Abstract
We previously showed that HIV infection leads to expansion of a rapidly proliferating pool (s1) of CD4 and CD8 T lymphocytes. In the current study, we used in vivo labeling with bromodeoxyuridine to characterize the kinetics of naive, memory, and activated (HLA-DR+/CD38+) subpopulations of CD4 and CD8 T lymphocytes, and to examine the relationship between kinetic parameters and baseline CD4 counts, HIV viral load, potential markers of microbial translocation, and cytokine levels. Activated cells showed the highest proliferation rates, followed by effector and central memory cells, with naive cells showing the lowest rates, for both CD4 and CD8 T cells. HIV viral load correlated with s1 of CD4 and CD8 effector memory cells, as well as CD8 naive cells, whereas CD4 cell counts correlated inversely with naive CD4 s1. Endotoxin levels showed a weak negative association with CD4 but not CD8 s1. INF-γ and TNF-α were associated with s1 for CD4 and CD8 cells, respectively. Thus, HIV is the primary driving force behind the activation and proliferation of most subsets of both CD4 and CD8 T lymphocytes, whereas naive CD4 cell proliferation likely represents a homeostatic response. Microbial translocation does not appear to play an important role in this proliferation.
Introduction
The hallmark of untreated HIV infection is an inexorable CD4 T-cell depletion in the majority of patients, yet the mechanisms leading to this depletion in vivo remain uncertain. Immune activation appears to play a major role in the pathogenesis of HIV infection and probably contributes to this CD4 depletion.1-4 One marker of immune activation, CD38 expression by CD4 and CD8 cells, has been shown to be an important prognostic marker for mortality or disease progression that is independent of viral load (VL).5,6
A major manifestation of immune activation is an increase in cell proliferation and death, especially of CD4 and CD8 T cells. In vivo labeling with bromodeoxyuridine (BrdU) provides a method for direct measurement of proliferation while allowing labeled cells to be followed longitudinally by flow cytometry. This leads to a more accurate determination of kinetic parameters, such as the decay rate and half-life of proliferating cells, than indirect methods, such as Ki67 staining provide.7-11 In a previously published study,11 we used BrdU labeling to identify rapidly and slowly proliferating subpopulations of CD4 and CD8 T cells and to demonstrate that plasma HIV levels were a primary determinant of lymphocyte turnover. These and other studies strongly suggested that increased cell turnover was a direct consequence of HIV infection and appeared to be a manifestation of immune activation rather than simply a homeostatic response to CD4 depletion.6,10,11
In the current study, we have extended these findings to a larger patient population to examine proliferation and decay kinetics in naive, memory, and activated (HLA-DR+/CD38+) subsets of CD4 and CD8 T cells, and to examine the relationship between these kinetic parameters and potential modulators of immune activation, including baseline CD4 T-cell numbers, plasma HIV VL, potential markers of microbial translocation, and serum or plasma cytokine levels.
Methods
Patients
A total of 41 HIV-1 infected patients, who were in various stages of infection and with no major clinical or laboratory abnormalities were enrolled in the study between January 1999 and November 2004. Ten patients were immunologic nonresponders (CD4 count < 300 cells/mm3 and VL < 50 copies/mL after > 1 year of highly active antiretroviral therapy [HAART]). Patients were excluded from participation if they were pregnant or breastfeeding, or receiving 5-fluorouracil. The study was approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board, and all patients provided written informed consent in accordance with the Declaration of Helsinki.
BrdU administration
BrdU was supplied initially by NeoPharm and subsequently by the National Cancer Institute and administered over 30 minutes by intravenous infusion at a dose of 200 mg/m2 as previously described.11 After the infusion, blood was collected at 4 or more time points a minimum of 1 day apart during the first week, at a median of 3 time points during the next 3 weeks, and then approximately monthly.
Flow cytometry
Ethylenediaminetetraacetic acid–treated whole blood was analyzed for surface markers and incorporated BrdU by flow cytometry as previously described.2,12 Cryo-preserved peripheral mononuclear cells were stained with phycoerythrin-labeled anti-Ki67 (clone B56; BD Biosciences PharMingen) as described.13
VL measurement
Plasma VL was measured by a bDNA assay with a lower detection limit of 50 HIV RNA copies/mL (Versant HIV-1 Version 3.0; Siemens). For statistical purposes, samples with a VL < 50 copies/mL were assigned a value of 50.
Plasma markers of microbial translocation and serum cytokine levels
Plasma levels of endotoxin were measured by the Limulus Amebocyte Assay (Lonza Switzerland), and plasma levels of soluble CD14 (sCD14; R&D Systems), lipopolysaccharide-binding protein (LBP; Cell Sciences), and endotoxin-core antibodies (EndoCAb; Cell Sciences) were measured by commercial ELISA kits as previously described.14,15 All samples except 1 were obtained within 1 week of the BrdU infusion. Serum IL-1β, IL-2, IL-5, IL-6, IL-10, INF-γ, and TNF-α levels were measured by a 7-plex commercial kit (Meso Scale Discovery). Serum IL-7 was measured by commercial ELISA (R&D Systems). Thirty-three samples were obtained within 1 month of the BrdU infusion, 4 within 2 months, and 4 beyond 2 months.
Modeling and statistical analysis
Baseline T-cell counts and baseline VL were defined as the arithmetic mean of the values obtained at the visits during the first 7 days after the BrdU infusion.
A previously developed semiempirical model was used to describe the kinetics of BrdU-labeled T cells in the blood.11 Briefly, the model considers that the kinetics of the fraction of labeled T cells after the peak of labeling consists of a 2-phase decay with significantly different slopes, suggesting the existence of 2 different subpopulations of cells each decaying with its own rate constant d (time−1). For each subpopulation, the model estimates the rate constant d (disappearance rate of the fraction of labeled cells in the blood), which reflects cell death, delabeling, and/or redistribution outside of the blood/lymphatic system, and the source rate of labeled cells, s (fraction of labeled cells entering the blood per unit time), which is proportional to the cell proliferation in the lymphoid tissue at the time of labeling. In addition, the model estimates the time required to reach the maximum concentration of labeled cells in the blood, τ, which correlates with the effects of trafficking from lymphoid tissues into the blood pool. The labeling data from each patient were fitted to the differential equations using the nonlinear least-squares Levenberg-Marquardt algorithm16 and were solved using Labview Version 8.0 (National Instruments).
The nonparametric Wilcoxon signed-rank and Wilcoxon rank-sum tests were used for paired and unpaired comparisons, respectively. The associations between kinetic and baseline parameters were assessed using the Spearman rank correlation coefficient (ρ) and by simple or multivariate linear regression analysis (with and without interaction of covariates). To account for the association between repeated measures from the 6 patients who received 2 BrdU infusions, the generalized estimating equation method17 was also applied. The predictive value, which is the proportion of variability explained by the regression model, was calculated for assessing goodness of fit of linear regressions. The predictive values of 2 nested models were compared using the F test. All P values were 2-sided and were not adjusted for multiple comparisons. P values < .05 were considered statistically significant.
Results
Patient characteristics
A total of 41 HIV-infected patients were enrolled in the study. At enrollment, the mean baseline CD4 count was 313 cells/μL, the mean baseline CD8 count was 959 cells/μL, and the mean baseline log-transformed VL was 3.24 log10 copies/mL (1739 copies/mL); 11 patients had VLs < 50 copies/mL. Twenty-seven of the 41 patients were receiving combination antiretroviral therapy: 19 patients with VL < 400 copies/mL and 8 patients with VL > 1000 copies/mL. Six patients received 2 infusions of BrdU; the first was while receiving no or ineffective antiretroviral therapy, and the second was 8 to 26 weeks after starting an effective HAART regimen. At the time of second infusion, 5 patients had VLs < 50 copies/mL. Some results for 17 patients, including the 6 with 2 BrdU infusions, have been previously reported.11,18 The patient's characteristics and the datasets available for each analysis are summarized in Table 1.
Baseline patient characteristics grouped by the datasets used for analysis
| Dataset . | No. of patients . | No. of kinetics . | Laboratory parameter . | Baseline values . | Mean age, y (range) . | Sex . | ||
|---|---|---|---|---|---|---|---|---|
| Mean . | Median . | Range . | ||||||
| Set 1 | 41 | 47* | CD4 (cells/μL) | 339 | 264 | 59-1013 | 44 (20-60) | 39 male |
| CD8 (cells/μL) | 951 | 808 | 180-2657 | 2 female | ||||
| log10VL (copies/mL) | 3.06 | 2.47 | < 1.70-6.15 | |||||
| Set 2† | 41 | 41 | CD4 (cells/μL) | 313 | 261 | 59-800 | 44 (20-60) | 39 male |
| CD8 (cells/μL) | 959 | 808 | 180-2657 | 2 female | ||||
| log10VL (copies/mL) | 3.24 | 3.06 | < 1.70-6.15 | |||||
| Set 3‡ | 22 | 22 | CD4 (cells/μL) | 300 | 235 | 78-636 | 44 (20-60) | 21 male |
| CD8 (cells/μL) | 976 | 739 | 180-2657 | 1 female | ||||
| log10VL (copies/mL) | 2.71 | 1.82 | < 1.70-6.15 | |||||
| % CD4: RA+27+ | 29 | 25 | 3-68 | |||||
| % CD4: RA−27+ | 51 | 50 | 28-78 | |||||
| % CD4: RA+27− | 5 | 5 | 1-21 | |||||
| % CD4: RA−27− | 15 | 11 | 1-35 | |||||
| % CD4: DR+38+ | 11 | 9 | 3-22 | |||||
| % CD8: RA+27+ | 30 | 27 | 6-83 | |||||
| % CD8: RA−27+ | 32 | 27 | 11-63 | |||||
| % CD8: RA+27− | 19 | 18 | 2-37 | |||||
| % CD8: RA−27− | 19 | 16 | 1-53 | |||||
| % CD8: DR+38+ | 19 | 16 | 2-55 | |||||
| Dataset . | No. of patients . | No. of kinetics . | Laboratory parameter . | Baseline values . | Mean age, y (range) . | Sex . | ||
|---|---|---|---|---|---|---|---|---|
| Mean . | Median . | Range . | ||||||
| Set 1 | 41 | 47* | CD4 (cells/μL) | 339 | 264 | 59-1013 | 44 (20-60) | 39 male |
| CD8 (cells/μL) | 951 | 808 | 180-2657 | 2 female | ||||
| log10VL (copies/mL) | 3.06 | 2.47 | < 1.70-6.15 | |||||
| Set 2† | 41 | 41 | CD4 (cells/μL) | 313 | 261 | 59-800 | 44 (20-60) | 39 male |
| CD8 (cells/μL) | 959 | 808 | 180-2657 | 2 female | ||||
| log10VL (copies/mL) | 3.24 | 3.06 | < 1.70-6.15 | |||||
| Set 3‡ | 22 | 22 | CD4 (cells/μL) | 300 | 235 | 78-636 | 44 (20-60) | 21 male |
| CD8 (cells/μL) | 976 | 739 | 180-2657 | 1 female | ||||
| log10VL (copies/mL) | 2.71 | 1.82 | < 1.70-6.15 | |||||
| % CD4: RA+27+ | 29 | 25 | 3-68 | |||||
| % CD4: RA−27+ | 51 | 50 | 28-78 | |||||
| % CD4: RA+27− | 5 | 5 | 1-21 | |||||
| % CD4: RA−27− | 15 | 11 | 1-35 | |||||
| % CD4: DR+38+ | 11 | 9 | 3-22 | |||||
| % CD8: RA+27+ | 30 | 27 | 6-83 | |||||
| % CD8: RA−27+ | 32 | 27 | 11-63 | |||||
| % CD8: RA+27− | 19 | 18 | 2-37 | |||||
| % CD8: RA−27− | 19 | 16 | 1-53 | |||||
| % CD8: DR+38+ | 19 | 16 | 2-55 | |||||
Six patients received a BrdU infusion before and after an effective HAART therapy.
First infusion for all 41 patients.
Patients for whom the BrdU kinetics of the subsets were also available.
Differential incorporation of BrdU in CD4 and CD8 T lymphocytes
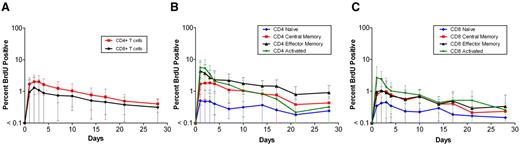
Data on BrdU incorporation in total CD4 and CD8 T-cell populations were available for all 41 patients (Figure 1A). Maximum BrdU incorporation for these cells ranged from 0.2% to 5.9% and usually occurred 1 to 3 days after the infusion. Of note, maximal percentage of labeled CD4 T cells was significantly higher than maximal percentage of labeled CD8 T cells (2.37% vs 1.46%, P = .002). There was a significant association between baseline VL and peak BrdU incorporation for both CD4 (ρ = 0.40, P = .009) and CD8 T cells (ρ = 0.60, P < .001). In addition, the baseline CD4 count correlated negatively with CD4 (ρ = −0.50, P < .001) but not CD8 (ρ = −0.09, P = .29) peak labeling. For 22 patients, each of whom received a single BrdU infusion, staining with CD45RA and CD27 was used to follow BrdU incorporation in naive (RA+27+), central memory (RA−27+) and effector memory (RA−27− plus RA+27−) subsets, and staining with HLA-DR and CD38 for following activated (HLA-DR+38+) subsets of both CD4 and CD8 T lymphocytes. The labeling in these subsets followed the same pattern seen with the whole population, peaking at days 1 to 4 after the infusion, followed by a gradual decline (Figure 1B-C). Higher peak labeling in CD4 compared with the respective CD8 subsets was observed for central memory (2.21% vs 1.20%, P = .002), effector memory (5.21% vs 2.00%, P < .001), and activated (8.01% vs 3.73%, P = .006) subsets, but not naive cells (0.76% vs 0.59%, P = .68).
Kinetics of BrdU-labeled lymphocytes in blood. Graphs represent the kinetics for total CD4 and CD8 T cells (A), and naive, central memory, effector memory, and activated subsets of CD4 (B) and CD8 (C) T cells. The x-axis represents the days following the BrdU infusion; and the y-axis, in log10 scale, the percentage of BrdU-positive cells as detected by flow cytometry. Data represent the geometric mean (± SD) for the first infusion for all 41 patients (panel A; Table 1, dataset 2) and the 22 patients for whom the BrdU kinetics of the subsets were also available (panels B-C; Table 1, dataset 3).
Kinetics of BrdU-labeled lymphocytes in blood. Graphs represent the kinetics for total CD4 and CD8 T cells (A), and naive, central memory, effector memory, and activated subsets of CD4 (B) and CD8 (C) T cells. The x-axis represents the days following the BrdU infusion; and the y-axis, in log10 scale, the percentage of BrdU-positive cells as detected by flow cytometry. Data represent the geometric mean (± SD) for the first infusion for all 41 patients (panel A; Table 1, dataset 2) and the 22 patients for whom the BrdU kinetics of the subsets were also available (panels B-C; Table 1, dataset 3).
Rapidly and slowly proliferating subpopulations of T lymphocytes
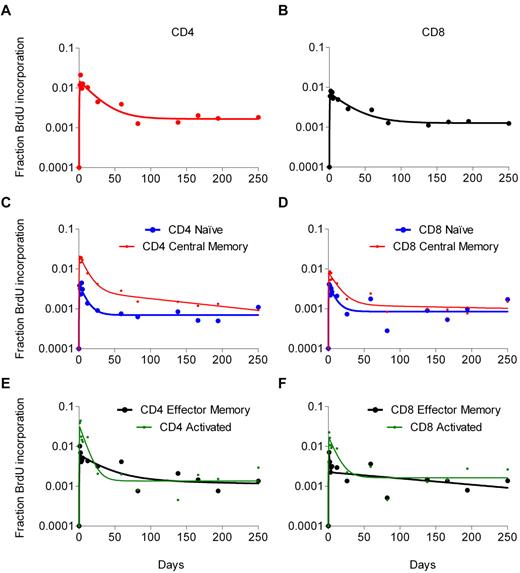
The relationship between lymphocyte turnover and immunologic or virologic parameters was examined for 47 BrdU infusions in 41 patients (Table 1). As previously reported, the decay in labeling of CD4 and CD8 T cells was best described by a semiempirical model that incorporates a 2-phase decay representing 2 populations of cells: a rapidly proliferating pool and a slowly proliferating pool.11 In that model, s is approximately proportional to the percentage of a given population that incorporated BrdU, and d represents the decay rate of labeled cells in that population. Thus, s1 and d1 are the kinetic parameters for the rapidly proliferating pool of cells, and s2 and d2 for the slowly proliferating pool. To understand the dynamics within the subsets, a similar analysis was performed for each subset. Figure 2 illustrates the 2-phase decay of BrdU label for total CD4 and CD8 T cells and their subsets for a single patient.
BrdU labeling kinetics of total CD4 and CD8 T cells and their subsets for a representative patient. (Left panels) Data from CD4 cells. (Right panels) Data from CD8 cells. The continuous lines represent the best fit of the experimental data (dots) to the model equations.
BrdU labeling kinetics of total CD4 and CD8 T cells and their subsets for a representative patient. (Left panels) Data from CD4 cells. (Right panels) Data from CD8 cells. The continuous lines represent the best fit of the experimental data (dots) to the model equations.
Analysis of the 47 sets of kinetics (Table 2) shows that, for the study population as a whole, CD4 s1 is significantly greater than CD8 s1 (0.012 day vs 0.009 day, P = .001). Moreover, CD4 s1 is highly correlated with CD8 s1 for the same patient (ρ = 0.66, P < .001). Similar to the peak labeling, s1 was greater in CD4 compared with respective CD8 subsets except naive cells (central memory, 0.014 day vs 0.007 day, P = .05; effector memory, 0.027 day vs 0.009 day, P < .001; activated, 0.079 day vs 0.049 day, P = .011; naive, 0.005 day vs 0.004 day, P = .69).
Estimated values of source rate (s), the decay constant (d), and the time to reach the peak of labeling (τ) and the calculated values of total source rate (s3), mean decay constant (dmean), and the half-life (T1/2) for total CD4 and CD8 T cells and subsets
| . | n . | s1,/day . | s2,/day . | d1,/day . | d2,/day . | τ,/day . | stotal,/day . | dmean,/day . | T1/2mean,/day . |
|---|---|---|---|---|---|---|---|---|---|
| CD4 | 47 | 0.012 (0.008-0.021) | 0.004 (0.002-0.008) | 0.197 (0.098-0.330) | 0.016 (0.005-0.033) | 1.354 (1.014-1.743) | 0.019 (0.013-0.027) | 0.127 (0.084-0.242) | 5.4 |
| CD8 | 46 | 0.009 (0.004-0.017) | 0.003 (0.001-0.004) | 0.206 (0.081-0.398) | 0.011 (0.001-0.027) | 1.564 (1.177-1.915) | 0.011 (0.006-0.021) | 0.181 (0.083-0.335) | 3.8 |
| CD4 naive | 22 | 0.005 (0.002-0.007) | 0.002 (0.001-0.003) | 0.200 (0.059-0.579) | 0.004 (0.000-0.013) | 1.114 (0.910-1.382) | 0.007 (0.004-0.009) | 0.167 (0.069-0.397) | 4.2 |
| CD4 central memory | 22 | 0.014 (0.010-0.019) | 0.004 (0.002-0.005) | 0.117 (0.087-0.152) | 0.007 (0.003-0.013) | 1.413 (1.082-1.598) | 0.018 (0.014-0.024) | 0.098 (0.078-0.124) | 7.1 |
| CD4 effector memory | 21 | 0.027 (0.011-0.058) | 0.007 (0.003-0.011) | 0.329 (0.227-0.752) | 0.013 (0.001-0.021) | 0.910 (0.742-1.044) | 0.035 (0.018-0.065) | 0.212 (0.176-0.604) | 3.3 |
| CD4 activated | 22 | 0.079 (0.060-0.119) | 0.007 (0.002-0.016) | 0.321 (0.206-0.632) | 0.013 (0.000-0.020) | 1.113 (0.827-1.311) | 0.084 (0.064-0.142) | 0.309 (0.195-0.562) | 2.2 |
