Abstract
Abstract 1468
INTRODUCTION. Many molecular biomarkers have proven to be useful for guiding therapy for AML patients, yet remain inadequate for accurately predicting clinical outcomes. Possible explanations may be that certain biomarkers are more informative for restricted subpopulations of AML patients, or that the results of biomarkers must be interpreted in combination. Another explanation may be that there are methodological limitations in the assays. Most AML biomarkers are currently examined in MNCs, which include non-leukemic cells (lymphocytes, monocytes, and erythroblasts) and leukemic blasts. In addition, cells within the AML blast population display a wide spectrum of differentiation stages. To evaluate the potential impact of the non-leukemic cells in AML samples on the expression of prognostic transcriptional biomarkers, we examined enriched populations of cells from diagnostic AML samples.
METHODS. Cryopreserved bone marrow (BM) and peripheral blood (PB) MNCs were obtained from 12 AML patients from the Leukemia Repository at the FHCRC. The cells were thawed and an aliquot was saved for future studies. Remaining cells were sorted for viable AML blasts using a combination of fluorochrome-conjugated antibodies (CD45, CD34, CD38, HLA-DR, and/or CD117) and DAPI. Additional sorts further differentiated the AML blasts into more or less mature (N=4 patients). RNA was extracted using Qiagen AllPrep RNA/DNA kit and reverse transcribed using AMV RT (Invitrogen). Expression of 22 potential prognostic biomarkers was evaluated using commercially available assays (Applied Biosystems) on ABI HT 7900 Fast Real-time PCR system. Fold differences in expression were computed by the comparative CT method, using GUSB to correct for RNA integrity and a pool of RNA from 10 PB from normal donors as a calibrator.
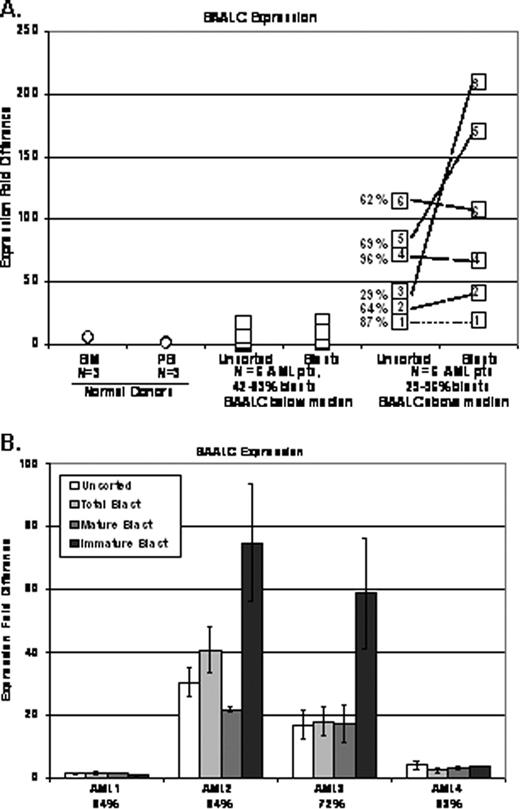
RESULTS. Analyses showed marked differences between expression in the unsorted MNCs and AML blasts. Transcript expression was generally the same or more pronounced in AML blasts than in MNCs (e.g., BAALC, Figure 1A). In addition, any transcript differences identified in two populations generally diminished and were less dramatic in AML samples with high blast percentages. However, this was not the case for all samples and/or biomarkers. For example, CEBPA showed similar expression levels in MNCs and AML blasts. Attempts to adjust the signal in MNCs by blast percentages typically did not yield similar transcriptional levels for many samples.
Similarly, the expression of examined prognostic biomarkers in subpopulations of AML blasts (mature vs. immature) showed a degree of heterogeneity among the patients. For some biomarkers, the immature population displayed a more pronounced fold difference in expression (e.g. BAALC, Figure1B), while for others, the expression was very similar in examined populations of most patients (e.g. CCNA1).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal