weinberg Foundation (S.A.G.), an American Society for Blood and Marrow Transplantation/Robert A. Good New Investigator Award (A.E.S.), and awards from the Prevent Cancer Foundation and When Everyone Survives Foundation (G.S.D.R.).
National Institutes of Health
Authorship
Contribution: D.M.B., A.E.S., and G.S.D.R. designed the research, performed the experiments, interpreted the data, and wrote the paper C.C., C.H.J., J.D.F., and D.T.T. provided critical reagents and designed the research; and S.A.G. provided critical reagents, interpreted the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gregor S. D. Reid, A5-162 CFRI Translational Research Building, 950 West 28th Avenue, Vancouver, BC V5Z 4H4, Canada; e-mail: grogreid@mail.ubc.ca.