| CD8 naive | 21 | 0.004 (0.002-0.006) | 0.001 (0.000-0.002) | 0.221 (0.063-0.426) | 0.002 (0.000-0.007) | 1.316 (0.902-2.061) | 0.006 (0.002-0.009) | 0.211 (0.121-0.386) | 3.3 |
| CD8 central memory | 22 | 0.007 (0.005-0.012) | 0.002 (0.001-0.004) | 0.107 (0.069-0.252) | 0.003 (0.000-0.011) | 1.421 (0.999-1.942) | 0.009 (0.006-0.017) | 0.123 (0.070-0.251) | 5.6 |
| CD8 effector memory | 22 | 0.009 (0.004-0.014) | 0.001 (0.000-0.003) | 0.140 (0.033-0.439) | 0.002 (0.000-0.010) | 0.855 (0.719-1.259) | 0.010 (0.004-0.017) | 0.196 (0.078-0.567) | 3.5 |
| CD8 activated | 22 | 0.049 (0.024-0.088) | 0.005 (0.002-0.007) | 0.410 (0.127-0.721) | 0.009 (0.001-0.017) | 0.796 (0.633-1.151) | 0.055 (0.028-0.096) | 0.413 (0.187-0.651) | 1.7 |
| . | n . | s1,/day . | s2,/day . | d1,/day . | d2,/day . | τ,/day . | stotal,/day . | dmean,/day . | T1/2mean,/day . |
|---|---|---|---|---|---|---|---|---|---|
| CD4 | 47 | 0.012 (0.008-0.021) | 0.004 (0.002-0.008) | 0.197 (0.098-0.330) | 0.016 (0.005-0.033) | 1.354 (1.014-1.743) | 0.019 (0.013-0.027) | 0.127 (0.084-0.242) | 5.4 |
| CD8 | 46 | 0.009 (0.004-0.017) | 0.003 (0.001-0.004) | 0.206 (0.081-0.398) | 0.011 (0.001-0.027) | 1.564 (1.177-1.915) | 0.011 (0.006-0.021) | 0.181 (0.083-0.335) | 3.8 |
| CD4 naive | 22 | 0.005 (0.002-0.007) | 0.002 (0.001-0.003) | 0.200 (0.059-0.579) | 0.004 (0.000-0.013) | 1.114 (0.910-1.382) | 0.007 (0.004-0.009) | 0.167 (0.069-0.397) | 4.2 |
| CD4 central memory | 22 | 0.014 (0.010-0.019) | 0.004 (0.002-0.005) | 0.117 (0.087-0.152) | 0.007 (0.003-0.013) | 1.413 (1.082-1.598) | 0.018 (0.014-0.024) | 0.098 (0.078-0.124) | 7.1 |
| CD4 effector memory | 21 | 0.027 (0.011-0.058) | 0.007 (0.003-0.011) | 0.329 (0.227-0.752) | 0.013 (0.001-0.021) | 0.910 (0.742-1.044) | 0.035 (0.018-0.065) | 0.212 (0.176-0.604) | 3.3 |
| CD4 activated | 22 | 0.079 (0.060-0.119) | 0.007 (0.002-0.016) | 0.321 (0.206-0.632) | 0.013 (0.000-0.020) | 1.113 (0.827-1.311) | 0.084 (0.064-0.142) | 0.309 (0.195-0.562) | 2.2 |
| CD8 naive | 21 | 0.004 (0.002-0.006) | 0.001 (0.000-0.002) | 0.221 (0.063-0.426) | 0.002 (0.000-0.007) | 1.316 (0.902-2.061) | 0.006 (0.002-0.009) | 0.211 (0.121-0.386) | 3.3 |
| CD8 central memory | 22 | 0.007 (0.005-0.012) | 0.002 (0.001-0.004) | 0.107 (0.069-0.252) | 0.003 (0.000-0.011) | 1.421 (0.999-1.942) | 0.009 (0.006-0.017) | 0.123 (0.070-0.251) | 5.6 |
| CD8 effector memory | 22 | 0.009 (0.004-0.014) | 0.001 (0.000-0.003) | 0.140 (0.033-0.439) | 0.002 (0.000-0.010) | 0.855 (0.719-1.259) | 0.010 (0.004-0.017) | 0.196 (0.078-0.567) | 3.5 |
| CD8 activated | 22 | 0.049 (0.024-0.088) | 0.005 (0.002-0.007) | 0.410 (0.127-0.721) | 0.009 (0.001-0.017) | 0.796 (0.633-1.151) | 0.055 (0.028-0.096) | 0.413 (0.187-0.651) | 1.7 |
Median (interquartile range) of the estimated source rate (s), the decay constant (d), the total source rate (stotal = s1 + s2), mean decay constant [dmean = (s1 × d1 + s2 × d2)/(s1 + s2)], and the half-life (T1/2 = 0.693/dmean) for the 2 subpopulations of T lymphocytes from a total of 47 BrdU infusions in 41 patients for CD4 and CD8 T cells (46 infusions for CD8 T cells); 1 patient, with unusually high CD8 T-cell proliferation after therapy (about 1 order of magnitude higher than the average, statistically an outlier) was excluded from the analysis; 22 infusions from 22 patients for CD4 and CD8 T-cell subsets; 1 patient with very high effector memory CD4 T-cell estimates (24-fold higher than the average) and 1 patient with very high naive CD8 T-cell estimates (38-fold higher than the average) were excluded from the analysis. Also shown are the estimates of time to reach the maximum concentration of labeled cells in the blood (τ). The values for half-life represent minimal estimates because the rate constant d includes the effects of delabeling and redistribution.
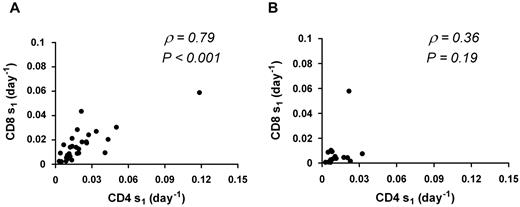
When the analysis was limited to the 31 sets of kinetics characterized by baseline VL > 50 copies/mL, results were similar: CD4 s1 is greater than CD8 s1 (0.014 day vs 0.013 day, P = .009), and within patients CD4 s1 is highly correlated with CD8 s1 (ρ = 0.79, P < .001). In contrast, no significant association between CD4 s1 and CD8 s1was seen for patients with VL < 50 copies/mL (ρ = 0.36, P = .19; n = 16; Figure 3).
Correlation between CD4 s1 and CD8 s1. Thirty-one sets of kinetics with VL > 50 copies/mL (A) and 16 sets of kinetics with VL < 50 copies/mL (B). ρ represents the Spearman rank correlation coefficient. A significant correlation is seen for patients with VLs > 50 copies/mL that is lost in patients with VL < 50 copies/mL.
Correlation between CD4 s1 and CD8 s1. Thirty-one sets of kinetics with VL > 50 copies/mL (A) and 16 sets of kinetics with VL < 50 copies/mL (B). ρ represents the Spearman rank correlation coefficient. A significant correlation is seen for patients with VLs > 50 copies/mL that is lost in patients with VL < 50 copies/mL.
Among the CD4 and CD8 T-cell subsets, CD4 s1 within the central memory pool and within the effector memory pool was 2- to 5-fold higher (P < .001) than CD4 s1 within the naive pool (Table 2; Figure 4). In addition, in both CD4 and CD8 T-cell populations, s1 of cells within the activated pool was significantly higher (P < .001) than s1 within the other subsets (Figure 4).
Comparison of size of the rapidly proliferating subpopulation (s1) of cells in CD4 and CD8 T lymphocyte subsets. Significant differences between groups are indicated by the lines above the groups, together with the P values. Bar (black) represents the median value. One patient from CD4 effector memory and a different patient from CD8 naive subset were excluded from the analysis because of unusually high model estimates, statistically an outlier.
Comparison of size of the rapidly proliferating subpopulation (s1) of cells in CD4 and CD8 T lymphocyte subsets. Significant differences between groups are indicated by the lines above the groups, together with the P values. Bar (black) represents the median value. One patient from CD4 effector memory and a different patient from CD8 naive subset were excluded from the analysis because of unusually high model estimates, statistically an outlier.
The time to reach the peak of labeling (τ) probably represents the time needed to complete cell division and transit from lymphoid tissue, where cell division primarily occurs, to the blood pool. No significant differences in τ were observed between total CD4 and total CD8 T cells. However, for both CD4 and CD8 T cells, τCentral Memory was significantly longer than τEffector Memory (P = < 0.001) or τActivated (P < .005), but no significant differences were observed between τNaive and τCentral Memory (Table 2)
There was no significant difference in the decay rates (d1) between rapidly proliferating CD4 and CD8 T cells, or in d1 between CD4 subsets and their respective CD8 subsets.
Peak labeling and total s of proliferating CD4 and CD8 T cell pools correlate with Ki67 expression
We compared the most commonly used surrogate marker for lymphocyte proliferation in vivo, Ki67, with the peak BrdU labeling and the kinetic parameters of the model. There was a positive correlation between the BrdU labeling peak and the percentage of Ki67-positive cells for both CD4 (ρ = 0.44, P = .008; n = 35) and CD8 (ρ = 0.67, P < .001; n = 34) T cells. Similar to peak BrdU labeling, the percentage of Ki67+ in CD4 T cells was higher than the percentage of Ki67+ in CD8 T cells (6.6% vs 5.9%, P = .048; n = 35). In addition, there was a positive correlation between the total s (stotal = s1 + s2) of the proliferating T cells and the percentage of cells positive for Ki67 for both CD4 (ρ = 0.57, P < .001) and CD8 cells (ρ = 0.78, P < .001).
Association of CD4 and CD8 T-cell proliferation with CD4 T-cell count and HIV VL
There was a significant association between baseline VL and s1 for both CD4 (ρ = 0.49, P < .001; n = 47) and CD8 T cells (ρ = 0.69, P < .001; n = 46). There were no significant correlations between baseline VL and s2 or either of the decay constants. In addition, there was a significant negative correlation between baseline CD4 count and CD4 (but not CD8) s1 (ρ = −0.38, P = .009; n = 47) and CD4 (but not CD8) s2 (ρ = −0.49, P < .001; n = 47). Because the CD4 count is typically correlated negatively with the VL, we estimated the relative contribution of VL and CD4 count through a multivariate analysis with s1 or s2 as the outcome of the model (Table 3). For CD4 s1 and CD8 s1, the predictive value of VL alone was 22.9% and 29.9%, respectively. Although the predictive value of CD4 s1 increased to 29.9% with the addition of CD4 count (P = .04), the increase was not significant for CD8 s1 (to 33.2%, P = .15). Similar results were seen for the subset of kinetics in patients with VL > 50 copies/mL, although the predictive value increased to 34% and 46%, respectively, in the multivariate analysis. These data suggest that CD4 T-cell proliferation (as defined by s1 or s2; data not shown) is a complex function driven by both homeostatic and virologic pressures, whereas for CD8 T cells, proliferation (as defined by s1) is primarily driven by VL. Neither VL nor the baseline CD4 T-cell count was significantly correlated with CD8 s2 (results not shown). Similar results were obtained when the multivariate analysis included the interaction of CD4 T-cell count and VL as an additional covariate. Finally, generalized estimating equation analysis, which takes into account the association between multiple infusions, also confirmed the above findings on the driving factors of CD4 and CD8 T-cell proliferation (results not shown). Multivariate analysis with d1 or d2 as the outcome showed no significant associations with baseline VL or baseline CD4 T-cell count for CD4 or CD8 T cells.
Multivariate linear regression analysis of s1 for total CD4 and total CD8 T cells with baseline CD4 T-cell count and baseline VL (log10) as covariates
| . | CD4 s1 . | CD8 s1 . | ||||
|---|---|---|---|---|---|---|
| Univariate . | Bivariate . | Univariate . | Bivariate . | |||
| Coefficient of CD4, × 103 (cells/μL) | −0.024 | −0.022 | −0.012 | — | −0.011 | |
| (P = .055) | (P = .041) | (P = .20) | (P = .151) | |||
| Coefficient of log10VL, × 103 (log10 copies/mL) | — | 6.116 | 5.997 | — | 5.076 | 5.046 |
| (P< .001) | (P< .001) | (P< .001) | (P< .001) | |||
| R2 | 0.079 | 0.229 | 0.299 | 0.037 | 0.299 | 0.332 |
| . | CD4 s1 . | CD8 s1 . | ||||
|---|---|---|---|---|---|---|
| Univariate . | Bivariate . | Univariate . | Bivariate . | |||
| Coefficient of CD4, × 103 (cells/μL) | −0.024 | −0.022 | −0.012 | — | −0.011 | |
| (P = .055) | (P = .041) | (P = .20) | (P = .151) | |||
| Coefficient of log10VL, × 103 (log10 copies/mL) | — | 6.116 | 5.997 | — | 5.076 | 5.046 |
| (P< .001) | (P< .001) | (P< .001) | (P< .001) | |||
| R2 | 0.079 | 0.229 | 0.299 | 0.037 | 0.299 | 0.332 |
— indicates not applicable.
Immunologic and virologic parameters contributing to the proliferation of naive, memory, and activated CD4 and CD8 T-cell subsets
In the CD4 T-cell subsets, there was a negative correlation between baseline CD4 T-cell counts (but not baseline VL) and CD4 naive s1 (ρ = −0.51, P = .014) and s2 (ρ = −0.45, P = .036); whereas in the CD8 T-cell subsets, there was a positive correlation between baseline VL (but not baseline CD4 T-cell count) and CD8 naive s1 (ρ = 0.49, P = .024). In addition, there was a positive correlation between baseline VL and CD8 effector memory s1 (ρ = 0.56, P = .006) and s2 (ρ = 0.54, P = .009). There were no other significant associations of VL or CD4 T-cell counts with either of the source rates in the remaining subsets of CD4 or CD8 T lymphocytes.
To better explain the differential effects in the subsets, we performed multivariate linear regression analysis using baseline CD4 counts and baseline VL as covariates and s1 as the outcome of the model (Table 4). Intriguingly, the CD4 count seems to modulate the proliferation of naive CD4 T cells (P = .037), whereas the VL seems to modulate naive CD8 T-cell proliferation (P = .002). VL is weakly associated with central memory CD4 (P = .033) but not central memory CD8 (P = .33) proliferation and is strongly associated with the proliferation of effector memory cells in both CD4 (P = .006) and CD8 (P < .001) T-cell populations. Although the percentage of cells expressing HLA-DR+38+ was positively correlated with VL as expected (% CD4: HLA-DR+38+, ρ = 0.45, P = .035; % CD8: HLA-DR+38+, ρ = 0.42, P = .054), neither covariate seems to influence the proliferation of activated cells within that pool for either CD4 or CD8 T cells.
Multivariate linear regression analysis of s1 for CD4 and CD8 T-cell subsets with baseline CD4 T-cell count and baseline VL (log10) as covariates
| . | CD4 naive s1 . | CD8 naive s1* . | CD4 CM s1 . | CD8 CM s1 . | CD4 EM s1† . | CD8 EM s1 . | CD4 Act s1 . | CD8 Act s1 . |
|---|---|---|---|---|---|---|---|---|
| Coefficient of CD4, × 103 (cells/μL) | −0.021 | −0.004 | −0.019 | −0.01 | −0.067 | −0.008 | −0.124 | −0.042 |
| (P = .037) | (P = .52) | (P = .16) | (P = .73) | (P = .06) | (P = .41) | (P = .12) | (P = .69) | |
| Coefficient of log10VL, × 103 (log10 copies/mL) | 0.901 | 2.526 | 3.522 | 3.291 | 12.733 | 4.613 | 5.516 | −7.615 |
| (P = .44) | (P = .002) | (P = .033) | (P = .33) | (P = .006) | (P< .001) | (P = .56) | (P = .55) | |
| R2 | 0.217 | 0.443 | 0.221 | 0.051 | 0.355 | 0.49 | 0.125 | 0.056 |
| . | CD4 naive s1 . | CD8 naive s1* . | CD4 CM s1 . | CD8 CM s1 . | CD4 EM s1† . | CD8 EM s1 . | CD4 Act s1 . | CD8 Act s1 . |
|---|---|---|---|---|---|---|---|---|
| Coefficient of CD4, × 103 (cells/μL) | −0.021 | −0.004 | −0.019 | −0.01 | −0.067 | −0.008 | −0.124 | −0.042 |
| (P = .037) | (P = .52) | (P = .16) | (P = .73) | (P = .06) | (P = .41) | (P = .12) | (P = .69) | |
| Coefficient of log10VL, × 103 (log10 copies/mL) | 0.901 | 2.526 | 3.522 | 3.291 | 12.733 | 4.613 | 5.516 | −7.615 |
| (P = .44) | (P = .002) | (P = .033) | (P = .33) | (P = .006) | (P< .001) | (P = .56) | (P = .55) | |
| R2 | 0.217 | 0.443 | 0.221 | 0.051 | 0.355 | 0.49 | 0.125 | 0.056 |
CM indicates central memory; EM, effector memory; and Act, activated.
One patient with very high naive CD8 T-cell estimate for s1 (38-fold higher than the average, statistically an outlier) was excluded from the CD8 naive analysis. Significant association with the VL is lost with the inclusion of this outlier.
One patient with very high effector memory CD4 T-cell estimate for s1 (24-fold higher than the average, statistically an outlier) was excluded from the CD4 EM analysis. Significant association with the VL is lost with the inclusion of this outlier.
Relationship between markers of microbial translocation and proliferation
Plasma levels of bacterial endotoxin and 3 other potential markers of microbial translocation, plasma lipopolysaccharide-binding protein, sCD14, and anti-endotoxin-core antibody (EndoCAb), were determined for a subset of 37 BrdU kinetics for which stored samples were available. There was no significant correlation between these parameters and baseline CD4 count or baseline VL. Further, no significant correlation was seen between CD4 or CD8 s1 and any of these markers. Each marker was then added as a third covariate to the multivariate analysis shown in Table 3. In these analyses (Table 5), VL remained the dominant predictor of both CD4 and CD8 proliferation (P ≤ .001), whereas endotoxin showed a weak negative correlation (P = .037) and EndoCAb a weak positive correlation (P = .046) only with CD4 s1.
Multivariate linear regression analysis of s1 for CD4 and CD8 T-cell subsets with baseline CD4 T-cell count, baseline VL (log10), and markers of microbial translocation as covariates
| . | CD4 s1 . | CD8 s1 . | ||||||
|---|---|---|---|---|---|---|---|---|
| Coefficient of CD4, × 103 (cells/μL) | −0.023 | −0.021 | −0.02 | −0.019 | −0.014 | −0.015 | −0.011 | −0.014 |
| (P = .049) | (P = .101) | (P = .115) | (P = .097) | (P = .131) | (P = .108) | (P = .2) | (P = .116) | |
| Coefficient of log10VL, × 103 (log10 copies/mL) | 8.128 | 8.084 | 8.062 | 7.039 | 6.183 | 6.203 | 6.094 | 5.607 |
| (P< .001) | (P< .001) | (P< .001) | (P = .001) | (P< .001) | (P< .001) | (P< .001) | (P< .001) | |
| Coefficient | ||||||||
| Endotoxin, × 103 (pg/mL) | −0.704 | — | — | — | 0.102 | — | — | — |
| (P = .037) | (P = .69) | |||||||
| LBP, × 103 (μg/mL) | — | 0.115 | — | — | — | 0.167 | — | — |
| (P = .78) | (P = .57) | |||||||
| sCD14, × 103 (μg/mL) EndoCAb, × 103 (μ/mL) | — | — | 0.501 | — | — | — | 9.772 | — |
| (P = .94) | (P = .065) | |||||||
| EndoCAb, × 103 (μ/mL) | — | — | — | 0.056 | — | — | — | 0.032 |
| (P = .046) | (P = .124) | |||||||
| R2 | 0.449 | 0.372 | 0.37 | 0.443 | 0.392 | 0.395 | 0.451 | 0.433 |
| . | CD4 s1 . | CD8 s1 . | ||||||
|---|---|---|---|---|---|---|---|---|
| Coefficient of CD4, × 103 (cells/μL) | −0.023 | −0.021 | −0.02 | −0.019 | −0.014 | −0.015 | −0.011 | −0.014 |
| (P = .049) | (P = .101) | (P = .115) | (P = .097) | (P = .131) | (P = .108) | (P = .2) | (P = .116) | |
| Coefficient of log10VL, × 103 (log10 copies/mL) | 8.128 | 8.084 | 8.062 | 7.039 | 6.183 | 6.203 | 6.094 | 5.607 |
| (P< .001) | (P< .001) | (P< .001) | (P = .001) | (P< .001) | (P< .001) | (P< .001) | (P< .001) | |
| Coefficient | ||||||||
| Endotoxin, × 103 (pg/mL) | −0.704 | — | — | — | 0.102 | — | — | — |
| (P = .037) | (P = .69) | |||||||
| LBP, × 103 (μg/mL) | — | 0.115 | — | — | — | 0.167 | — | — |
| (P = .78) | (P = .57) | |||||||
| sCD14, × 103 (μg/mL) EndoCAb, × 103 (μ/mL) | — | — | 0.501 | — | — | — | 9.772 | — |
| (P = .94) | (P = .065) | |||||||
| EndoCAb, × 103 (μ/mL) | — | — | — | 0.056 | — | — | — | 0.032 |
| (P = .046) | (P = .124) | |||||||
| R2 | 0.449 | 0.372 | 0.37 | 0.443 | 0.392 | 0.395 | 0.451 | 0.433 |
— indicates not applicable.
We next undertook to determine whether microbial translocation played a role in subgroups of patients either with viremia or with viral replication controlled by HAART. For the 24 kinetics where the baseline VL was > 50 copies/mL, EndoCAb levels correlated positively with both CD4 (ρ = 0.42, P = .042) and CD8 (ρ = 0.51, P = .011) s1. In multivariate analysis, none of the microbial translocation indicators remained significant for CD8 proliferation, with VL alone (P ≤ .001) explaining 59.1% of variance in CD8 s1; for CD4 cells, VL remained the most significant correlate (P < .005), whereas endotoxin again showed a weak negative correlation (P = .031) with CD4 s1. In the 13 patients with VL < 50 copies/mL, none of the microbial translocation parameters correlated with CD4 s1 or CD8 s1. Interestingly, for CD4 cells only, sCD14 showed a strong correlation with stotal (ρ = 0.73, P = .005, n = 13). In this population, sCD14 also showed a strong correlation with the percentage of activated CD4 (ρ = 0.79, P = .036) and CD8 cells (ρ = 0.86, P = .014), although the number of patients analyzed is small (n = 7).
Although no significant association was found between s1 in the subsets and the markers of microbial translocation, we further investigated the adjusted effect of the markers of microbial translocation on s1 of CD4 or CD8 T-cell subsets by adding each parameter as a third covariate to the multivariate analysis shown in Table 4. In both CD4 and CD8 T-cell subsets, none of the markers of microbial translocation were significantly associated with the proliferation (s1); the covariates that remained (or were marginally) significant were the covariates that showed significance in Table 4.
Relationship between plasma cytokine levels and proliferation
To investigate the potential role of various cytokines known to be associated with T-cell proliferation and immune activation, serum levels of IL-7, IL-1β, IL-2, IL-5, IL-6, IL-10, IFN-gamma, and TNF-α were determined for 41 kinetics for which samples were available. A nonsignificant inverse correlation was observed between baseline CD4 counts and serum IL-7 levels (ρ = −0.27, P = .085, n = 41). Moreover, there were no significant associations between IL-7 levels and any of the kinetic parameters of the model for either CD4 or CD8 T cells.
IFN-γ trended toward a negative association with baseline CD4 counts (ρ = −0.29, P = .065) and TNF-α trended toward a positive correlation with baseline VL (ρ = 0.3, P = .059). Only IFN-γ was associated with CD4 s1 (ρ = 0.31, P = .048, n = 41) with the significance improving slightly when only kinetics where VL more than 50 copies/mL were considered (ρ = 0.38, P = .043, n = 29). Only TNF-α was significantly associated with CD8 s1 (ρ = 0.39, P = .014, n = 40). The significance again increased when only kinetics where VL > 50 copies/mL were considered (ρ = 0.58, P < .001, n = 29). Because TNF-α is also correlated with the VL, multivariate analysis with CD8 s1 as output was performed with TNF-α and VL as covariates. The association between CD8 s1 and TNF-α was lost when adjusted for VL (P = .92).
Clinical events
No major toxicities were associated with the BrdU infusions, which were well tolerated. Of 53 patients who received BrdU under the current protocol (41 are included in the current report), 7 patients developed new or recurrent malignancies, including squamous cell carcinoma of the anus, squamous cell carcinoma of the tongue, Hodgkin lymphoma, non-Hodgkin large B-cell lymphoma, new or recurrent basal cell carcinoma of the skin, recurrent Bowen disease, and progressive Kaposi sarcoma in a patient with previously stable disease. These events occurred 8 to 171 weeks after a BrdU infusion. A Data and Safety Monitoring Board review requested by the study team concluded that, although BrdU could not be definitively associated with the development of malignancies in this clinical trial, an increased risk associated with BrdU infusion could not be ruled out. Based on the Data and Safety Monitoring Board recommendations, no additional patients were enrolled and no further BrdU was administered under this protocol. All patients were followed for 5 years after their last BrdU infusion, and no additional malignancies were identified during this follow-up period. However, 3 patients who continued to be followed in our clinic under other protocols developed (one each) squamous cell carcinoma of the anorectal region, squamous cell carcinoma of the neck, and ocular B-cell lymphoma (which responded to antiretroviral therapy alone) 301 to 420 weeks after their last BrdU infusion.
Discussion
Using in vivo labeling of proliferating cells, we have demonstrated that plasma HIV VL is strongly associated with s1 of central and effector memory CD4 cells, as well as naive and effector memory CD8 cells, whereas CD4 cell number is inversely associated with s1 of naive CD4 cells. Our results expand on prior observations based on in vivo BrdU and deuterium labeling, as well as ex vivo BrdU labeling and Ki67 staining,1,2,10,11,19-21 by better defining kinetics in naive, memory, and activated subpopulations of T cells, and identifying potential modulators of proliferation. These data support the concept that 2 independent factors, VL and CD4 depletion, are playing an important role in lymphocyte turnover in the setting of HIV infection.20,21 Moreover, based on these data, endotoxin does not appear to be an important factor in CD4 or CD8 proliferation in this setting.
The primary driving force of turnover in most CD4 and CD8 memory cell subsets as well as CD8 naive cells is HIV itself. Immune responses, either innate or adaptive, appear to play an important role, as evidenced by the association of CD8 s1 with TNF-α levels. In multivariate analysis, the significance of TNF-α significance was lost, whereas HIV remained significant, suggesting that HIV, rather than another factor, such as endotoxin, is driving TNF-α production. TLR engagement may be an important mechanism by which HIV mediates immune activation, given that HIV encodes ligands of TLR 7 and TLR 8 that can lead to TNF-α production as well as CD8 T-cell activation in vitro.22 Type I interferons may also be playing an important role,21,23-25 but we did not measure them in the current study. Given the strong correlation between CD4 and CD8 s1, it is probable that the same set of factors is driving proliferation in both T-cell subsets.
The primary driving force of turnover in naive CD4 cells is not HIV, but rather, is CD4 depletion; this thus likely represents homeostatic proliferation rather than immune-mediated activation. Whereas one prior study using Ki67 staining found a decrease in turnover soon after the initiation of HAART and before the normalization of CD4 cell numbers, suggesting that proliferation is not a homeostatic response,1 our results are not inconsistent with these. In the latter study, CD4 cell numbers had increased substantially by the first post-HAART measurement (4 weeks); moreover, Ki67 staining at that point remained elevated in many patients, especially those with low CD4 counts. We have previously reported a decrease in proliferation of CD45RO− (nominally naive) cells 3 to 6 months after starting HAART but also found an inverse correlation between change in proliferation and change in recovery of naive CD4+ T cells, consistent with homeostatic mechanisms.18 A recent study using ex vivo labeling with BrdU or 5-ethynyl-2′-deoxyuridine also found an inverse correlation between naive CD4 cell proliferation and CD4 cell count, but no relationship with plasma VL.21
Interestingly, although IL-7 levels correlated inversely with baseline CD4 counts, there was no correlation between IL-7 levels and any of the kinetic parameters. This suggests that increased IL-7 levels are a consequence of CD4 depletion but that IL-7 may not be the primary effector of CD4 reconstitution in this setting. Alternatively, plasma levels may not adequately reflect IL-7 levels present in the tissue microenvironment.
Although plasma endotoxin has been postulated to contribute to HIV pathogenesis,15 it appears to play no significant role in the increased proliferation of CD4 and CD8 cells seen in HIV infection. Among the potential markers of microbial translocation, there were weak associations of s1 with endotoxin and EndoCab levels, but each was in a direction opposite to that previously reported, with a negative correlation for endotoxin and a positive one for EndoCab.15 Thus, increased endotoxin levels are not associated with increased proliferation, which is a dynamic marker of HIV-associated immune activation.11 Although sCD14 showed little correlation with kinetic parameters overall, in patients with VLs less than 50 copies/mL, it was strongly associated with the overall proliferation (stotal) of CD4 cells, and the percentage of activated CD4 and CD8 cells. Given the absence of correlation with other markers of microbial translocation, this suggests that sCD14 is a more general marker for monocyte activation rather than a specific marker for endotoxin-mediated activation, or, alternatively, is an acute phase reactant.26,27 Consistent with this, a recent microarray study found that the gene expression profile of monocytes in HIV-infected patients most closely resembled that seen in vitro after stimulation with INF-α, rather than stimulation with either IFN-γ or endotoxin.28
The proportion of CD4 cells in the rapidly proliferating pool (s1) was consistently greater than that of CD8 cells, in agreement with previous observations.20,29 In concordance with this, peak levels of labeling were also greater for CD4 cells. These findings suggest that the proliferation is not primarily antigen induced because CD8 cells show a much greater expansion than CD4 cells in response to viral infections, such as lymphocytic choriomeningitis virus.30 It is unlikely to represent combined homeostatic and immune-mediated proliferation because memory and activated cells, for which we did not identify a homeostatic component, showed a similar pattern. Why CD4 cells would respond with greater proliferation is uncertain but may be related to the specific profile of cytokines or other immunoregulatory molecules that are up-regulated as part of the immune activation associated with HIV. This relentless high proliferation rate may be an important contributor to the ongoing CD4 depletion that is characteristic of untreated HIV infection.
Ki67 is commonly used as a surrogate for proliferation, although some studies have found that it may be a poor marker in the setting of HIV infection.31 In the current study, Ki67 correlated better with overall proliferation (stotal) than with s1 and correlated better with CD8 than CD4 proliferation. In multivariate analysis where Ki67 was substituted for s1 in those patients for whom both Ki67 staining and in vivo BrdU labeling were available, the associations with VL or CD4 count decreased substantially and the predictive values were approximately halved compared with those with s1 output (data not shown). Thus, Ki67 is an acceptable, although not perfect, marker for in vivo proliferation, but measurement of Ki67 does not give information about the dynamics and correlates of the rapidly and slowly proliferating pools of cells.
Intriguingly, whereas effector memory proliferation (s1) for both CD4 and CD8 cells correlated strongly with VL, proliferation of activated cells (HLA-DR+38+) was unrelated to VL, although the percentage of activated CD4 and CD8 cells did show a correlation. These data suggest that HIV is driving the activation of both populations; however, once activated, proliferation proceeds independently of HIV, possibly as a direct biologic consequence of activation, given that activated cells showed the highest levels of proliferation. Alternatively, other factors may be driving proliferation, although neither cytokine levels nor markers of microbial translocation correlated with s1 in these cells.
We stopped administration of BrdU under this protocol after the occurrence of new or recurrent malignancies in 7 patients during the 5-year follow-up period; 3 additional malignancies occurred 6 to 8 years after BrdU administration. The relationship of these malignancies to BrdU administration is uncertain, given the small number of cases and the diversity of malignancies. BrdU had previously been used as a labeling agent in National Cancer Institute-sponsored studies in > 4000 patients, with no reported increase in malignancies. The malignancies seen included basal cell carcinoma, the most common cancer of humans, as well as malignancies seen with increased frequency in HIV-infected patients, including Kaposi sarcoma, non-Hodgkin and Hodgkin lymphoma, and anal carcinoma.32-35 The latter occur at frequencies ranging from 0.03% to 0.3% per year, with a higher risk in patients with lower CD4 counts and higher VLs.32,35 Given the uncertainty of the association between malignancies and BrdU administration, the Data and Safety Monitoring Board reviewing the study recommended that, if future protocols use BrdU, the informed consent should state that there may be an increased risk of malignancies and should include a summary of the data from the current study. We concur with this recommendation and advise caution in using BrdU as a labeling agent in this population.
In conclusion, in vivo labeling with BrdU has demonstrated that HIV is the primary factor leading to activation and proliferation of most subsets of CD4 and CD8 T cells, although in naive CD4 cells homeostatic proliferation is an important driving force. Endotoxin does not appear to play an important role in CD4 or CD8 proliferation. Although current options for in vivo labeling are limited, ex vivo labeling with BrdU or 5-ethynyl-2′-deoxyuridine, can provide an alternative method for further studying the mechanisms contributing to activation and proliferation in the context of HIV infection.20
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients for their willingness to participate in these studies and caregivers in the National Institute of Allergy and Infectious Diseases–Critical Care Medicine Department HIV/AIDS program for their ongoing efforts.
This work was supported by the Intramural Research Programs of the National Institutes of Health Clinical Center and the National Institute of Allergy and Infectious Diseases and supported in part by the National Cancer Institute, National Institutes of Health (contract HHSN261200800001E).
National Institutes of Health
Authorship
Contribution: J.A.K., R.A.L., I.S., H.C.L., and M.D.M. contributed to conception and design of the research; R.A.L., J.W.A., P.I.L., A.R., I.S., R.S., and J.A.K. contributed to acquisition of data; S.S. and M.D.M. generated model estimates; S.S., C.-Y.H., M.D.M., I.S., J.A.K., and J.R. analyzed and interpreted data; S.S., J.A.K., M.D.M., and C.-Y.H. wrote the manuscript; and all authors reviewed the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sharat Srinivasula, 6700A Rockledge Dr, Rm 5227, Bethesda, MD 20817; e-mail: srinivasulas@mail.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal