Abstract
Perforin (Prf1) and granzyme B (GzmB) are essential effector molecules for natural killer (NK)–cell cytotoxicity, but how Prf1 and GzmB expression is regulated during arming of NK cells is poorly defined. We show that human microRNA (miR)–27a* is a negative regulator of NK-cell cytotoxicity by silencing Prf1 and GzmB expression. Human miR-27a* specifically bound to the 3′ untranslated regions of Prf1 and GzmB, down-regulating expression in both resting and activated NK cells, and it functioned as a fine-tuner for homeostasis of the net amount of the effector proteins. Consistent with miR-27a* having an inhibitory role, knockdown of miR-27a* in NK cells dramatically increased cytotoxicity in vitro and decreased tumor growth in a human tumor xenograft model. Thus, NK-cell cytotoxicity is regulated, in part, by microRNA, and modulating endogenous miR-27a* levels in NK cells represents a potential immunotherapeutic strategy.
Introduction
Natural killer (NK) cells play key roles in innate and adaptive immune responses during early host defense against infectious pathogens and tumors via 2 major mechanisms: contact-dependent cytotoxicity and cytokine production for immune modulation.1-4 Target-cell death is primarily mediated via the granule-exocytosis pathway. NK cells are armed by functional cytotoxic granules containing perforin (Prf1) and granzymes, essential effector molecules for NK-cell cytotoxicity as shown in knockout mice,4,5 and are triggered to mediate effector activity by receptor ligation. Prf1 facilitates the delivery of granzymes into the cytosol of the target cell, and GzmB, the best-characterized granzyme, cleaves several procaspases, BID, inhibitor of caspase-activated DNase, and other intracellular substrates to initiate the classic apoptotic pathways.6-9
Many of the studies of Prf1 and GzmB expression in NK cells have suggested the possible involvement of posttranscriptional regulation. Recently, studies using murine NK cells have shown that acquisition of murine NK-cell cytotoxicity requires the translation of a pre-existing pool of Prf1 and GzmB mRNAs.4 Despite high basal levels of Prf1 and GzmB mRNA, little protein expression is observed under resting conditions in many types of NK cells, whereas expression of both proteins is up-regulated during activation.4,10,11 These observations are consistent with a posttranscriptional mechanism operating to allow NK cells to be poised for but to prevent translation before activation, such as silencing by microRNAs.12,13
microRNAs are an abundant class of endogenous small noncoding RNAs (19-22 nt) generated by sequential processing of primary miRNA transcripts by the ribonuclease Drosha in the nucleus and Dicer1 in the cytoplasm, both of which are essential enzymes in the miRNA biogenesis pathway. In mammals, mature miRNAs are integrated into an RNA-inducing silencing complex, including Argonaute 2 (Ago2), a required endonuclease in the RNA interference pathway, and they associate with 3′ untranslated regions (UTRs) of specific target mRNAs to down-regulate gene expression by targeting mRNAs for translational suppression or mRNA degradation.13-17 The involvement of miRNA in immune responses and the development of immune cells from hematopoietic stem cells have been widely investigated by manipulation of specific miRNA levels13,18 or by disruption of molecules involved in biogenesis and activity of all miRNAs, such as Arg,19 Drosha,20 and Dicer.21-24 Recently, characterization of NK cells from mice with conditional deletion of Dicer and DiGeorge syndrome critical region 8 were reported, with evidence of impairments in NK-cell activation, survival, and function during viral infection.24 These genetic studies have suggested miRNAs play essential roles in immune cell development and function.13,14,25
Despite evidence for a broad impact in regulation of immune function, the molecular mechanism, importance, and biologic significance of miRNAs in NK-cell biology remains poorly understood.25-27 Furthermore, as the potential to use the activity of NK cells for therapeutic applications has become more evident,2,28 identifying target molecules that can be modulated to enhance NK-cell cytotoxicity could become potentially useful. Here, we show a novel mechanism by which NK-cell cytotoxicity is regulated by microRNA and its potential therapeutic applications.
Methods
Cell preparation and culture
In vitro–differentiated mature NK (mNKs) cells were generated and differentiated from a starting population of purified CD34+ cells isolated from human umbilical cord blood (UCB).29-31 The percentage of CD56+/CD3− cells was > 90% of the total cells after in vitro differentiation, with ∼ 95% of the gated lymphocyte population CD56+/CD122+ mNK cells. Primary human NK cells were obtained from UCB by an ACCUSPIN System-Histopaque-1077 (Sigma-Aldrich) density separation. NK cells were then enriched by negative selection using a MACS NK cell isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. Enriched NK-cell populations were > 93% CD56+/CD3− and < 5% CD3+. The human NK-92 immortal cell line was grown in α-MEM (Invitrogen), supplemented with 20% heat-inactivated FBS, 2mM l-glutamine, 100 units/mL IL-2 (PeproTech), and streptomycin/penicillin as described previously.30 Human K562 erythroleukemia cells were grown in RPMI-1640 containing 10% FBS and streptomycin/penicillin. Human SW620 colon cancer cells (ATCC CCL-227) were purchased from ATCC. Cells were maintained at 37°C in humidified air (5% CO2) and grown in RPMI-1640 (Invitrogen) supplemented with 10% FBS (HyClone Laboratories), 2mM l-glutamine (Sigma-Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen).
Transfection
Premade small interfering (si)RNAs against human Dicer1, Ago2, Prf1, and GzmB (ON-TARGETplus SMARTpool), microRNA mimics, and microRNA hairpin inhibitors were purchased from Dharmacon RNA Technologies. Transfections of NK cells with small RNAs such as small interfering RAN (siRNA), microRNA mimics, and microRNA inhibitors were performed by nucleofection using an Amaxa Human CD34 Cell Nucleofector kit (Lonza).30 In brief, 100 μL of one nucleofection sample contained 4 × 106 mNK cells and 100nM (final concentration) siRNA or miRNA mimics or miRNA inhibitor. These cells were subjected to nucleofection using program U-08 and then subjected to immunoblot analysis, real-time quantitative (q)PCR using PCR Thermal Cycler DICE (Takara), and standard chromium release. Transfections of human embryonic kidney (HEK)293FT cells were performed by overlaying the cells with a precipitate of Metafectene (Biontex) according to the manufacturer's instructions.
Lentiviral preparation and transduction
For lentiviral transduction of NK-92 cells, the pMIRNA1-GFP control vector and pMIR-27a* vector expressing pre–miR-27a* were purchased from System Biosciences. Lentiviruses were produced using a third-generation packaging system (pMDLg/pRRE, pRSV-Rev, and pMD2.G) in HEK293T cells as described previously.30
Human tumor xenograft model
Female BALB/c nude mice (BALB/c-nu/nu, 5 weeks old) were purchased from SLC (Hamahatsu) and approved by the Institutional Animal Care and Use Committee of Korea Research Institute of Bioscience and Biotechnology. Human SW620 cells (6 × 106 cells) were injected subcutaneously into BALB/c nude mice. mNK cells (2.8 × 106 cells) transfected with Ctrl_miR, miR-27a*, and miR-27a* inhibitor or vehicle PBS were injected intravenously twice at 2 hours after SW620 implantation and at day 7. Doxorubicin (2 mg/kg) was administered to cohorts of mice intraperitoneally every other day through day 20. Tumor volume and tumor growth inhibition rate were calculated.
Statistical analysis
Experimental results are expressed as means ± SEM. Statistically significant differences between groups were determined using a 2-tailed unpaired Student t test, where a P < .05 is considered significant (but for values in Figure 7B-C, analyses were performed using nonparametric measures and Mann-Whitney-Wilcoxon [Mann-Whitney rank sum] test).
Results
Involvement of microRNA in Prf1 and GzmB expression in human NK cells
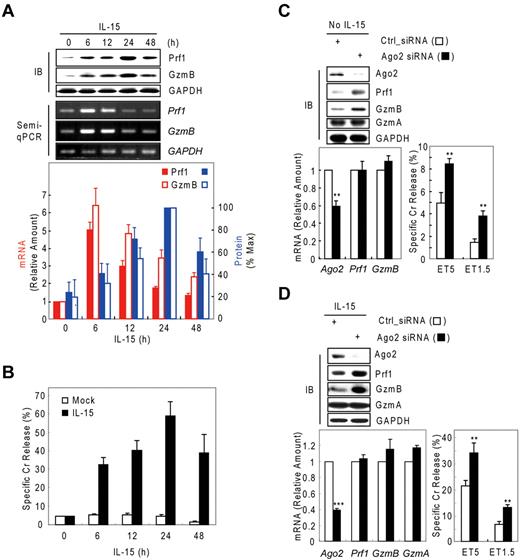
Although IL-15–activated NK-cell cytotoxicity is closely associated with up-regulation of Prf1 and GzmB proteins,4,32-34 little is known about the mechanism regulating such arming of NK cells. Thus, we analyzed resting and IL-15–activated human mNK (in vitro–differentiated human mNK) cells for GzmB and Prf1 expression. GzmB and Prf1 protein expression were some detectable in resting mNK cells but dramatically induced after IL-15 treatment (Figure 1A top) and peaked at 24 hours, with expression coupled to NK-cell killing ability (Figure 1B). By contrast, Prf1 and GzmB mRNA levels were not correlated with protein expression. GzmB and Prf1 mRNA were at high levels in resting cells, transcriptionally increased with activation and peaked at 6 hours after IL-15 treatment. A time lag was observed between the increase in Prf1/GzmB mRNA levels and the increase in protein levels (Figure 1A bottom), implying that posttranscriptional regulation could be involved in expression of these effector molecules. Similar intervals between increased mRNA and protein expression of Prf1 and GzmB also were observed in NK-92 cells after IL-15 treatment (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Possible involvement of microRNA in Prf1 and GzmB expression in human NK cells. (A-B) In vitro–differentiated human mNK cells were freshly incubated in the absence (mock) or presence of IL-15 (30 ng/mL) after 24-hour deprivation of IL-15. The cells were harvested at the times (0, 6, 12, 24, and 48 hours) indicated relative to IL-15 addition. (A) Kinetic analysis of Prf1 and GzmB expression in human NK cells during activation. (Top) Immunoblot (IB) analyses. (Middle) DNAs amplified by semi-qPCR were stained with ethidium bromide and photographed using N-Trans illuminator. (Bottom) Real-time qPCR (red bars) and band intensities of Prf1 and GzmB proteins (top) were normalized and plotted as a percentage of the maximum (100%; blue bars). The quantitative data are expressed as the mean value of independent measurements from 3 separate experiments (mean ± SEM). (B) Kinetic analysis of cytotoxicity in human NK cells during activation. (C-D) Suppression of Prf1 and GzmB expression by microRNA. Transfected mNK cells with Ago2 and Dicer1 siRNAs were incubated in the absence (C) or presence (D) of IL-15, respectively, and then subjected to IB analysis, real-time qPCR, and standard chromium (Cr) release assay at effector to target cell ratio 5:1 (ET5) and ET1.5. Data are representative of 3 independent experiments (mean ± SEM of triplicates; Student t test, **P < .01 and ***P < .001 vs Ctrl_siRNA).
Possible involvement of microRNA in Prf1 and GzmB expression in human NK cells. (A-B) In vitro–differentiated human mNK cells were freshly incubated in the absence (mock) or presence of IL-15 (30 ng/mL) after 24-hour deprivation of IL-15. The cells were harvested at the times (0, 6, 12, 24, and 48 hours) indicated relative to IL-15 addition. (A) Kinetic analysis of Prf1 and GzmB expression in human NK cells during activation. (Top) Immunoblot (IB) analyses. (Middle) DNAs amplified by semi-qPCR were stained with ethidium bromide and photographed using N-Trans illuminator. (Bottom) Real-time qPCR (red bars) and band intensities of Prf1 and GzmB proteins (top) were normalized and plotted as a percentage of the maximum (100%; blue bars). The quantitative data are expressed as the mean value of independent measurements from 3 separate experiments (mean ± SEM). (B) Kinetic analysis of cytotoxicity in human NK cells during activation. (C-D) Suppression of Prf1 and GzmB expression by microRNA. Transfected mNK cells with Ago2 and Dicer1 siRNAs were incubated in the absence (C) or presence (D) of IL-15, respectively, and then subjected to IB analysis, real-time qPCR, and standard chromium (Cr) release assay at effector to target cell ratio 5:1 (ET5) and ET1.5. Data are representative of 3 independent experiments (mean ± SEM of triplicates; Student t test, **P < .01 and ***P < .001 vs Ctrl_siRNA).
We hypothesized that the observed suppression of Prf1 and GzmB expressions during rest and activation may be because of microRNA-mediated gene silencing. To test this hypothesis, we performed knockdown experiments of Dicer121-23 , Ago219 , or both in resting and IL-15–activated NK cells. In resting mNK cells, transfection with Ago2 siRNA dramatically increased the amount of Prf1 and GzmB protein compared with control siRNA (Ctrl_siRNA)–transfected mNK cells without changing their mRNA levels and was concomitant with up-regulation of NK-cell cytotoxicity (Figure 1C). Because mNK cells up-regulated expression of Ago2 and Dicer1 (supplemental Figure 2A-B) on activation, we used siRNAs to down-regulate the expression of Ago2 or Dicer1 in IL-15–activated NK cells. mNK cells transfected with Ago2 (Figure 1D) or Dicer1 (supplemental Figure 2C) siRNA showed increased Prf1 and GzmB protein expression despite little difference in the measured mRNA levels, and again this was associated with an increase in NK-cell cytotoxicity. In contrast, GzmA mRNA and protein expression was not affected by blocking miRNA biogenesis and interference pathways. These effects of blocking Dicer1 on Prf1 and GzmB protein expression and NK-cell cytotoxicity also were seen in NK-92 cells (supplemental Figure 1). These data suggest cellular miRNAs down-regulate Prf1 and GzmB expression in both resting and activated human NK cells.
Human miR-27a* targets both Prf1 and GzmB expression
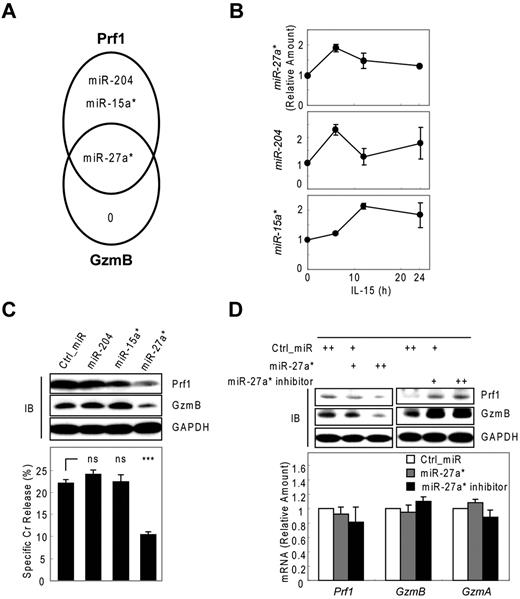
To identify miRNAs that suppress Prf1 and GzmB expression in both rest and activation, we used microarrays to analyze microRNAs derived from activated human mNK cells. All microarray data are available in the Gene Expression Omnibus under accession GSE32411. Based on these kinetic analyses of Prf1 and GzmB expression (Figure 1A; supplemental Figure 3A), we made an assumption that candidate miRNAs involved in Prf1 and GzmB suppression in rest and activation might not only be expressed at some levels under resting conditions but also might show a decrease particularly at the time of the greatest increase in protein expression after activation (∼ 24 hours after IL-15 treatment). To encompass the potentially relevant candidate miRNAs, we included ones either increased when Prf1 and GzmB mRNA is dramatically increased on activation (∼ 6 hours after IL-15 treatment) to account for the delay in protein expression until 24 hours (supplemental Figure 3B model a) or decreased after activation based on the assumption that the delay in protein expression might reflect the presence of excess unbound miRNA in the resting state (supplemental Figure 3B model b). Therefore, we analyzed differential expression of known human miRNAs in resting and activated human mNK cells. Initial screening identified 25 miRNAs showing at least 1.5-fold increase or decrease in response to IL-15 (supplemental Table 1). To determine whether these miRNAs contributed to Prf1 and GzmB suppression, we performed in silico sequence complementarity analysis of the 25 miRNAs against Prf1 and GzmB mRNAs with an initial focus on the crucial seed sequence using 3 miRNA target prediction algorithms: Microcosm, TargetScan, and miRanda. Only 3 of the IL-15–induced miRNAs (miR-15a*, miR-204, and miR-27a*) were predicted by any of these programs, and all 3 had profiles consistent with the model a (supplemental Figure 3B). Interestingly, miR-27a* was identified as a possible suppressor of both Prf1 and GzmB mRNA (Figure 2A). Next, we validated the microarray data and analyzed expression kinetics of the 3 candidate miRNAs during NK-cell activation using real-time qPCR (Figure 2B). Peak induction was observed within 12 hours, similar to the peak expression of the putative target mRNAs. The fold-induction of the microRNAs was much smaller than that of targets Prf1 and GzmB mRNAs on activation (Figure 1A), suggesting that the candidate miRNAs could serve as a fine tuner of the target gene expression during activation.
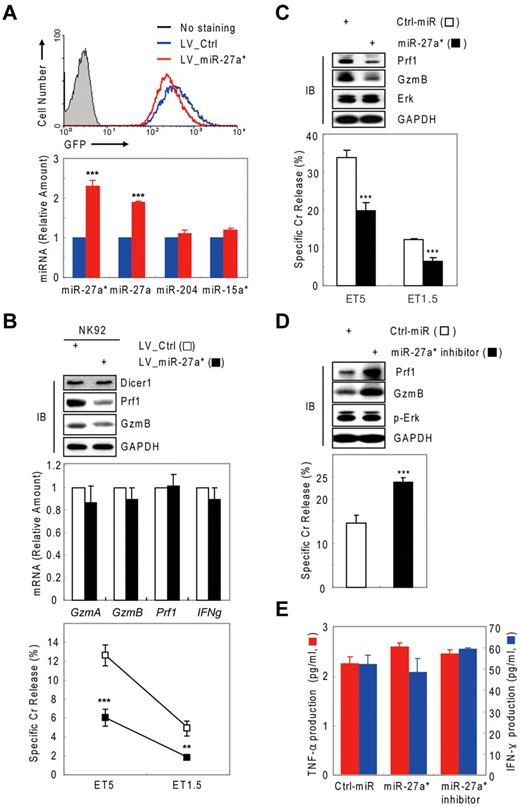
Human miR-27a* suppresses both Prf1 and GzmB expression. (A) Venn diagram of the predictions of human Prf1- and GzmB-targeting miRNAs yielding one common hit miR-27a*. (B) Validation of microarray data with real-time qPCR and kinetic analysis of candidate miRNAs during NK-cell activation (0, 6, 12, and 24 hours). (C) Screening of functional miRNA(s) in suppression of Prf1 and GzmB protein expressions. One hundred microliters of 1 nucleofection sample contained 4 × 106 mNK cells and 100nM (final concentration) of miRNA mimics and was subjected to nucleofection. (Top) IB analysis. (Bottom) Standard Cr release assay at ET5. (D) Regulation of Prf1 and GzmB expression by miR-27a*. mNK cells were transfected with increasing concentrations (50 and 100nM) of miR-27a* or miR-27a* inhibitor. The transfected cells were incubated in the presence of IL-15 (30 ng/mL) for 24 hours and then further incubated with freshly added IL-15 (30 ng/mL) for 24 hours. Finally, cells were subjected to IB analysis and real-time qPCR. (Top) IB analysis. (Bottom) Real-time qPCR analysis. Data are representative of 3 independent experiments (mean ± SEM of triplicates; ns, not significant; Student t test, P > .05 and ***P < .001 vs Ctrl_miR).
Human miR-27a* suppresses both Prf1 and GzmB expression. (A) Venn diagram of the predictions of human Prf1- and GzmB-targeting miRNAs yielding one common hit miR-27a*. (B) Validation of microarray data with real-time qPCR and kinetic analysis of candidate miRNAs during NK-cell activation (0, 6, 12, and 24 hours). (C) Screening of functional miRNA(s) in suppression of Prf1 and GzmB protein expressions. One hundred microliters of 1 nucleofection sample contained 4 × 106 mNK cells and 100nM (final concentration) of miRNA mimics and was subjected to nucleofection. (Top) IB analysis. (Bottom) Standard Cr release assay at ET5. (D) Regulation of Prf1 and GzmB expression by miR-27a*. mNK cells were transfected with increasing concentrations (50 and 100nM) of miR-27a* or miR-27a* inhibitor. The transfected cells were incubated in the presence of IL-15 (30 ng/mL) for 24 hours and then further incubated with freshly added IL-15 (30 ng/mL) for 24 hours. Finally, cells were subjected to IB analysis and real-time qPCR. (Top) IB analysis. (Bottom) Real-time qPCR analysis. Data are representative of 3 independent experiments (mean ± SEM of triplicates; ns, not significant; Student t test, P > .05 and ***P < .001 vs Ctrl_miR).
Next, we evaluated whether the 3 miRNAs with sequence matches to the 3′ UTRs of Prf1, GzmB, or both can inhibit Prf1 and GzmB expression by transfecting synthetic miRNA mimics corresponding to these miRNAs into mNK cells during IL-15 treatment. Introduction of miR-27a* into mNK cells reduced Prf1 and GzmB protein levels by ∼ 60% and concurrently reduced NK-cell cytotoxicity by ∼ 50%. In contrast, miR-15a* and miR-204 had little effect on Prf1 or GzmB expression and NK-cell cytotoxicity (Figure 2C). To confirm whether Prf1 and GzmB expression was regulated by an endogenous version of miR-27a*, we transfected a miR-27a* inhibitor into mNK cells.35 Dramatic enhancement of Prf1 and GzmB expression was observed (Figure 2D top panel), suggesting miR-27a* endogenously suppressed Prf1 and GzmB expression in mNK cells. The overexpression and knockdown of miR-27a* by transfecting mNK cells was confirmed by qPCR (supplemental Figure 3C-D), showing little effect on changes of Prf1 and GzmB mRNA levels (Figure 2D bottom panel). These results suggested that human miR-27a* is a negative regulator of Prf1 and GzmB expressions during activation.
To further test whether miR-27a* specifically targets both Prf1 and GzmB, we performed reporter assays in cell culture (Figure 3A). Serotonin N-acetyltransferase (AANAT) as a reporter protein is unstable in the absence of protein kinase A activation (half-life, ∼ 3.5 minutes; supplemental Figure 4A), making it a useful reporter for de novo protein synthesis.36 Whereas overexpression of miR-27a* in HEK-293FT cells dramatically reduced in a dose-dependent manner the expression of an AANAT reporter gene construct containing the wild-type Prf1 3′ UTR or GzmB 3′ UTR, ectopic expression of control miRNA (Ctrl_miR) had no significant effect on the expression of these reporters (supplemental Figure 4B-C). Moreover, point mutations of miR-27a* induced recovery of reporter gene expression without changing AANAT mRNA levels (Figure 3B). To demonstrate that the effect of miR-27a* on the AANAT reporter gene is dependent on the miR-27a* binding site, we generated reporter constructs containing deletions or mutations of the 3′ UTRs where miR-27a* is predicted to bind (Figure 3A bottom panel). Although overexpression of miR-27a* dramatically reduced the expression of AANAT gene in the Prf1_D1 reporter, which still contained the binding site, recovery of reporter gene expression was observed with Prf1_D2, which lacks the putative miR-27a* binding site (Figure 3C). Moreover, introduction of 3′ UTR mutations (Prf1_M and GzmB_M) to make the sequence complementary to the miR-27a* mutant (miR-27a*M) resulted in suppression by miR-27a*M but not by miR-27a* (Figure 3D). For these experiments, reporter AANAT mRNA levels were normalized to NeoR mRNA as an internal control and showed little difference among samples. Furthermore, little differences in the decay rate of reporter mRNAs containing Prf1 or GzmB 3′ UTR were observed using an actinomycin D chasing assay, either with or without miR-27a* overexpression (supplemental Figure 4D-E), indicating that miR-27a* suppresses Prf1 and GzmB expression by translational inhibition. Taken together, these data suggest miR-27a* down-regulates both Prf1 and GzmB expression by specifically targeting their 3′ UTR sequences.
Human miR-27a* specifically targets both Prf1 and GzmB 3′ UTR sequence. (A top) Predicted miR-27a* binding sites. (Bottom) Schematic diagrams of Prf1 and GzmB 3′ UTRs (wild type [WT]) or their deletions (D1 and D2) and mutants (M). Point mutations are bolded. Numbers indicate positions of nucleotides in the 3′ UTRs. Crosses indicate the mutation depicted in the top panel. (B-D) Reporter assay using IB analysis. HEK-293FT cell lines were cotransfected with combinations of reporter plasmids containing WT, D, or M forms of Prf1 and GzmB 3′ UTRs and miR-27a* or its mutated miRNA (miR-27a*M). Reporter AANAT mRNA levels were normalized to NeoR mRNA level as an internal control of the vector. Net amount of translated AANAT protein was determined by IB. GAPDH served as loading control. Data are representative of 4 independent experiments (mean ± SEM of triplicates).
Human miR-27a* specifically targets both Prf1 and GzmB 3′ UTR sequence. (A top) Predicted miR-27a* binding sites. (Bottom) Schematic diagrams of Prf1 and GzmB 3′ UTRs (wild type [WT]) or their deletions (D1 and D2) and mutants (M). Point mutations are bolded. Numbers indicate positions of nucleotides in the 3′ UTRs. Crosses indicate the mutation depicted in the top panel. (B-D) Reporter assay using IB analysis. HEK-293FT cell lines were cotransfected with combinations of reporter plasmids containing WT, D, or M forms of Prf1 and GzmB 3′ UTRs and miR-27a* or its mutated miRNA (miR-27a*M). Reporter AANAT mRNA levels were normalized to NeoR mRNA level as an internal control of the vector. Net amount of translated AANAT protein was determined by IB. GAPDH served as loading control. Data are representative of 4 independent experiments (mean ± SEM of triplicates).
Human miR-27a* down-regulates NK-cell cytotoxicity through targeting Prf1 and GzmB
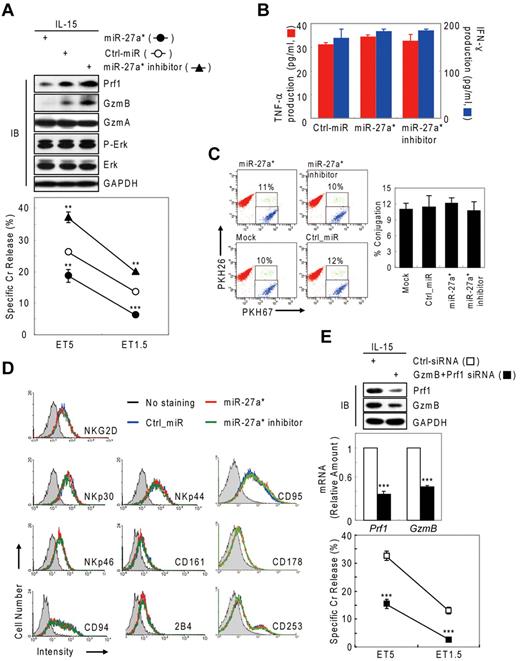
Next, we tested whether negative regulation of NK-cell cytotoxicity by miR-27a* was because of selective reduction of Prf1 and GzmB rather than other genes or other processes associated with NK-cell activation, including priming, immune synapse formation, receptor signaling, cytotoxicity, and cytokine production. Introduction of miR-27a* and miR-27a* inhibitor did not affect the amount or phosphorylation state of ERK and GzmA (Figure 4A top panel) or cytokine production such as IFN-γ and TNF-α (Figure 4B), indicators of activated NK cells,1,6 but it did decrease and increase both Prf1 and GzmB expression, respectively, as reflected in the killing capacity in an NK-cell dose-dependent manner (Figure 4A bottom panel). To assess formation of an immunologic synapse, a conjugation assay was performed to detect adhesion of NK cells to target cells using 2-color flow cytometry, and miR-27a* had no effect on NK-cell conjugation to K-562 cells (Figure 4C). Next, we investigated the level of other effectors and activating and inhibitory receptors on transfected mNK cells that can play a role in cytotoxicity. The overexpression or knockdown of miR-27a* has little effect on expression of stimulatory (NKG2D, NKp30, NKp44, and NKp46) or inhibitory (CD161, CD94, and CD244) receptors and death molecules (Fas/CD95, FasL/CD178, and TRAIL/CD253; Figure 4D). To formally determine whether specific suppression of Prf1 and GzmB protein by miR-27a* is linked to modulating NK-cell cytotoxicity, we alternatively reduced Prf1 and GzmB protein levels using siRNAs. mNK cells transfected with a pool of siRNAs against Prf1, GzmB, or both showed dramatic decrease of their protein levels, coupled with down-regulated NK-cell cytotoxicity (Figure 4E), suggesting both Prf1 and GzmB proteins are essential factors for determining NK-cell killing capacity. Taken together, these results demonstrate that miR-27a* down-regulates NK-cell cytotoxicity through targeting effector Prf1 and GzmB expression.
Human miR-27a* down-regulates NK-cell cytotoxicity by silencing Prf1 and GzmB. (A-D) Overexpression and knockdown of miR-27a* induce decrease and increase of NK-cell cytotoxicity, respectively. mNK cells transfected with Ctrl_miR, miR-27a, or miR-27a inhibitor were analyzed for IB (A top), standard Cr release assay (A bottom), ELISA for de novo synthesis of TNF-α (B red), and IFN-γ (B blue), conjugation assay using 2-color FACS cytometry (C), and FACS analysis from the CD56+-gated lymphocyte population (D). Data are representative of 3 independent experiments. The quantitative data are expressed as the mean value of independent measurements from 3 separate experiments (mean ± SEM; Student t test, **P < .01 and ***P < .001 vs Ctrl_miR). (E) Level of Prf1 and GzmB protein is an essential regulator of NK-cell killing capacity. (Top) IB analysis. (Middle) Real-time qPCR. (Bottom) Standard Cr release assay. Data are representative of 4 independent experiments (mean ± SEM of triplicates; Student t test, ***P < .001 vs Ctrl_siRNA.
Human miR-27a* down-regulates NK-cell cytotoxicity by silencing Prf1 and GzmB. (A-D) Overexpression and knockdown of miR-27a* induce decrease and increase of NK-cell cytotoxicity, respectively. mNK cells transfected with Ctrl_miR, miR-27a, or miR-27a inhibitor were analyzed for IB (A top), standard Cr release assay (A bottom), ELISA for de novo synthesis of TNF-α (B red), and IFN-γ (B blue), conjugation assay using 2-color FACS cytometry (C), and FACS analysis from the CD56+-gated lymphocyte population (D). Data are representative of 3 independent experiments. The quantitative data are expressed as the mean value of independent measurements from 3 separate experiments (mean ± SEM; Student t test, **P < .01 and ***P < .001 vs Ctrl_miR). (E) Level of Prf1 and GzmB protein is an essential regulator of NK-cell killing capacity. (Top) IB analysis. (Middle) Real-time qPCR. (Bottom) Standard Cr release assay. Data are representative of 4 independent experiments (mean ± SEM of triplicates; Student t test, ***P < .001 vs Ctrl_siRNA.
Human miR-27a* serves as a regulator of Prf1 and GzmB protein expression in both resting and activated NK cells
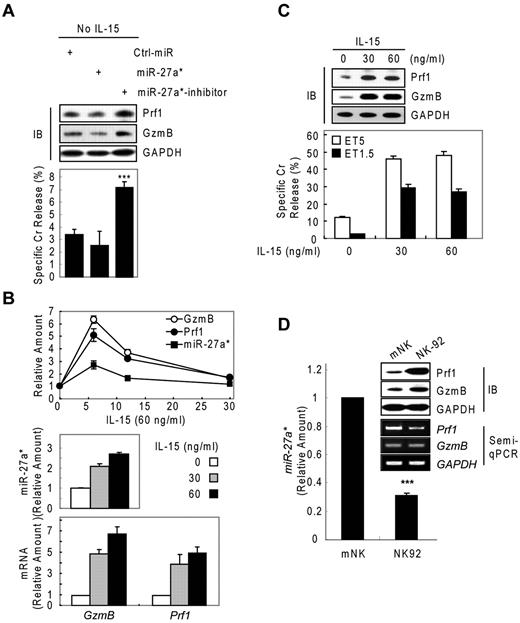
We next asked about the biologic role of miR-27a* in NK cells. We performed knockdown and dose-dependency experiments in resting and IL-15–activated NK cells, respectively. In resting mNK cells, transfection with miR-27a* inhibitor increased Prf1 and GzmB ∼ 2-fold compared with Ctrl_miR–transfected mNK cells, with a concomitant up-regulation of NK-cell cytotoxicity. By contrast, miR-27a*–transfected resting mNK cells showed little change in Prf1 and GzmB expression and cytotoxicity (Figure 5A), suggesting endogenous levels of miR-27a* are adequately suppress basal Prf1 and GzmB mRNA levels to maintain low protein levels at rest.
Human miR-27a* serves as a regulator of Prf1 and GzmB protein expression. (A) Basal suppression of GzmB and Prf1 expression by miR-27a* under resting conditions. (Top) IB analysis. (Bottom) Cr release assay at ET5. (B-C) miR-27a* regulates the level of Prf1 and GzmB protein during activation. mNK cells were freshly incubated in the presence of IL-15 (60 ng/mL) after 24-hour deprivation of IL-15. The cells were harvested at the times (0, 6, 12, and 30 hours) indicated relative to treatment of 60 ng/mL IL-15 and analyzed for kinetics of Prf1/GzmB mRNA and miR-27a* (B top). mNK cells were left untreated or treated for 6 hours with various concentrations of IL-15 (0, 30, and 60 ng/mL) after 24-hour deprivation of IL-15. miR-27a* (B middle) and Prf1/GzmB mRNA (B bottom) were analyzed by real-time qPCR. Concurrently, the mNK cells were subjected to IB analysis (C top) and Cr release assay (C bottom). (D) High expression of Prf1 and GzmB protein in NK-92 is because of low level of endogenous miR-27a*. (Top) IB analysis. (Middle) Semi-qPCR analysis. (Bottom) Real-time qPCR. Data are representative of 3 independent experiments (mean ± SEM of triplicates; Student t test, ***P < .001 vs Ctrl_miR.
Human miR-27a* serves as a regulator of Prf1 and GzmB protein expression. (A) Basal suppression of GzmB and Prf1 expression by miR-27a* under resting conditions. (Top) IB analysis. (Bottom) Cr release assay at ET5. (B-C) miR-27a* regulates the level of Prf1 and GzmB protein during activation. mNK cells were freshly incubated in the presence of IL-15 (60 ng/mL) after 24-hour deprivation of IL-15. The cells were harvested at the times (0, 6, 12, and 30 hours) indicated relative to treatment of 60 ng/mL IL-15 and analyzed for kinetics of Prf1/GzmB mRNA and miR-27a* (B top). mNK cells were left untreated or treated for 6 hours with various concentrations of IL-15 (0, 30, and 60 ng/mL) after 24-hour deprivation of IL-15. miR-27a* (B middle) and Prf1/GzmB mRNA (B bottom) were analyzed by real-time qPCR. Concurrently, the mNK cells were subjected to IB analysis (C top) and Cr release assay (C bottom). (D) High expression of Prf1 and GzmB protein in NK-92 is because of low level of endogenous miR-27a*. (Top) IB analysis. (Middle) Semi-qPCR analysis. (Bottom) Real-time qPCR. Data are representative of 3 independent experiments (mean ± SEM of triplicates; Student t test, ***P < .001 vs Ctrl_miR.
On activation with increasing doses of IL-15 (0, 30, and 60 ng/mL), mNK cells up-regulated expression of Prf1 mRNA, GzmB mRNA, and miR-27a* in a dose-dependent manner, showing peak at 6 hours after IL-15 treatment (Figures 1A and 2B and Figure 5B top panel). The induction of Prf1 and GzmB mRNA was higher than that of miR-27a* by ∼ 2-fold (Figure 5B), and this relative increase in Prf1 and GzmB mRNA levels presumably exceeded the mRNA quantity that can be titrated by the levels of miR-27a*, because Prf1 and GzmB proteins were dramatically up-regulated after IL-15 treatment. Although further increases in mRNA levels were observed with a higher dose of IL-15 (60 ng/mL), this increase seemed to be associated with a proportional increase in miR-27a* (Figure 5B middle and bottom panels) and did not result in a further increase in the levels of Prf1 and GzmB proteins, consistent with the absence of a further increase in NK-cell cytotoxicity (Figure 5C). This role for miR-27a* as a regulator of Prf1 and GzmB levels in NK cells was confirmed in the different types of NK cells. NK-92 cells showed higher basal levels of Prf1 and GzmB protein compared with the levels in mNK cells, despite the fact that the mRNA levels in NK-92 cells were decreased or similar to those in mNK cells, and this difference was associated with NK-92 cells having less endogenous miR-27a* than mNK cells (Figure 5D).
We next examined the effect of miR-27a* on NK-cell cytotoxicity in a more physiologic situation in which a hairpin for miR-27a*/miR-27a as a pre-microRNA is overexpressed, therefore mimicking the transcriptional induction of miR-27a* by IL-15. NK-92 cells were transduced by a lentivirus expressing pre-miR-27a*/27a and GFP (LV_miR-27a*; supplemental Figure 5A-B), sorted by GFP signal intensity (Figure 6A top panel), and expression in sorted cells was confirmed by real-time qPCR. In NK-92 cells, transduction with LV_miR-27a* specifically increased both miR-27a* and miR-27a levels by ∼ 2-fold compared with cells transduced with a control lentivirus (LV_Ctrl), with no changes detected in other miRNAs, such as miR-204 and miR-15a* (Figure 6A bottom panel). The enhanced level of miR-27a* in NK-92 cells (LV_miR-27a*) decreased Prf1 and GzmB protein levels without changes of their mRNA levels, and it concomitantly reduced NK-cell cytotoxicity (Figure 6B). LV_miR-27a*–transduced NK-92 cells showed little or no differences in cytokine production (supplemental Figure 5C), receptor expression (supplemental Figure 5D), or conjugation to target K-562 cells (data not shown), compared with LV_Ctrl–transduced NK-92 cells. Similar results were obtained with miR-27a*–transfected mNK cells.
Human miR-27a* is a negative regulator of NK-cell cytotoxicity. (A top) Sorting of the GFP-positive NK-92 cells expressing pre-miR-27a*/miR-27a. (Bottom) Real-time qPCR for miRNAs. (B) NK-92 cells expressing pre-miR-27a*/miR-27a show decreased NK-cell cytotoxicity. (Top) IB analysis. (Middle) Real-time qPCR for mRNAs. (Bottom) Cr release assay. Data are representative of 3 independent experiments (mean ± SEM of triplicates; Student t test, **P < .01 and ***P < .001 vs LV_Ctrl. (C-E) miR-27a* negatively regulates NK-cell cytotoxicity in the primary NK cells. Primary human NK cells transfected with Ctrl-miR or miR-27a* (C) or miR-27a* inhibitor (D) were subjected to IB analysis; Cr release assay at ET5, ET1.5, or both; and ELISA for TNF-α (red) and IFN-γ (blue, E). Data are representative of 2 independent experiments (mean ± SEM of triplicates; Student t test, ***P < .001 vs Ctrl_miR).
Human miR-27a* is a negative regulator of NK-cell cytotoxicity. (A top) Sorting of the GFP-positive NK-92 cells expressing pre-miR-27a*/miR-27a. (Bottom) Real-time qPCR for miRNAs. (B) NK-92 cells expressing pre-miR-27a*/miR-27a show decreased NK-cell cytotoxicity. (Top) IB analysis. (Middle) Real-time qPCR for mRNAs. (Bottom) Cr release assay. Data are representative of 3 independent experiments (mean ± SEM of triplicates; Student t test, **P < .01 and ***P < .001 vs LV_Ctrl. (C-E) miR-27a* negatively regulates NK-cell cytotoxicity in the primary NK cells. Primary human NK cells transfected with Ctrl-miR or miR-27a* (C) or miR-27a* inhibitor (D) were subjected to IB analysis; Cr release assay at ET5, ET1.5, or both; and ELISA for TNF-α (red) and IFN-γ (blue, E). Data are representative of 2 independent experiments (mean ± SEM of triplicates; Student t test, ***P < .001 vs Ctrl_miR).
To rule out the possibility that the negative role of miR-27a* in Prf1 and GzmB expression is limited to in vitro differentiated human mNK or human NK-92 cells, we transfected miR-27a* into primary NK cells (CD3−/CD56+/CD16+) purified from human UCB. Similar to mNK cells, miR-27a*–transfected primary NK cells showed less cytotoxicity concomitant with suppression of Prf1 and GzmB (Figure 6C). By contrast, miR-27a* inhibitor-transfected primary NK cells showed the opposite results (Figure 6D). Furthermore, primary NK cells transfected with miR-27a* and miR-27a* inhibitor showed no changes in cytokine production (Figure 6E) and the level of death molecules such as Fas/CD95, FasL/CD178, and TRAIL/CD253 (data not shown). Thus, our data suggest miR-27a* might be a general regulator of Prf1 and GzmB expression in human NK cells.
Manipulation of NK-cell cytotoxicity by human miR-27a* in a tumor model
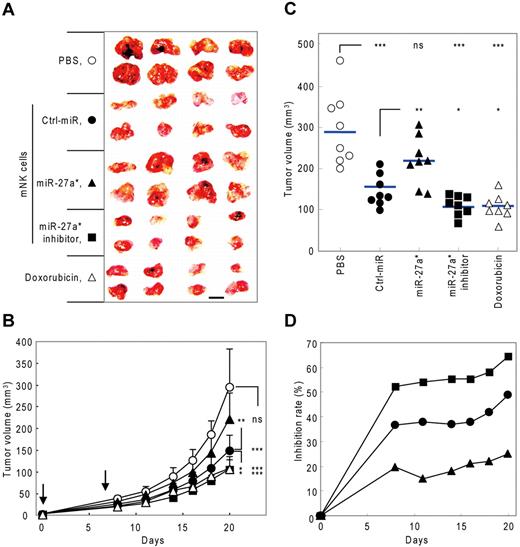
To evaluate in vivo the negative regulatory role of miR-27a* in NK-cell cytotoxicity by targeting both Prf1 and GzmB, we examined the effect of mNK cells transfected with miR-27a* or miR-27a* inhibitor on tumor growth in a human tumor xenograft model. We first determined the kinetics of miR-27a* expression in mNK cells transfected with miR-27a* and miR-27a* inhibitor. mNK cells transfected with miR-27a* or miR-27a* inhibitor showed overexpression or knockdown, respectively, of miR-27a* levels in vitro that was still discernible by day 7 after transfection (supplemental Figure 6A). Kinetic analysis of NK-cell killing capacity in miR-27a* inhibitor-transfected mNK cells revealed that Ctrl_miR–transfected mNK cells showed a gradual decrease in NK-cell cytotoxicity over 10 days after transfection, and mNK cells transfected with the miR-27a* inhibitor exhibited higher cytotoxicity than Ctrl_miR–transfected mNK cells at all time points through day 7 after transfection, with the largest differences early at day 2 when total cytotoxic activity was greatest (supplemental Figure 6B). Therefore, at 2 hours after tumor transplantation and then, based on the cytotoxicity data, again at 7 days, mice received intravenously as therapy 3 kinds of mNK cells, vehicle PBS as a negative control, or doxorubicin chemotherapy as a positive control.
Subcutaneous implantation of SW620 cells, a human colon cancer cell line, resulted in rapidly growing tumors in the PBS control group, with an average tumor volume of 270 ± 48 mm3 by day 20 after injection. Although tumor growth was inhibited by mNK cells, the inhibitory effect of mNK cells was significantly decreased by miR-27a* overexpression. In contrast, transfection of mNK cells with miR-27a* inhibitor resulted in enhanced antitumor activity, achieving results similar to doxorubicin chemotherapy (Figure 7A-C). At day 20, treatment of mice with Ctrl_miR–transfected mNK cells had inhibited tumor growth by 40%, with miR-27a*–overexpressed mNK cells being less effective, with 22.5% reduction in tumor, and with miR-27a*–knockdown mNK cells being more effective, with 58% reduction (Figure 7D), suggesting miR-27a* plays a negative role in regulating NK-cell cytotoxicity in vivo. Although doxorubicin chemotherapy had clear antitumor activity, it also resulted in systemic toxicity, as reflected by a significant loss of body weight, and this toxicity was not observed with mNK-cell treatment (supplemental Figure 6C), which might be an advantage of such cancer immunotherapy regimens.2,28 In addition, no signs of organ toxicity related to NK-cell therapy were observed on autopsy of the mice (data not shown). Taken together, these results suggest that human miR-27a* inhibits NK-cell cytotoxicity against tumors in vivo and imply that microRNA in NK cells may be a potential targetable molecule, particularly using strategies that more permanently disrupt miR-27a* activity, for enhancing therapy with NK cells.
Manipulation of NK-cell cytotoxicity by miR-27a* in human tumor xenograft model. (A-C) Tumor growth changes in nude mice bearing human tumor xenografts after treatment of modified mNK cells. (A) At the end of experiment (day 20), tumors were removed and photographed. Scale bar represents 1 cm. Tumor volume changes were measured until day 20 (days 0, 8, 11, 14, 16, 18, and 20). (B) Downward arrows indicate that mice were treated with mNK cells at that time point (days 0 and 7). (C) Scatter plots showing tumor volume of individual mice at day 20. Horizontal lines indicate average of tumor volume of each mouse (n = 8). (D) Time course of inhibition rate (vs PBS-treated group) of tumor volume by treatment with manipulated mNK cells. Tumor volumes were measured using a caliper and estimated by the formula: length (mm) × width (mm) × height (mm)/2. The tumor growth inhibition rate was calculated using the following equation: % inhibition = 100% − (tumor volume of test group/tumor volume of PBS group) × 100% (Mann-Whitney rank sum test, ns, P > .05, *P < .05, **P < .01, and ***P < .001 vs vehicle PBS or Ctrl-miR).
Manipulation of NK-cell cytotoxicity by miR-27a* in human tumor xenograft model. (A-C) Tumor growth changes in nude mice bearing human tumor xenografts after treatment of modified mNK cells. (A) At the end of experiment (day 20), tumors were removed and photographed. Scale bar represents 1 cm. Tumor volume changes were measured until day 20 (days 0, 8, 11, 14, 16, 18, and 20). (B) Downward arrows indicate that mice were treated with mNK cells at that time point (days 0 and 7). (C) Scatter plots showing tumor volume of individual mice at day 20. Horizontal lines indicate average of tumor volume of each mouse (n = 8). (D) Time course of inhibition rate (vs PBS-treated group) of tumor volume by treatment with manipulated mNK cells. Tumor volumes were measured using a caliper and estimated by the formula: length (mm) × width (mm) × height (mm)/2. The tumor growth inhibition rate was calculated using the following equation: % inhibition = 100% − (tumor volume of test group/tumor volume of PBS group) × 100% (Mann-Whitney rank sum test, ns, P > .05, *P < .05, **P < .01, and ***P < .001 vs vehicle PBS or Ctrl-miR).
Discussion
The net amount of protein is controlled by the balance between protein synthesis and degradation. Regulation of translation is an important mechanism that enables spatiotemporal modulation of protein levels.27,37 Fine-tuning of translation by miRNAs is not only useful to maintain homeostatic protein levels but also allows for rapid and sensitive responses to environmental stimuli.12,25 Regulation of translation by miRNAs has been widely investigated in the function and development of most immune cells,13,14,25,26 with thus far the exception of NK cells. Here, we demonstrate that human miR-27a* is an essential player in regulating homeostatic NK-cell cytotoxicity by (1) specifically targeting the 3′ UTR sequences of Prf1 and GzmB, (2) suppressing Prf1 and GzmB expression in both resting and activated states, (3) functioning as a fine controller of the net amount of the target proteins in human NK cells, and (4) negatively regulating NK-cell cytotoxic activity. This example of microRNA-mediated gene expression defines a new paradigm for regulation of NK-cell cytotoxicity in NK cells. Consistent with miR-27a* having an inhibitory role, knockdown of miR-27a* in mNK cells exhibited enhanced antitumor activity in the human xenograft model, suggesting this molecule might be selectively targeted therapeutically.
In contrast to the high basal level of GzmA protein, Prf1 and GzmB protein expression was low in resting cells and up-regulated after activation. Although analyses of expression of Prf1 and GzmB in effector CD8+ cytolytic T lymphocytes have shown that control is maintained at the level of transcription,4,38,39 in this report we show that human Prf1 and GzmB expression in NK cells is regulated by translational suppression as well as transcriptional induction. In resting NK cells, Prf1 and GzmB protein levels are low, with basal levels of human Prf1 and GzmB mRNA high but suppressed by endogenous miR-27a*. A time lag was observed between the increase in Prf1 and GzmB mRNA levels and the increase in protein levels during activation. In early NK-cell activation, Prf1 and GzmB mRNA and miR-27a* were simultaneously up-regulated, but the fold induction of Prf1 and GzmB mRNA was higher than that of miR-27a*. Thus, the higher up-regulation of Prf1 and GzmB mRNA seems to saturate the suppressive capacity of miR-27a* and ultimately results in increased Prf1 and GzmB protein expression. Protein levels of Prf1 and GzmB gradually increased and reach a maximum after 24 hours of IL-15 treatment, coincident with the kinetics of NK-cell cytotoxicity. The different populations of human NK cells used in our experiments showed differences in the basal levels of Prf1 and GzmB protein, and these differences could be because of differences in the endogenous amount of miR-27a*, GzmB mRNA, or Prf1 mRNA (Figure 5D). Thus, this expression of Prf1 and GzmB in NK cells could be explained by a balance between transcriptional induction and suppression mediated by a fine-tuner such as miR-27a*.
miR-27a* is a regulator that simultaneously targets both Prf1 and GzmB expression and endogenously modulates NK-cell cytotoxicity. However, we could not fully exclude the possibility that miR-27a* targets additional cytotoxicity-regulating genes or processes not examined in this study, because miRNAs do have a broad range of target mRNAs. In fact, miR-27a*–transfected mNK cells showed a slight decrease in SH2-containing inositol phosphatase SHP-1 and Src kinase expression (data not shown), both known to be involved in NK-cell activation.6,40 Moreover, in vitro knockdown of microRNAs by silencing Dicer1 and Ago2 resulted in a dramatic increase in NK-cell cytotoxicity, suggesting many other miRNAs are probably involved in regulating NK-cell cytotoxicity, including priming,33 trafficking,41 immune synapse formation,40 receptor signaling,1 and arming with effector molecules.6 As with B and T cells, the molecular action mechanism, impact, and biologic roles of miRNAs in NK-cell biology requires further investigation.
Although it is not clear why Prf1 and GzmB protein expression is suppressed by miR-27a* at both rest and activation, we can speculate such miRNA-mediated fine-tuning of Prf1 and GzmB expression has some possible advantages. By having high levels of mRNA present at rest and during activation, NK cells are poised for a quick response to stimuli with production of Prf1 and GzmB proteins, while circumventing the risk of expressing unnecessarily high levels of potentially toxic proteins at rest and early activation, as suggested by GzmB leakage-induced cell death that has been reported in T and NK cells shortly after activation.7,42,43 In mice, IL-15–mediated priming of NK cells occurs by contact with dendritic cells in draining lymph nodes that starts a cascade leading to full activation. Such full activation of NK cells occurs when primed NK cells then respond to localized tumor cells or infected cells.33 Similarly, a time lag between mRNA peak and protein peak by IL-15 treatment may reflect a sequential process required for full activation in human NK cells. High levels of Prf1 and GzmB protein are not required early during activation; therefore, a lag in Prf1 and GzmB protein expression could be advantageous for proper timing control of effector functions.
Several microRNAs involved in signaling for NK-cell cytotoxicity have been found in target cells,44-46 with the potential target genes including the NKG2D-activating ligands MHC class I polypeptide-related sequence A (MICA) and MICB, and the immunoinhibitory molecule B7-H3 (CD276). Expression of MICA and MICB seems to be regulated by cellular miRNAs that maintain protein expression below a defined threshold and facilitate acute up-regulation of MICA and MICB during cellular stress.44,45 Many tumors overexpress the MICA- and MICB-regulating miRNAs, down-regulate the CD276-regulating miR-29,46 or both. Thus, miRNAs have been implicated in escape of target cells from NK-cell immune surveillance by regulating ligand expression. Recently, the miRNA transcriptomes of resting and cytokine-activated primary mouse NK cells was defined and reported for bioinformatics analysis.47 This study suggested that, in mouse NK cells, miR-223 has the potential to regulate expression of GzmB, but this could not be the case with human NK cells because the 3′ UTRs of human GzmB and Prf1 lack the miRNA-targeting site for miR-223. In the course of experiments, we had a query whether the role of miR-27a* in Prf1 and GzmB expression is conserved among species. We examined in silico mouse Prf1 and GzmB 3′ UTRs for miR-27a* binding sites, and both mouse GzmB and Prf1 mRNAs had conserved miR-27a* binding sites in their 3′ UTRs. Overexpression of miR-27a* dramatically decreased expression of reporter genes containing mouse Prf1 and GzmB 3′ UTR (data not shown), suggesting miR-27a* might be a common regulator of Prf1 and GzmB expression among species.
NK cells have been shown to mediate therapeutic effects in human clinical trials,2 but efforts to modulate NK-cell cytolytic capacity against human cancer have not been successful, suggesting novel targets that regulate NK-cell cytotoxicity are needed.28 One approach that is being pursued is to manipulate NK-cell signaling, especially the balance, sensitivity, and cross-talk between stimulatory and inhibitory NK receptor signals.2,5 However, as shown in knockout mice,2,4,5 NK-cell cytotoxicity requires the effector molecules Prf1 and GzmB, and the molecular insight into the role of microRNAs that specifically regulate NK-cell killing provided by our study suggest it also may be possible to enhance NK-cell–based immunotherapy against human cancer by modulating microRNA expression in NK cells.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Global Research Laboratory grant funded by the Korean Foundation of International Cooperation of Science and Technology (FGM1400711-10), the Korea Research Institute of Bioscience and Biotechnology Research Initiative Program, and a National Institutes of Health grant (CA33084).
National Institutes of Health
Authorship
Contribution: T.-D.K. designed experiments, performed research, analyzed results, and wrote the manuscript; S.U.L. conducted experiments; H.-N.S. and J.W.K. interpreted results; S.Y., S.H.L., and S.R.Y. contributed technical support; H.M.K., S.-K.P., and C.W.L. conducted experiments; P.D.G. assisted in experimental design and provided critical comments; and I.C. supervised the project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Inpyo Choi, Cell Therapy Research Center, Korea Research Institute of Bioscience and Biotechnology, 125 Gwahak-ro, Yuseong, Daejeon 305-806, Republic of Korea; e-mail: ipchoi@kribb.re.kr.

![Figure 3. Human miR-27a* specifically targets both Prf1 and GzmB 3′ UTR sequence. (A top) Predicted miR-27a* binding sites. (Bottom) Schematic diagrams of Prf1 and GzmB 3′ UTRs (wild type [WT]) or their deletions (D1 and D2) and mutants (M). Point mutations are bolded. Numbers indicate positions of nucleotides in the 3′ UTRs. Crosses indicate the mutation depicted in the top panel. (B-D) Reporter assay using IB analysis. HEK-293FT cell lines were cotransfected with combinations of reporter plasmids containing WT, D, or M forms of Prf1 and GzmB 3′ UTRs and miR-27a* or its mutated miRNA (miR-27a*M). Reporter AANAT mRNA levels were normalized to NeoR mRNA level as an internal control of the vector. Net amount of translated AANAT protein was determined by IB. GAPDH served as loading control. Data are representative of 4 independent experiments (mean ± SEM of triplicates).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/20/10.1182_blood-2011-04-347526/4/m_zh89991182080003.jpeg?Expires=1767755461&Signature=XPqZbv1tbQaK7gFUDPPcg-s0CfcSssc4lY7paJS9C0xbtYWITTDcaKF5nX3WUX~ECEBSMULSKBzvQ5UR~LEGKpGkYEPNvc6QEaNI5J4N62ShJTBag8f2ztz6yI9DFXaF-vFhauB0RR-xflywa0f0Sym2rQSQ4a8OfXKQ9DusMg70U5zQbAIVLBVxB5gKjCvXNVfD~ia5SS5OjqC1FGj3WJFOu-Zhrr58nU0hRn5IJHVWacfhKSi9wxZdfwCTPCiw3pbRS71bxVs4DLcElYYU8JqLe2yedWPB3ojJjGk~YBTQ0V6VrDvnFaGDT9h0gbFUF2MBWsbP4ZxXYaZEm5pztw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal