Abstract
Abstract 430
25–30% of patients with advanced stage classical Hodgkin lymphoma (cHL) develop progressive disease despite treatment with ABVD. While more intensive upfront treatments, such as BEACOPP, result in a higher failure free survival, this comes at the expense of increased toxicity and morbidity. 10–15% of patients with advanced stage cHL will ultimately succumb to the disease, with similar outcomes using different upfront regimens likely reflecting the planned use of second-line high dose therapy. The International Prognostic Factors Project Score, a prognostic tool developed using freedom from progression of disease as the outcome, has been used in clinical practice and has guided clinical trials. We hypothesized that biological factors, as measured by gene expression profiling in pretreatment formalin fixed paraffin embedded tissue (FFPET) samples, might provide a robust predictor of overall survival (OS).
NanoString technology was used to quantitate 261 mRNA species in diagnostic FFPET samples from patients in the Intergroup E2496 trial, which compares ABVD with Stanford V chemotherapy in locally advanced and advanced stage cHL. The 261 genes included individual genes and genes that represent cell and cellular process signatures that have been associated with outcome in cHL. Total RNA was extracted from whole section scrolls from the 309 FFPET blocks available. A penalized binary logistic regression model with leave-one-out cross validation was used to produce a parsimonious predictive model for OS. A threshold was selected that maximized the Chi square of the Log Rank (Mantel-Cox) test between the identified low and high risk groups. An independent cohort of patients with advanced stage cHL enriched for treatment failure, consisting of 59 patients from British Columbia treated with ABVD and 71 patients from Portugal treated with Stanford V, was used to test the generated model and threshold.
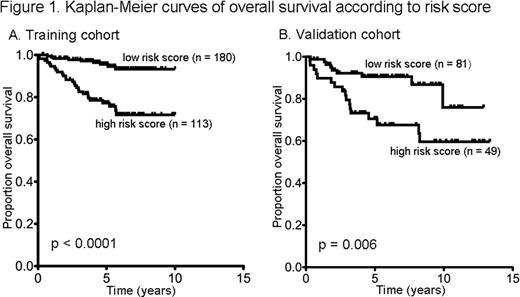
Gene expression of adequate quality was produced in 293 of the 309 samples (95%). At a median follow up of 5.3 years, 36 (12%) of the 293 patients had died. 51 genes were differentially expressed (t-test p < 0.05) with respect to OS, including genes that are part of macrophage, cytotoxic/NK cell and apoptosis signatures. A predictive model for OS was produced with an area under the curve (AUC) of the receiver operating characteristic (ROC) curve of 0.73. The predictor identified a high-risk group, comprising 39% of the cohort, with a 77% estimated 5-year OS compared with 96% in the low risk group (Figure 1A, log-rank p < 0.0001). The predictor was validated in the independent cohort giving an AUC of the ROC curve of 0.73 and an estimated 5-year OS of 71% in the high-risk group compared with 91% in the low risk group (Figure 1B, log-rank p = 0.006).
We have produced a parsimonious gene expression-based predictor of OS in cHL, developed in, and applicable to, widely available FFPET using NanoString technology. The predictor identifies, at diagnosis, a clinically meaningful proportion of patients at significantly higher risk of death when treated with ABVD or Stanford V. Further evaluation of the gene expression signature with alternative treatments, ideally in the context of clinical trials, is needed to understand the impact of more intensive cytotoxic therapies versus novel targeted approaches.
Horning:Genentech: Employment, Equity Ownership. Connors:Seattle Genetics: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal