Abstract
Mesenchymal stromal cells (MSCs) are an essential cell type of the hematopoietic microenvironment. Concerns have been raised about the possibility that MSCs undergo malignant transformation. Several studies, including one from our own group, have shown the presence of cytogenetic abnormalities in MSCs from leukemia patients. The aim of the present study was to compare genetic aberrations in hematopoietic cells (HCs) and MSCs of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) patients. Cytogenetic aberrations were detected in HCs from 25 of 51 AML patients (49%) and 16 of 43 MDS patients (37%). Mutations of the FLT3 and NPM1 genes were detected in leukemic blasts in 12 (23%) and 8 (16%) AML patients, respectively. Chromosomal aberrations in MSCs were detected in 15 of 94 MDS/AML patients (16%). No chromosomal abnormalities were identified in MSCs of 36 healthy subjects. We demonstrate herein that MSCs have distinct genetic abnormalities compared with leukemic blasts. We also analyzed the main characteristics of patients with MSCs carrying chromosomal aberrations. In view of these data, the genetic alterations in MSCs may constitute a particular mechanism of leukemogenesis.
Introduction
Myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) are clonal disorders affecting pluripotent stem cells and are characterized by ineffective hematopoiesis.1 Most MDS and AML patients harbor cytogenetic and molecular defects that identify entities with peculiar biologic and clinical data and distinct therapeutic responses.2-7 Overall, approximately 40%-70% of de novo MDS, 50%-60% of de novo AML, and 80%-95% of secondary MDS/AML (s-MDS/s-AML) patients display chromosomal aberrations. In de novo MDS, chromosomal lesions consist of deletion or numerical defects with a frequency of 50% and 10%, respectively, whereas chromosomal translocations account for only 4% of lesions.1,5 In contrast, structural chromosomal rearrangements are the most common cytogenetic abnormalities in de novo AML, with an incidence of ∼ 40%.1-4,6,7 Recent large collaborative studies have demonstrated the importance of cytogenetic aberrations for the prognosis of MDS/AML patients.2,3,5,6
Increasing evidence suggests that mutations of multiple genes mediate the pathogenesis and progression of MDS and AML.4,8-10 Molecular analysis has revealed recurrent genetic markers in > 85% of AML patients with normal karyotype. These genetically differentiated subtypes are associated with diverse biologic characteristics and distinct clinical profiles.8-12 Mutations of the FMS-like tyrosine kinase-3 (FLT3) gene, including both point mutations within the tyrosine kinase domain (TKD) and internal tandem duplication (ITD) in the juxtamembrane domain, are the most common genetic alterations detected in AML. FLT3-ITD, which occurs with a frequency of 35%-45% in normal karyotype AML, has an adverse impact on prognosis. A few incidences of FLT3-TKD mutations represented by single nucleotide base exchanges have also been reported.8-11 AML is associated with mutation of the nucleophosmin-1 gene (NPM1), which causes aberrant cytoplasmic expression of nucleoplasmin and accounts for approximately one-third of adult AML, but 55% of normal karyotype AML.8-10,12
Chromosomal aberrations and genetic mutations play a pivotal role in the pathogenesis of AML and MDS. According to a proposed multistep pathogenesis of leukemia, after the initial damage of the progenitor cell, several additional alterations may affect these cells, providing them with a growth advantage.1,13 In addition, the hematopoietic microenvironment (HM) is involved in the pathophysiology.14 The HM controls the formation of blood cells through the production and secretion of cytokines and extracellular matrix molecules.15 The central component of HM are BM mesenchymal stromal cells (MSCs). These cells have been defined as primitive, multipotent, undifferentiated cells capable of self-renewal, and they have the ability to give rise to different cell lineages. The HM can regulate hematopoiesis by interacting directly with hematopoietic cells (HCs) and/or by secreting regulatory molecules that influence, in a positive or negative manner, the growth of HCs.15 Whether MSC alterations influence hematologic disorders, and how such alterations contribute to the progression of the disease, remain controversial. Several studies have proposed that important quantitative and functional alterations occur in the MSCs of patients with different hematologic disorders.16,17 MSCs seem to have a relevant role in AML because they prevent spontaneous and induced apoptosis and may attenuate chemotherapy-induced cell death. This possibility has been confirmed by the finding that cocultivation of a leukemic cell line with the murine stroma cell line MS-5 can block apoptosis.18 MDS/AML may arise in an abnormal HM, resulting in the generation of multiple populations with varying initiation events. More recently, using transgenic mice, Raaijmakers et al showed that genetic alteration of cells in the BM microenvironment could induce MDS with ineffective hematopoiesis and dysmorphic HCs and with occasional transformation to AML.19 The leukemic cells showed distinct genetic abnormalities compared with those in the BM microenvironment.19 Previously, Walkley et al demonstrated that dysfunction of the RB protein or retinoic acid receptor ϒ in the BM microenvironment could contribute to the development of preleukemic myeloproliferative disease.20
Differences have been reported between the MSCs of leukemia patients and those of healthy donors.17,18 However, the presence of cytogenetic abnormalities in the MSCs of patients with hematologic disorders is controversial. Conventional cytogenetic, FISH, and array-based comparative genomic hybridization studies aiming to establish whether the MSCs of leukemia patients harbor genetic aberrations have reported conflicting results. Only a few studies, including one from our own group, have shown the presence of cytogenetic abnormalities in MSCs of a significant proportion of leukemia patients.21-24 In addition, previously published data have documented a distinctive gene-expression profile of MSCs of MDS and AML patients compared with MSCs of healthy donors using microarray analysis.25 Other investigators have reported conflicting results.26 To gain insight into these questions and to compare genetic aberrations in HCs and MSCs, we performed cytogenetic, FISH, and RT-PCR analyses of MSCs of 94 patients and 36 healthy donors. We also investigated the most typical mutations (FLT3-ITD, FLT3-TKD, and NPM1) in leukemic cells and MSCs of AML patients and characterized the main clinical characteristics and outcome of patients with MSC aberrations.
Methods
Patient characteristics
A total of 43 MDS patients, 51 AML patients, and 36 healthy donors were included in this study. All patients were treated at University Clinic Charité–Campus Benjamin Franklin from August 2006 to January 2010. The patient characteristics are summarized in Table 1. The male/female ratio was 69/25 and the median age was 61 years (range, 19-84 years). Diagnoses were established according to World Health Organization (WHO) criteria.27 s-AML was diagnosed in 19 of 51 (37%) AML patients. Normal BM samples were obtained from 36 healthy donors (17 males and 19 females) with a median age of 48 years (range, 21-86 years). Written informed consent was obtained from all patients and donors in accordance with the Declaration of Helsinki and the ethical guidelines of the Charité University School of Medicine, which approved this study.
MSC harvest, culture conditions, and characterization
Mononuclear cells (MNCs) were isolated from BM samples using Ficoll-Paque Plus (Amersham Biosciences) during initial diagnosis. MNCs were cultured in IMDM (GIBCO-BRL) supplemented with 20% FBS (GIBCO-BRL), glutamine, and antibiotics/antimycotics (Biochrom). The MSCs were collected after 3-4 passages for subsequent cytogenetic analyses, FISH studies, and RNA and genomic DNA isolation. The following mAbs were used to characterize cultured cells and demonstrate their nonhematopoietic origin: anti-CD19, anti-CD29, anti-CD33, anti-CD34, anti-CD45, anti-CD73, and anti-CD90 (BD Biosciences/Pharmingen); anti-CD14 (Beckman Coulter); and anti-CD105 (Caltag). Analysis of cell surface molecules was performed as described previously.21,28
Cytogenetic analysis and FISH
Chromosome studies of HCs and MSCs were performed as described previously.21 We analyzed a median of 26 metaphases (range, 15-50) in all samples. The karyotype was described according to the International System for Human Cytogenetic Nomenclature (ISCN 2009).29 To confirm chromosomal aberrations identified by G-banding in both HCs and MSCs, FISH with appropriate specific fluorescent probes was performed as described previously21 using commercially available probes. The following probes were used: LSI EGR1 (5q31), D5S23 (5p15.2), LSI WSR (7q11), LSI D7S522 (7q31), D7S740 (7q22)/7q35, CEP7, CEP4, CEP5, CEP8, LSIPML/RARA, LSI SBFB, LSI MLL, Lib1, and Lib2 (Abbott); LSI 1p36/LSI 1q25, and CEPXSG/CEPYSO (Vysis); and RP11-175G14 (1p31)/RP11-34K3 (2q33)/red and RP11-93E22 (2q37.1)/RP4 770C6 (1p13.2)/green (BlueGnome). Procedures were performed according to standard Abbott/Vysis protocols. In total, 200 HC nuclei and 500 MSC nuclei were scored.
Molecular studies: DNA and RNA preparation and PCR to detect t(11;19), FLT3, and NPM1 mutations
Genomic DNA and RNA were isolated from both MNCs and MSCs using TRIzol reagent (Invitrogen) according to the manufacturer's recommendations. RT-PCR was performed simultaneously for HCs and MSCs of 3 AML patients carrying a translocation t(11;19) in accordance with methods described previously.30 We also investigated the NPM1 and FLT3 mutation status in both HCs and MSCs of 51 AML patients. PCR amplification of NPM1 exon 12 was the same as those used previously.31 PCR products were sequenced with the primer NPM1-R2 using the ABI Ready Reaction Dye Terminator Cycle Sequencing Kit and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). The ITD and D835 regions of the FLT3 gene were amplified using the FLT3 Mutation Detection Kit (InVivoScribe Technologies) according to the manufacturer's recommendations. The D835 PCR product was digested with EcoRV (Fermentas). The presence of mutations was determined by 2% agarose gel electrophoresis.
Statistics
Central tendency of the data was measured as the median, and the dispersion of values around the median was expressed as the range. Median follow-up was calculated according to recommended criteria.32 Comparisons between different groups were made using the Student t test and the 2-sided exact Fisher test (dichotomous variables). P < .05 was considered significant. The overall survival (OS) and disease-free survival (DFS) were estimated using the Kaplan-Meier method. Kaplan-Meier life tables were constructed for survival data and were compared by the log-rank test. Cox regression analysis was performed to identify prognostic variables for OS. For multivariate analysis of prognostic factors, a proportional hazard regression model was used. Stepwise forward selection was performed. All calculations were performed using RASW statistics software Version 18 (http://www.winwrap.com).
Results
Karyotype analysis and FISH of HCs
Cytogenetic aberrations were detected in 16 MDS (37%) and 25 AML (49%) patients (Table1). All refractory anemia (RA) patients showed normal karyotype. The majority of patients with RA with excess blasts (RAEB II) demonstrated different chromosomal markers. Complex karyotype abnormalities were detected in 4 MDS and 5 AML patients. Translocations t(15;17), t(8;21), and inv(16) were detected in 4 AML patients with the appropriate forms of leukemia. The following aberrations of 11q23 were found in 7 AML patients: t(4;11), t(11;19), and del(11)(q23). We confirmed the aberration in all cases by FISH and RT-PCR. In all patients with normal karyotypes, FISH was performed with specific probes for chromosomes 5, 7, 8, 11, 16, and 20. No aberrations were identified in 200 interphase nuclei.
Cytogenetic and clinical characteristics of patients
| Diagnosis . | Cytogenetic data (HCs) . | n . | Median age, y (range) . |
|---|---|---|---|
| RA | 46,XX/46,XY | 5 | 64 (49-80) |
| RCMD | 46,XX/46,XY | 6 | 62 (52-76) |
| −Y | 2 | 61 (56-67) | |
| del(20)(q11q13) | 1 | 70 | |
| inv(7)(q21q31) | 1 | 70 | |
| RCMD-RS | 46,XX/46,XY | 7 | 68 (58-83) |
| −X | 1 | 63 | |
| RAEB I | 46,XX/46,XY | 6 | 63 (53-70) |
| del(11)(q23) | 1 | 63 | |
| Complex aberrations | 1 | 84 | |
| RAEB II | 46,XX/46,XY | 3 | 63 (36-78) |
| Complex aberrations | 3 | 74 (68-84) | |
| −7/7q− alone | 2 | 47 (23-71) | |
| +8 | 2 | 70 (69-71) | |
| +11 | 1 | 80 | |
| 5q Syndrome | del(5)(q13q31) | 1 | 64 |
| AML M0 | 46,XY | 1 | 68 |
| AML M1 | 46,XX/46,XY | 4 | 65 (58-74) |
| Complex aberrations | 3 | 53 (30-66) | |
| 11q23 aberrations | 2 | 57 (52-62) | |
| t(3;8)(q26;q24) | 1 | 49 | |
| AML M2 | 46,XX/46,XY | 6 | 63 (51-69) |
| t(8;21)(q22;q22) | 1 | 46 | |
| AML M3 | t(15;17)(q22;q21) | 2 | 64 (54-75) |
| AML M4 | 46,XX/46,XY | 10 | 55 (27-74) |
| inv(16)(p13;q22) | 1 | 57 | |
| 11q23 aberrations | 3 | 60 (39-71) | |
| −7/7q− alone | 3 | 76 (65-83) | |
| +8 | 1 | 59 | |
| Complex aberrations | 1 | 74 | |
| AML M5 | 46,XX/46,XY | 4 | 57 (51-63) |
| 11q23 aberrations | 1 | 26-55 | |
| +8 | 1 | 33 | |
| del(1)(p35),+21 | 1 | 69 | |
| AML M6 | 46,XY | 1 | 60 |
| Complex aberrations | 1 | 32 | |
| +8 | 1 | 19 | |
| del(9)(q22) | 1 | 71 | |
| del(5)(q13q31) | 1 | 56 |
| Diagnosis . | Cytogenetic data (HCs) . | n . | Median age, y (range) . |
|---|---|---|---|
| RA | 46,XX/46,XY | 5 | 64 (49-80) |
| RCMD | 46,XX/46,XY | 6 | 62 (52-76) |
| −Y | 2 | 61 (56-67) | |
| del(20)(q11q13) | 1 | 70 | |
| inv(7)(q21q31) | 1 | 70 | |
| RCMD-RS | 46,XX/46,XY | 7 | 68 (58-83) |
| −X | 1 | 63 | |
| RAEB I | 46,XX/46,XY | 6 | 63 (53-70) |
| del(11)(q23) | 1 | 63 | |
| Complex aberrations | 1 | 84 | |
| RAEB II | 46,XX/46,XY | 3 | 63 (36-78) |
| Complex aberrations | 3 | 74 (68-84) | |
| −7/7q− alone | 2 | 47 (23-71) | |
| +8 | 2 | 70 (69-71) | |
| +11 | 1 | 80 | |
| 5q Syndrome | del(5)(q13q31) | 1 | 64 |
| AML M0 | 46,XY | 1 | 68 |
| AML M1 | 46,XX/46,XY | 4 | 65 (58-74) |
| Complex aberrations | 3 | 53 (30-66) | |
| 11q23 aberrations | 2 | 57 (52-62) | |
| t(3;8)(q26;q24) | 1 | 49 | |
| AML M2 | 46,XX/46,XY | 6 | 63 (51-69) |
| t(8;21)(q22;q22) | 1 | 46 | |
| AML M3 | t(15;17)(q22;q21) | 2 | 64 (54-75) |
| AML M4 | 46,XX/46,XY | 10 | 55 (27-74) |
| inv(16)(p13;q22) | 1 | 57 | |
| 11q23 aberrations | 3 | 60 (39-71) | |
| −7/7q− alone | 3 | 76 (65-83) | |
| +8 | 1 | 59 | |
| Complex aberrations | 1 | 74 | |
| AML M5 | 46,XX/46,XY | 4 | 57 (51-63) |
| 11q23 aberrations | 1 | 26-55 | |
| +8 | 1 | 33 | |
| del(1)(p35),+21 | 1 | 69 | |
| AML M6 | 46,XY | 1 | 60 |
| Complex aberrations | 1 | 32 | |
| +8 | 1 | 19 | |
| del(9)(q22) | 1 | 71 | |
| del(5)(q13q31) | 1 | 56 |
RCMD indicates refractory cytopenia with multilineage dysplasia; and RCMD-RS, RCMD with ringed sideroblasts.
Karyotype analysis and FISH of MSCs
Cytogenetic analysis of MSCs was successfully performed in 94 cases. We detected chromosomal abnormalities in 15 of 94 (16%) MSCs (Table 2). Cytogenetic aberrations were detected in MSCs of 10 AML and 5 MDS patients.
Chromosomal aberrations in MSCs
| Patient no.* . | Diagnosis . | Cytogenetic markers . | |
|---|---|---|---|
| HCs . | MSCs . | ||
| 0806 | AML M2 | 46,XX[25] | t(1;2)(p32;q31)[15] |
| 1282 | AML M5 | 47,XY,+8,t(11;19)(q23;p13.1)[25] | t(1;6)(p32;p12)[23] |
| 0734 | AML M1 | 46,XX,t(11;19)(q23;p13.1)[21]/46,XX[4] | del(7)(q11.2q32)[9] |
| 0717 | RA | 46,XX[25] | del(7)(q22)[3] |
| 0214 | s-AML M6 | 46,XY,del(5)(q13q32)[25] | del(7)(q22)[3] |
| 1068 | AML M5 | 46,XY[25] | t(3;20)(p13;p11.2)[15] |
| 0600 | RAEB II | 47,XY,+8[25] | inv(X)(q12p22)[16] |
| 1150 | s-AML M4 | 45,XY,−7[20]/46,XY[5] | del(11)(q23)[2] |
| 0589 | AML M4 | 46,XY[25] | del(13)(q12q22)[6] |
| 0204 | s-AML M1 | 46,XY,add(1)(p36),−4,der(5),der(7), der(9),add(12)(p13),der(17),+21[25] | del(15)(q14)[4] |
| 1398 | s-AML M2 | 46,XY[25], FLT3-ITD mutation | +5[4] |
| 0816 | RAEB II | 46,XX,del(7)(q31)[25] | +5[14] |
| 0645 | RCMD-RS | 46,XY[25] | −4[6] |
| 0752 | s-AML M1 | 46,XX,del(5)(q22q35)[8]/47,XX,idem, +21[3]/48,XX,idem,+21,+mar[6] | −X[12] |
| 0789 | RAEB I | 46,XY[25] | −Y[6] |
| Patient no.* . | Diagnosis . | Cytogenetic markers . | |
|---|---|---|---|
| HCs . | MSCs . | ||
| 0806 | AML M2 | 46,XX[25] | t(1;2)(p32;q31)[15] |
| 1282 | AML M5 | 47,XY,+8,t(11;19)(q23;p13.1)[25] | t(1;6)(p32;p12)[23] |
| 0734 | AML M1 | 46,XX,t(11;19)(q23;p13.1)[21]/46,XX[4] | del(7)(q11.2q32)[9] |
| 0717 | RA | 46,XX[25] | del(7)(q22)[3] |
| 0214 | s-AML M6 | 46,XY,del(5)(q13q32)[25] | del(7)(q22)[3] |
| 1068 | AML M5 | 46,XY[25] | t(3;20)(p13;p11.2)[15] |
| 0600 | RAEB II | 47,XY,+8[25] | inv(X)(q12p22)[16] |
| 1150 | s-AML M4 | 45,XY,−7[20]/46,XY[5] | del(11)(q23)[2] |
| 0589 | AML M4 | 46,XY[25] | del(13)(q12q22)[6] |
| 0204 | s-AML M1 | 46,XY,add(1)(p36),−4,der(5),der(7), der(9),add(12)(p13),der(17),+21[25] | del(15)(q14)[4] |
| 1398 | s-AML M2 | 46,XY[25], FLT3-ITD mutation | +5[4] |
| 0816 | RAEB II | 46,XX,del(7)(q31)[25] | +5[14] |
| 0645 | RCMD-RS | 46,XY[25] | −4[6] |
| 0752 | s-AML M1 | 46,XX,del(5)(q22q35)[8]/47,XX,idem, +21[3]/48,XX,idem,+21,+mar[6] | −X[12] |
| 0789 | RAEB I | 46,XY[25] | −Y[6] |
RCMD indicates refractory cytopenia with multilineage dysplasia; and RCMD-RS, RCMD with ringed sideroblasts.
For the patient sample.
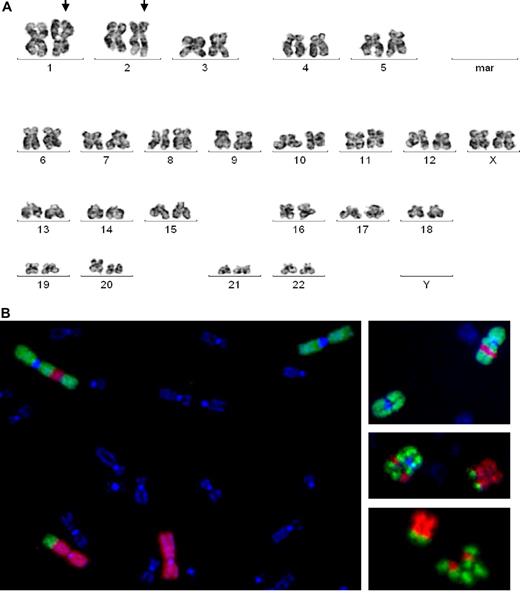
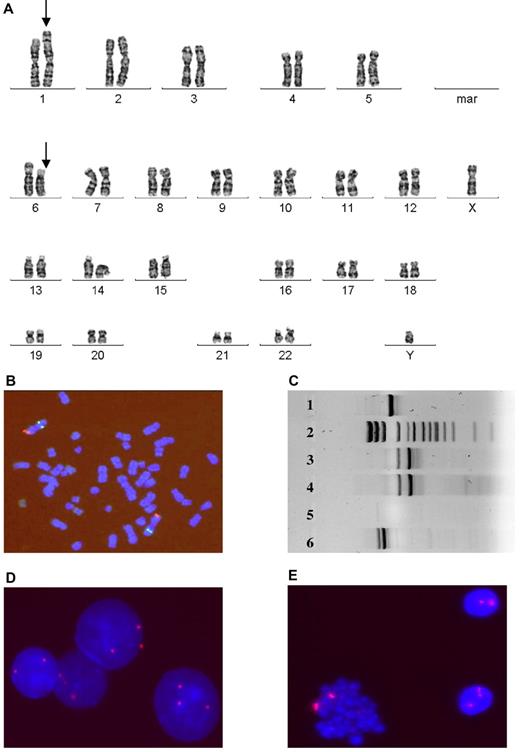
An AML patient (N0806) with normal HC karyotype showed MSC markers involving chromosomes 1 and 2: der(1;2)(1qter→1q42::2q31→2qter::1p32→1q42) and der(1;2)(p32;31) (Figure 1). Translocation was detected in 3 metaphases using G-banding and in an additional 12 cells by FISH with library probes for whole chromosomes 1 and 2. For detailed and precise analysis of the aberrant chromosomal loci, we performed FISH with BAC probes for 1p13, 1p31, 2q37.1, and 2q33. One AML patient (N1282) carried a t(11;19)(q23;p13.3) translocation in HCs. We used RT-PCR and FISH to examine this aberration in both HCs and MSCs; t(11;19) was detected in HCs only (Figure 2). Although we did not detect t(11;19) in MSCs, we detected a different chromosomal abnormality, t(1;6)(p32;p12), in 23 of 30 analyzed MSCs. The absence of t(1;6) in HCs was confirmed by FISH (Figure 2). Another patient with t(11;19)(q23;p13.3) in HCs (N0734) carried del(7)(q11.2q32) in MSCs. The aberration was detected in 9 MSC mitoses and was confirmed by FISH. An RA patient with normal HC karyotype (N0717) and an AML patient with del(5)(q13q32) in HCs (N0214) displayed del(7)(q22) in MSCs. All HC and MSC aberrations were verified by FISH (Figure 3). Another AML patient (N1068) with a normal HC karyotype displayed t(3;20)(p13;p11.2) in MSCs. A total of 23 MSC metaphases were analyzed, and t(3;20) was detected in 15. All HC mitoses were cytogenetically normal. Another patient (N0600) with trisomy 8 in HCs demonstrated inv(X)(q12p22) in MSCs. We established this aberration in 16 metaphases using conventional cytogenetic techniques and in an additional 32 cells with probes for Xp11.1-q11.1.We identified trisomy 8 in HCs and 2 chromosomes 8 in MSCs by FISH. In an s-AML patient (N1150) with monosomy 7 in HCs, we also identified del(11)(q23) in MSCs. We verified monosomy 7 in HCs but not in MSCs and del(11q) in MSCs but not in HCs by FISH (Figure 3). In a patient (N0589) with a normal HC karyotype, 6 MSC metaphases displayed del(13)(q12q22). A patient with normal HC karyotype (N0204) displayed del(15)(q14) in 4 MSC mitoses.
Cytogenetic aberrations in MSC N0806. (A) G-banding image demonstrates translocation of chromosomes 1 and 2 (arrows). (B) FISH with library probes for whole chromosomes 1 and 2 confirms that a fragment of 1p (green) is translocated to 2q (red) and part of 2q (red) is inserted into 1p (green).
Cytogenetic aberrations in MSC N0806. (A) G-banding image demonstrates translocation of chromosomes 1 and 2 (arrows). (B) FISH with library probes for whole chromosomes 1 and 2 confirms that a fragment of 1p (green) is translocated to 2q (red) and part of 2q (red) is inserted into 1p (green).
MSC (1282) demonstrates distinct aberrations compared with markers detected in HCs. (A) G-banding image shows translocation t(1;6)(p32;p12) (arrows) in MSCs. (B) FISH image with probes 1p36(red)/1q25(green) demonstrates the absence of this aberration in HCs. (C) RT-PCR for t(11;19)(q23;p13.3) demonstrates the aberration in HCs (lane 4), but not in MSCs (lane 5). Lane 3 is a positive control for the MLL-ENL fusion gene. Lanes 1 and 6 illustrate 2 alternative MLL-ENL fusion genes, and lane 2 displays the size standard. FISH image with CEP probes for chromosome 8 confirms trisomy 8 in HCs (D) and the absence of aberrations in MSCs (E).
MSC (1282) demonstrates distinct aberrations compared with markers detected in HCs. (A) G-banding image shows translocation t(1;6)(p32;p12) (arrows) in MSCs. (B) FISH image with probes 1p36(red)/1q25(green) demonstrates the absence of this aberration in HCs. (C) RT-PCR for t(11;19)(q23;p13.3) demonstrates the aberration in HCs (lane 4), but not in MSCs (lane 5). Lane 3 is a positive control for the MLL-ENL fusion gene. Lanes 1 and 6 illustrate 2 alternative MLL-ENL fusion genes, and lane 2 displays the size standard. FISH image with CEP probes for chromosome 8 confirms trisomy 8 in HCs (D) and the absence of aberrations in MSCs (E).
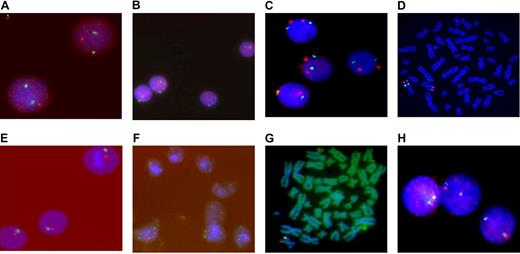
Results of FISH of HCs and MSCs. FISH of HCs and MSCs from a patient (N0214) with s-AML M6 (A-D). (A) FISH image with probes LSI EGR1 (5q31, red)/D5S23 (5p15.2, green) showing del(5q) in interphase nuclei from leukemic blasts. (B) FISH image of MSCs with the same probes demonstrates 2 signals for chromosome 5. (C) FISH image with probes D7S740 (7q22, green)/(7q35, red) demonstrates del(7)(q22) in MSCs and 2 signals for corresponding chromosomes in HCs (D). FISH of HCs and MSCs of a patient (N1150) with s-AML M4 (E-J): (E) FISH image with probes LSI WBS (7q11, red) and LSI D7S522 (7q31, green) demonstrates monosomy 7 in HCs. (F) MSCs shows 2 signals for chromosome 7. (G) MSC metaphase with 1 split signal from MLL DC probe confirms del(11)(q23). (H) Interphase nuclei of HCs demonstrate 2 normal chromosomes 11.
Results of FISH of HCs and MSCs. FISH of HCs and MSCs from a patient (N0214) with s-AML M6 (A-D). (A) FISH image with probes LSI EGR1 (5q31, red)/D5S23 (5p15.2, green) showing del(5q) in interphase nuclei from leukemic blasts. (B) FISH image of MSCs with the same probes demonstrates 2 signals for chromosome 5. (C) FISH image with probes D7S740 (7q22, green)/(7q35, red) demonstrates del(7)(q22) in MSCs and 2 signals for corresponding chromosomes in HCs (D). FISH of HCs and MSCs of a patient (N1150) with s-AML M4 (E-J): (E) FISH image with probes LSI WBS (7q11, red) and LSI D7S522 (7q31, green) demonstrates monosomy 7 in HCs. (F) MSCs shows 2 signals for chromosome 7. (G) MSC metaphase with 1 split signal from MLL DC probe confirms del(11)(q23). (H) Interphase nuclei of HCs demonstrate 2 normal chromosomes 11.
Numerical chromosomal aberrations, such as gain of chromosomes 5 and loss of chromosomes 4, X, and Y, was detected in the MSCs of 2 AML and 3 MDS patients (Table 2). Trisomy 5 was detected in 2 MSCs of patients with s-AML (N1398) and RAEB II (N0816). These patients were characterized by del(7)(q31) and by normal karyotype in HCs. Loss of X or Y chromosome was detected in MSC cultures of 1 patient with RAEB I (N0789) and normal HC karyotype and in 1 patient with s-AML (N0752) and complex cytogenetic aberrations in HCs. Monosomy 4 was detected in MSCs of a patient with refractory cytopenia with multilineage dysplasia and ringed sideroblasts (N0645) and normal HC karyotype. FISH with appropriate probes for chromosomes 4, 5, X, and Y confirmed these findings.
Karyotype aberrations in MSCs were detected in cases with normal (7 patients) and aberrant HC karyotypes (8 patients). To determine whether the MSCs from these patients were devoid of chromosomal aberrations identified in HCs, we performed FISH and RT-PCR on 40 MSC cultures. Monosomy or deletions of chromosomes 5 and 7 were detected in the HCs of 7 and 10 patients, respectively. In the MSCs of these patients, 2 signals for chromosomes 5 and 7 were detected (Figure 3). Deletions, trisomy, or monosomies 8, 9, 11, 20, 21, and X were revealed in HCs of 14 patients. We detected 2 signals for the corresponding chromosomes in MSCs of these patients by FISH. Translocations t(15;17), t(8;21), t(4;11), t(9;11), and inv(16) were detected in HCs of 9 AML patients. We used FISH and RT-PCR to identify these aberrations in HCs and MSCs. None of the MSC samples showed split signals for the corresponding translocations (Figure 2).
Characteristics of patients with MSC chromosomal aberrations
The main characteristics of the patients who displayed clonal MSC aberrations are presented in Table 3. The age of the patients ranged from 26-83 years (median, 59 years). Of the 15 patients with cytogenetic MSC abnormalities, 7 were less than 65 years of age. The required number of passages and culture time were similar for patient groups with aberrant or normal MSC karyotypes and healthy individuals (control group). No statistical difference was observed between the 3 groups in terms of age or sex. Of 10 AML patients, 5 had s-AML. Eight of the 15 patients (53%) with abnormal MSC karyotype and 33 of 79 (42%) with normal MSC karyotype had aberrant HC karyotypes. Adverse prognostic cytogenetic abnormalities according to the Cancer and Leukemia Group B (CALGB) cooperative group2 were more frequently identified in the HCs of patients with aberrant MSCs (P < .05). All patients with favorable cytogenetic markers, such as translocation t(8;21) inv(16)/t(16;16), and t(15;17), in HCs had no cytogenetic aberrations in MSCs.
Comparative characteristics of patients with cytogenetic aberrations in MSCs, patients with a normal MSC karyotype, and a control group of healthy donors
| . | MSCs with aberrations (n = 15) . | MSCs with normal karyotype (n = 79) . | Healthy donors (n = 36) . |
|---|---|---|---|
| Median age, y (range) | 59 (26-83) | 62 (19-84) | 48 (21-86) |
| Sex, male/female ratio | 10/5 | 59/20 | 17/19 |
| MSC culture time, median (range) | 30 (23-49) | 34 (18-76) | 33 (22-57) |
| Number of MSC passages, median (range) | 4 (3-5) | 4 (3-8) | 4 (3-8) |
| s-AML, n (%) | 5 (50%)* | 14 (34%)* | |
| Cytogenetic aberrations in HCs, n (%) | 8 (53%)* | 33 (42%)* | |
| Unfavorable prognostic genetic and cytogenetic aberrations in HCs, n (%) | 8 (53%)† | 15 (19%)† | |
| Overall mortality, n (%) | 9 (60%)† | 23 (29%)† | |
| Leukemia-related mortality, n (%) | 4 (44%)† | 6 (26%)† |
| . | MSCs with aberrations (n = 15) . | MSCs with normal karyotype (n = 79) . | Healthy donors (n = 36) . |
|---|---|---|---|
| Median age, y (range) | 59 (26-83) | 62 (19-84) | 48 (21-86) |
| Sex, male/female ratio | 10/5 | 59/20 | 17/19 |
| MSC culture time, median (range) | 30 (23-49) | 34 (18-76) | 33 (22-57) |
| Number of MSC passages, median (range) | 4 (3-5) | 4 (3-8) | 4 (3-8) |
| s-AML, n (%) | 5 (50%)* | 14 (34%)* | |
| Cytogenetic aberrations in HCs, n (%) | 8 (53%)* | 33 (42%)* | |
| Unfavorable prognostic genetic and cytogenetic aberrations in HCs, n (%) | 8 (53%)† | 15 (19%)† | |
| Overall mortality, n (%) | 9 (60%)† | 23 (29%)† | |
| Leukemia-related mortality, n (%) | 4 (44%)† | 6 (26%)† |
Statistical analysis was performed using Student t test and unpaired 2-sided exact Fisher test. Unfavorable prognostic genetic and cytogenetic aberrations are complex aberrations (3 or more aberrations pro metaphase), monosomy 5 or 7, del(5q) in AML patients, t(11;19)(q11;p13.1), and FLT3 mutation.2 Other abbreviations are explained in Table 1.
Not significant.
P < .05.
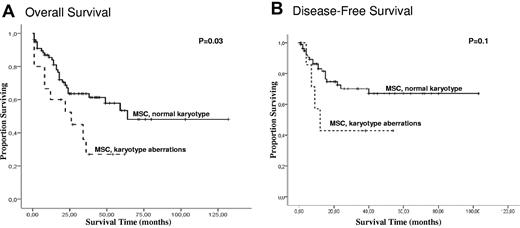
The median follow-up time among patients with and without chromosomal aberrations in MSC was 19 and 33 months, respectively. No statistical differences were observed with regard to complete remission or relapse rates between patients with and without aberrations in MSCs (data not shown). Four (40%) AML patients with aberrations in MSCs and 9 (22%) with a normal MSC karyotype demonstrated primary resistance to standard therapy. Of 5 MDS patients with aberrations in MSCs, 4 had stable disease without progression to AML. One MDS patient died during progression to AML. Of 15 AML/MDS patients with aberrations in MSCs, 9 (60%) died (Table 3). Overall mortality and leukemia-related mortality were more frequently identified in patients with aberrations in MSCs (P = .009 and P = .001, respectively). As shown in Figure 4, the presence of chromosomal aberrations in MSCs was associated with inferior OS and DFS. The median OS was 23 months in patients with aberrations in MSCs (95% confidence interval [95% CI]: 16-40 and 54-87 months) and 32 months in patients with normal MSC karyotype (95% CI: 61-92 and 15-38 months; P = .03). No significant difference in DFS was observed between these groups (95% CI: 10-44 and 77-66 months in patients with aberrations in MSCs versus 95% CI: 62-84 and 52-41 months in patients with a normal MSC karyotype; P = .1). We next investigated whether the stromal cytogenetic aberrations were linked to the HC karyotype. Multivariate analysis was performed to investigate whether MSC abnormalities represented an independent prognostic factor. We included several known risk factors in the model (age, s-AML, FAB, cytogenetics in HCs) and MSCs aberrations. Interestingly, there was a direct relationship: in 37 cases with favorable and intermediate cytogenetics in HCs, only 1 patient with stromal aberrations was observed; conversely, all 8 patients with unfavorable cytogenetics displayed stromal aberrations. Within the largest subgroup of 49 patients with normal karyotype in HCs, stromal aberrations were observed in only 6 patients, and no prognostic impact for OS within this subgroup was seen. As expected from this observed link between HC and MSC aberrations, the stromal aberrations were not an independent prognostic factor for OS in Cox multivariate regression analysis when the HC karyotype was also considered as variable.
Survival analysis for 94 patients according to the cytogenetic data in MSCs. Kaplan-Meier curves for OS (A) and DFS (B).
Survival analysis for 94 patients according to the cytogenetic data in MSCs. Kaplan-Meier curves for OS (A) and DFS (B).
Mutation analysis in HCs and MSCs of AML patients
Different types of mutations were detected in 15 patients. An ITD or a TKD mutation of the FLT3 gene was identified in 12 and 4 cases, respectively (29% and 10%), and NPM1 mutation types A, B, and D were identified in 8 cases (19%). Two of 12 FLT3-ITD-positive and 2 of 8 NPM1-positive patients showed chromosomal aberrations in MSCs. To determine whether the MSCs of AML patients could harbor FLT3 and NPM1 mutations, MSCs of all AML patients were investigated by PCR and DNA sequencing. We detected no mutations in MSCs of FLT3/NPM1–positive or FLT3/NPM1–negative AML patients (Figure 5).
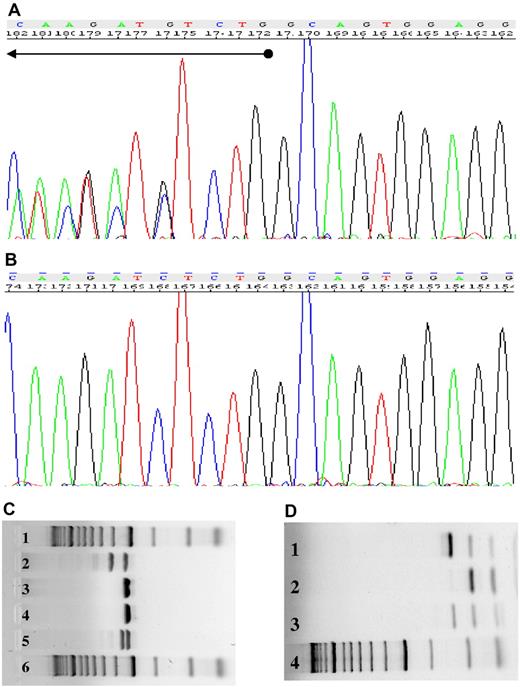
Absence of FLT3 and NPM1 mutations in MSCs. (A) The DNA sequence derived from the HCs of patient 1093 demonstrates mutation of the NPM1 gene. (B) MSCs of patient 1093 display the wild-type NPM1 gene. (C) The FLT3-ITD mutation was detected in the HCs of patient 0757 (lane 2), but not in the MSCs (lane 3). Lane 4 displays the negative control, lane 5 the positive control, and lanes 1 and 6 the size standards. (D) The FLT3-D835 mutation was observed in the HCs of patient 0281 (lane 1), but not in the MSCs (lane 2). Lanes 3 and 4 present the positive control and size standard, respectively.
Absence of FLT3 and NPM1 mutations in MSCs. (A) The DNA sequence derived from the HCs of patient 1093 demonstrates mutation of the NPM1 gene. (B) MSCs of patient 1093 display the wild-type NPM1 gene. (C) The FLT3-ITD mutation was detected in the HCs of patient 0757 (lane 2), but not in the MSCs (lane 3). Lane 4 displays the negative control, lane 5 the positive control, and lanes 1 and 6 the size standards. (D) The FLT3-D835 mutation was observed in the HCs of patient 0281 (lane 1), but not in the MSCs (lane 2). Lanes 3 and 4 present the positive control and size standard, respectively.
Cytogenetic analysis of MSCs of healthy donors
We performed cytogenetic analysis of MSCs of 36 healthy volunteers (median age, 48 years; range, 21-86 years). The median number of passages was 4 (range, 3-8). The culture time ranged from 22 to 57 days. We analyzed 30 mitoses in all cases. As anticipated, chromosome analysis of MSCs of healthy donors displayed normal diploid karyotype, with no aneuploidy, polyploidy, or structural abnormalities.
Discussion
During the last few years, a great deal of interest has been generated about MSCs. Although the supposition that the BM microenvironment in leukemia is the key determinant of malignant progression is now well accepted, it is unclear whether this role of the BM microenvironment is supported by genomic alterations of MSCs that can coevolve during leukemogenesis. The development of MDS and AML is a complex process, and a multistep model has been proposed.1,13 In this model, an abnormal hematopoietic clone would interact with an altered BM microenvironment. Genetically altered stromal cells would undergo clonal selection for MSCs that can modify leukemic cell–stromal cell interactions and promote leukemic growth. In a mouse model, selection of genetic changes in the stroma have been found to occur in MDS, with occasional transformation to AML.19 In fact, several studies have reported important quantitative and functional alterations in the MSCs of leukemia patients.14-20 Some independent studies, including one from our own group, have documented the existence of genomic alterations in the stroma of leukemia patients.21-25 Different studies have shown the extensive variability of the aberrations, such as hypodiploidy, balanced and unbalanced translocations, and whole chromosome gains and deletions.21-25 To gain more insight into this field, we focused on the cytogenetic characterization of MSCs and HCs of 94 AML and MDS patients in the present study. With this aim, we performed conventional cytogenetic, FISH, RT-PCR, and DNA sequencing to explore genomic alterations in MSCs of a large number of patients, and compared these with aberrations in leukemic blasts. As a control, we analyzed the MSCs of 36 healthy donors. The chromosome analysis of HCs revealed clonal abnormalities in 41 of 94 patients. This finding is in agreement with previous reports indicating that a significant number of MDS and AML patients harbor cytogenetic alterations.2-7 Chromosomal analysis of MSCs revealed karyotype abnormalities in 15 of 94 (16%) patients. To confirm the aberrations in MSCs, FISH was performed using the appropriate probes (Figures 1,Figure 2–3). We observed that MSC chromosomal aberrations were detected more often in AML patients than in MDS patients (10 and 5 cases, respectively). In contrast to previous studies, we included in the analysis only clonal aberrations that were detected in at least 2 (structural aberrations or trisomy) or 3 (monosomy) metaphase cells according to the ISCN recommendations.29 In 5 MSC cultures, chromosome aberrations were detected in a small number of analyzed metaphases (2-5 cells). However, in 10 patients, cytogenetically abnormal MSC clones were more frequent and were involved in 6-23 aberrant metaphases. We detected structural aberrations (translocation and inversion) in 4 patients, partial deletions in 6 MSC cultures, and numerical aberrations in 5 patients. Structural aberrations were detected more often in the MSCs of AML patients (Table 2). In AML patients with aberrations in MSCs, translocations and partial deletions were detected in 8 of 10 MSC cultures. On the contrary, in the MDS group with MSC aberrations, gain or loss of whole chromosomes was detected in 3 of 5 MSC cultures. Chromosomes 1 and 7 were more frequently involved in MSC structural aberrations. In our previous study,21 we also observed MSC aberrations frequently involving chromosomes 1 and 7. Two MSC cultures from AML and MDS patients displayed clonal trisomy 5. Klaus et al demonstrated gain of chromosome 5 in MSC cultures from 3 MDS patients.24 In contrast to previous studies, we demonstrated a significant number of clonal structural aberrations (ie, not only numerical aberrations) in MSCs of AML/MDS patients. It should be emphasized that genomic aberrations in MSCs were identified in patients with normal HCs (n = 7) and cytogenetically aberrant HCs (n = 8). No cytogenetic markers in MSCs repeated aberrations identified in HCs. Because there were no associations between chromosomal aberrations in HCs and MSCs, we can state that MSCs were devoid of residue HCs. In 8 patients, independent cytogenetic clones were identified in HCs and MSCs. We performed FISH or RT-PCR on all cells to confirm different cytogenetic markers in leukemic blasts and MSCs. Furthermore, we demonstrated that no AML patients with FLT3 and/or NPM1 mutations in HCs displayed mutations in MSCs.
The loss of one sex chromosome was identified in 2 MSC cultures obtained from a 65-year-old and a 66-year-old patient. It has been suggested that loss of sex chromosomes increases with age.33 In our study, 49 patients (52%) were younger and 45 patients (48%) were older than 65 years. Cytogenetic aberrations in MSCs were seen in 7 patients from the first group and in 8 patients from the second group. Furthermore, all MSC cultures of healthy individuals were cytogenetically normal. The age of the donors included in our study ranged from 21-86 years. These observations are in agreement with data from other investigators,21-23,34,35 who reported normal diploid karyotype without aneuploidy or polyploidy in MSCs from healthy donors or patients with no hematopoietic disorders.
Because we detected cytogenetic aberrations in the MSCs of 10 AML and 5 MDS patients, we compared some characteristics of patients with MSC cytogenetic aberrations (Table 3). Chromosomal markers in MSCs were more often associated with adverse prognostic chromosomal markers in HCs. Moreover, we detected no chromosomal aberrations in MSCs from patients with favorable cytogenetics in HCs. The incidence of s-AML tended to be higher among AML patients with a abnormal MSC karyotype.
We found a correlation between MSC aberration and OS in our cohort of patients. There are several potential explanations for this observation. It cannot be excluded that the inferior outcome that we have mentioned for patients with MSC aberrations is associated with not only MSCs but also with HC cytogenetic abnormalities. Because we detected cytogenetic aberrations in MSCs almost exclusively in several patients with normal chromosomal markers in HCs and in all patients with unfavorable chromosomal markers, the relation of MSCs to OS expectedly was not independent, but rather was directly associated with HC cytogenetic abnormalities. Within the group of patients with normal karyotype, MSC aberrations were not prognostic for OS; however, this subgroup analysis would have to be confirmed in a larger cohort of patients. Previously published data have documented functional and genetic aberrations in MSCs,21-25 but there are no published data regarding the clinical or prognostic characteristics of patients displaying MSC genetic markers. In the present study, we did not investigate the functional effects of chromosomal abnormalities in MSCs or their effects on HCs. Future studies are needed to assess the functional integrity of leukemia-derived MSCs and, more importantly, the interaction between abnormal HCs and MSCs, which may be crucial to disease biology. Although hematologic malignancies are believed to arise from a stem or progenitor cell abnormality, a primary MSC defect may also lead to or support a hematologic malignancy.34 Until recently, there was little evidence on the role of primary stromal abnormalities in the pathogenesis of hematologic neoplasms. Based on published studies in mouse models, the microenvironment has now been shown to induce malignancy.19,20,36 Because the microenvironment supports MDS and AML clones as a result of reciprocal interactions, abnormalities in MSCs worsen those intrinsic to the neoplastic cells.1
Our results illustrate that chromosomal abnormalities in MSCs were detected in a subset of MDS and AML patients and were mostly associated with unfavorable cytogenetic abnormalities in HCs. The MSC cytogenetic aberrations differed from the HC chromosomal markers of the same subjects. Furthermore, the MSCs of AML patients with FLT3 and/or NPM1 mutations were devoid of these mutations. In both MDS and AML patients, the genetic defect in leukemic blasts developed in cells equipped with self-renewal potential and unique stem cell properties. The detection of genetic alterations in MSCs suggests that unstable MSCs may facilitate the expansion of malignant cells. In view of these data, genetic alterations in MSCs may be a particular mechanism of leukemogenesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: O. Blau designed research, analyzed the data, and wrote the manuscript; C.D.B. designed the research and critically reviewed the manuscript; W.-K.H., S.M., and E.T designed the research; G.T., F.N., T.B., S.T., O. Benlasfer, E.S., A.S, and M.M. analyzed the data; U.K. provided excellent help with the statistical analyses and critically reviewed the manuscript; and I.W.B. designed the research, contributed intellectually, and critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Olga Blau, MD, PhD, Department of Hematology and Oncology, Charité University School of Medicine, Hindenburgdamm 30, D-12200 Berlin, Germany; e-mail: olga.blau@charite.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal