Abstract
Abstract 1464
Aberrant DNA methylation has been shown as an important mechanism in the progression from myelodysplasia (MDS) to acute myeloid leukemia (AML), leading to use of the demethylating agents, 5-azacitidine and decitabine, for treatment of both disorders. While these drugs produce responses, the ability to distinguish potential responders from potential non-responders remains limited. The purpose of this work was to bring new technology to study this problem. Recent studies demonstrated that promoter DNA methylation is not randomly distributed in AML blasts but rather is highly organized and associated with biologically distinct AML subtypes. Because cytogenetics at presentation is the most important prognostic factors in predicting response to therapy, remission duration and overall survival in AML, we aimed to identify differentially methylated genomic regions (DMR) in cytogenetically defined risk groups of AML. Published literature suggested that the new comprehensive high-throughput array-based relative methylation analysis (CHARM) has the highest sensitivity and specificity among all the array-based genome profiling methods and should be the most accurate means to identify methylation markers. It is a customized NimbleGen HD2 array of tiled 50mer-probes typically separated by 30–40 bases covering approximately 4.6 million CpG sites across the genome. This assay is highly quantitative for approximately 100,000 independent CpG sites.
| . | Number of DMRs . | Number with p-value < 0.01 . |
|---|---|---|
| Normal blood VS at risk | 3768 | 311 |
| Low-risk VS mid-risk | 1565 | 30 |
| Low-risk VS high-risk | 2475 | 73 |
| Mid-risk VS high-risk | 2651 | 107 |
| . | Number of DMRs . | Number with p-value < 0.01 . |
|---|---|---|
| Normal blood VS at risk | 3768 | 311 |
| Low-risk VS mid-risk | 1565 | 30 |
| Low-risk VS high-risk | 2475 | 73 |
| Mid-risk VS high-risk | 2651 | 107 |
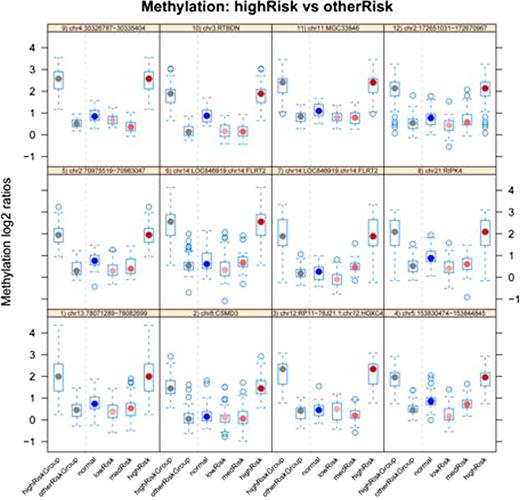
Results from the CHARM analysis on AML patients stratified by cytogenetic risk, highlighting comparison between the high-risk and other risk groups. The top twelve differentially methylated regions ( DMRs) with p -value < 0.01 are shown. Box-and-whisker plots to the left of the dashed vertical line in each panel present the log2 methylation ratio of the high-risk group and the other two risk groups combined, in a single region of differential methylation. To the right of the dashed line the high-risk group is compared with each of the other groups. Each panel's header text identifies the genomic region.
Results from the CHARM analysis on AML patients stratified by cytogenetic risk, highlighting comparison between the high-risk and other risk groups. The top twelve differentially methylated regions ( DMRs) with p -value < 0.01 are shown. Box-and-whisker plots to the left of the dashed vertical line in each panel present the log2 methylation ratio of the high-risk group and the other two risk groups combined, in a single region of differential methylation. To the right of the dashed line the high-risk group is compared with each of the other groups. Each panel's header text identifies the genomic region.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal