Abstract
Familial hemophagocytic lymphohistiocytosis (FHL) is an autosomal recessive, often-fatal hyperinflammatory disorder. Mutations in PRF1, UNC13D, STX11, and STXBP2 are causative of FHL2, 3, 4, and 5, respectively. In a majority of suspected FHL patients from Northern Europe, sequencing of exons and splice sites of such genes required for lymphocyte cytotoxicity revealed no or only monoallelic UNC13D mutations. Here, in 21 patients, we describe 2 pathogenic, noncoding aberrations of UNC13D. The first is a point mutation localized in an evolutionarily conserved region of intron 1. This mutation selectively impairs UNC13D transcription in lymphocytes, abolishing Munc13-4 expression. The second is a 253-kb inversion straddling UNC13D, affecting the 3′-end of the transcript and likewise abolishing Munc13-4 expression. Carriership of the intron 1 mutation was found in patients across Europe, whereas carriership of the inversion was limited to Northern Europe. Notably, the latter aberration represents the first description of an autosomal recessive human disease caused by an inversion. These findings implicate an intronic sequence in cell-type specific expression of Munc13-4 and signify variations outside exons and splice sites as a common cause of FHL3. Based on these data, we propose a strategy for targeted sequencing of evolutionary conserved noncoding regions for the diagnosis of primary immunodeficiencies.
Introduction
Hemophagocytic lymphohistiocytosis (HLH) is a hyperinflammatory disorder characterized by unremitting fever, hepatosplenomegaly, hyperferritinemia, cytopenia, and sometimes hemophagocytosis.1-4 HLH can be divided into primary and secondary forms.4-6 Without a known familial history or a molecular diagnosis, it is difficult to distinguish between such primary and secondary HLH. For the diagnosis of HLH, at least 5 of the 8 defined criteria need to be fulfilled.7 First-line treatment of HLH is directed at controlling the hyperinflammation caused by aberrant immune activation.7 However, the only cure for primary HLH is hematopoietic stem cell transplantation.8 Thus, for treatment, it is important to obtain a rapid molecular diagnosis.
Familial hemophagocytic lymphohistiocytosis (FHL), which has an autosomal recessive inheritance, has so far been linked to 5 different loci. No gene has yet been associated with FHL1, whereas mutations in PRF1, UNC13D, STX11, and STXBP2 are causative of FHL2, 3, 4, and 5, respectively.9-13 These genes all encode proteins required for lymphocyte cytotoxicity,14-16 and defective NK cell cytotoxicity is one of the 8 diagnostic criteria for HLH.7 In addition, mutations in UNC13D and STXBP2, encoding Munc13-4 and syntaxin binding protein 2, respectively, also impair platelet granule release.17,18 Furthermore, patients with X-linked lymphoproliferative syndrome (XLP) may present with severe Epstein-Barr virus infections, hypogammaglobulinemia, lymphoma, or HLH.19 Mutations in SH2D1A and XIAP are associated with XLP1 and XLP2, respectively.20,21 Although the syndromes share clinical phenotypes, recent reports emphasize differences between the syndromes, such as the frequent development of lymphoma in XLP1 patients in contrast to XLP2 patients.22,23 In addition, Griscelli syndrome type 2 and Chediaki-Higashi syndrome, both with autosomal recessive inheritance and caused by mutations in RAB27A and LYST, respectively, are associated with development of HLH in addition to manifesting partial albinism.24,25
The relative frequency of mutations in various genes causative of FHL differs among ethnicities.26,27 With the advance of knowledge concerning genetic associations with FHL, a molecular diagnosis can be attained for an increasing proportion of patients. In this regard, Côte et al recently reported that only 10% of the FHL cases they have studied are without a molecular diagnosis.13 However, in our and other cohorts of infants from Northern Europe with HLH and defective NK cell cytotoxicity,26 sequencing of exons and exon-intron boundaries in the genes associated with HLH have not provided a molecular diagnosis for many patients.
In this study, we describe 2 different aberrations in UNC13D that together can explain the majority of the FHL cases in Scandinavia as well as several other cases across Europe. These findings improve diagnostics and carrier testing and may also offer the possibility of prenatal testing for a number of families. Moreover, they also define an intronic sequence involved in transcriptional control of Munc13-4 expression in lymphocytes.
Methods
Patients and controls
In this study, Swedish infants presenting with a known familial history of HLH and/or defective cytotoxicity and/or degranulation between December 2005 and January 2011 were included. In addition, the study also encompassed all infants at our unit identified with UNC13D mutations from Sweden, Denmark, Norway, Finland, Slovenia, and Croatia. The studies were approved by the ethics committee at the Karolinska Institutet. Written consent was obtained from all families in accordance with the Declaration of Helsinki.
DNA extraction, amplification, and sequence analysis
Genomic DNA was isolated from peripheral blood or cultured fibroblasts according to standard procedures. Specific primers were designed and used for amplification of selected regions by PCR. The amplified products were subsequently used for direct sequencing according to standard procedures (BigDye Version 3.1; Applied Biosystems). Sequencing reactions were analyzed by capillary electrophoresis (ABI 3730 Genetic Analyzer; Applied Biosystems). Data were analyzed using SeqScape software (Version 2.5; Applied Biosystems) and a UNC13D reference sequence template (NCBI Accession NM_199242.2). Primers, PCR conditions, and sequencing reaction conditions are available on request.
RNA extraction, cDNA synthesis, amplification, and analyses
Total RNA was extracted from WBCs (RNeasy, QIAGEN), and cDNA was synthesized with oligo(dT)20 primed reverse transcription (SuperScript III, Invitrogen) according to the manufacturer's protocol. For evaluation of UNC13D splicing, 9 primer pairs were used for amplification of overlapping cDNA fragments. For 3′-rapid amplification of cDNA ends PCR, cDNA was synthesized using oligo(dT)20 primers marked with an M13R-tag. A gene specific forward primer and a M13R reverse primer were used for amplification of the 3′-end of the UNC13D transcript. Amplified products were separated by agarose gel electrophoresis, extracted (QIAquick gel extraction kit; QIAGEN), and cloned (TOPO-TA cloning kit, Invitrogen). Plasmid DNA was isolated (GeneJET plasmid miniprep kit, Fermentas) and sequenced.
Allele-specific quantitative RT-PCR of isolated cell populations
PBMCs were isolated by density gradient centrifugation. Specific cell populations were consecutively isolated by magnetic positive selection using anti–CD14, anti–CD4, anti–CD8, and anti–CD56 mAb-coated beads, respectively (Miltenyi Biotec). Purity of the isolated cell populations was assessed by flow cytometry using fluorochrome-conjugated mAbs against CD3 (clone UHCT1; eBioscience), CD4 (clone 53.5; Invitrogen), CD8 (clone 3B5; Invitrogen), CD14 (clone MϕP9; BD Bioscience), CD45 (clone 2D1; BD Bioscience), and CD56 (clone NCAM16.2; BD Bioscience). RNA was isolated (TRIzol; Invitrogen) and cDNA synthesized as described. For quantitative real-time PCR quantification of allele-specific transcription, 2 different reverse primers that could discriminate a single nucleotide polymorphism, c.888G > C located in exon 11, were designed. The specificity of these primers was evaluated by agarose gel electrophoresis and by examining real-time PCR dissociations curves. Relative quantification was assessed using the standard curve method. All samples were run in triplicates. For each reaction, 100 ng cDNA was mixed with 10 μL of Power SYBR Green PCR master mix (Applied Biosystems) and 10 pmol of forward and reverse primers in a total volume of 20 μL. The reactions were analyzed by real-time PCR (ABI 7900 HT; Applied Biosystems) and analyzed with SDS software (Version 2.2.1; Applied Biosystems).
Western blot analysis of Munc13-4 expression
PBMCs from patients and healthy controls were lysed in lysis buffer (20mM Tris, pH 7.4, 1mM EDTA, 1% Triton X-100, 150mM NaCl) with protease inhibitors (Roche Diagnostics). Cells were disrupted and centrifuged for 15 minutes at 14 000g. Protein content was determined by the Bradford assay (Bio-Rad), and 50 μg protein was loaded per lane and analyzed using SDS-PAGE separation and Western blotting (NuPAGE; Invitrogen). The rabbit polyclonal antibodies to Munc13-4 (Protein Technologies Group, raised against amino acids 1-236) and ERK1 (Santa Cruz Biotechnology) were used for Western blotting.
Analysis of cytotoxic lymphocyte function
For assessment of NK cell-mediated cytotoxicity, a standard 4-hour 51Cr assay was used.28 PBMCs were isolated by density gradient centrifugation and used as effector cells either after incubation overnight in complete medium (RPMI 1640 supplemented with 10% FBS, l-glutamine, penicillin, and streptomycin, all from Invitrogen) or after activation with IL-2 (Proleukin; Novartis) for 36 to 60 hours. 51Cr-labeled K562 target cells (ATCC) were used as target cells. Lymphocyte cytotoxicity was calculated as lytic units (LU) at 25% lysis. A value < 10 LU was considered pathologic.7 NK cell degranulation was assessed by flow cytometric quantification of induction of CD107a surface expression on CD3−CD56+ gated cells in response to incubation with K562 target cells, as previously described.29
Genotyping of microsatellite markers
PCR was performed according to standard procedures with fluorescently labeled primers. PCR products were separated by electrophoresis on an ABI 3730 DNA Analyzer (Applied Biosystems) with GeneScan 400HD Rox (Applied Biosystems) as size standard and analyzed with Peak Scanner Version 1.0 software (Applied Biosystems).
Results
HLH patients
Between December 2005 and January 2011, 13 infants from Sweden (age, 0-3 years) came to our attention who fulfilled the current criteria for HLH, specifically including defective NK cell cytotoxicity.7 Within this group, one infant (of Afghani origin) carried a novel, homozygous PRF1 mutation, c.658G > C (p.Gly220Arg), whereas another infant (of Somali origin) carried a novel homozygous UNC13D mutation, c.2381delT (p.Leu794ArgfsX2). A pair of twins carried heterozygous compound mutations in RAB27A, and another infant carried a heterozygous compound mutation in PRF1, all with mutations that have been described.28,30 Of the remaining 8 patients, 2 infants had monoallelic mutations in UNC13D that have been described.26,31 Hence, the majority (62%) of these infants from Sweden were without a molecular diagnosis after sequencing of the coding region and splice sites of genes associated with FHL. Through sequencing other infants referred to us from Europe with HLH and defective NK cell cytotoxicity, we identified an additional 5 infants from 4 families with monoallelic UNC13D mutations (Table 1). Two brothers carried a novel missense mutation, c.2642T > C, p.Leu881Pro predicted to be pathogenic using 2 different protein prediction programs (PolyPhen, SIFT32,33 ), whereas the other mutations are previously described.10,26,31 Notably, a German study also reported 6 infants who developed HLH in which monoallelic UNC13D mutations were identified.26 This prompted us to search for mutations in UNC13D outside of exons and splice sites.
Clinical and laboratory findings in patients with the UNC13D intron 1 mutation, c.118-308C > T, and/or the 253-kb inversion
| . | Patient A . | Patient B:1 . | Patient B:2 . | Patient C . | Patient D . | Patient E . | Patient F . | Patient G . | Patient H . | Patient I . | . | Patient J:1 . | Patient J:2 . | Patient K . | Patient L . | Patient M . | Patient N . | Patient O:1 . | Patient O:2 . | Patient P . | Patient Q . | Patient R . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethnical origin | Croatia | Slovenia | Slovenia | Sweden | Denmark/Ukraine | Croatia | Sweden | Denmark | Sweden | Finland | Ethnical origin | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden/USA | Sweden | Norway |
| Familial disease | No | Yes | Yes | No | No | No | No | No | No | Familial disease | Yes | Yes | No | Yes | No | No | Yes | Yes | No | No | No | |
| Parental consanguinity | No | No | No | No | No | No | No | No | No | No | Parental consanguinity | No | No | No | No | No | No | No | No | No | ||
| Sex | Male | Male | Male | Male | Male | Female | Male | Female | Female | Male | Sex | Female | Female | Female | Female | Male | Female | Male | Female | Female | Female | Male |
| Allele 1 | 753 + 1G, splice error | 2642T > C, Leu881Pro | 2642T > C, Leu881Pro | 532delC, Gln178ArgfsX71 | 2346_2349delGGAG, Arg782SerfsX12 | 118-308C > T | Inversion | Inversion | Inversion | Inversion | Allele 1 | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | ||
| Allele 2 | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 2346_2349delGGAG, Arg782SerfsX12 | 2176C > T, Gln726X | Allele 2 | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | ||
| Age at diagnosis of HLH, days | 55 | 195 | 229 | 57 | 150 | 133 | 87 | 198 | 104 | 148 | Age at diagnosis of HLH, days | 365 | 1 | 79 | 459 | 104 | 127 | 56 | 134 | 73 | 3 | 388 |
| Fever | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Fever | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Splenomegaly | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Splenomegaly | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Hepatomegaly | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Hepatomegaly | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Hb, g/L | 100 | 115 | 105 | 70 | 61 | 81 | 60 | 95 | 46 | 97 | Hb, g/L | 83 | 138 | 62 | 100 | 87 | 100 | 58 | 65 | 82 | 61 | 62 |
| Neutrophils, × 109/L | 0.3 | 0.8 | 1.0 | 1.0 | 0.4 | 0.0 | 0.2 | 0.6 | 0.4 | 0.9 | Neutrophils, × 109/L | 2.9 | 2.9 | 1.4 | 0.8 | 0.7 | 0.4 | 0.1 | 0.7 | 0.2 | 0.3 | 0.8 |
| Platelets, × 109/L | 28 | 143 | 34 | 13 | 22 | 3 | 31 | 72 | 17 | 59 | Platelets, × 109/L | 38 | 15 | 32 | 15 | 12 | 37 | 7 | 76 | 18 | 10 | 33 |
| Triglycerides, mM | 2.8 | 12 | 4.5 | 2.3 | 7.3 | 3.9 | 2.4 | 6.1 | 4.2 | 5.4 | Triglycerides, mM | 3.0 | 2.7 | 11 | 2.6 | 3.4 | 2.7 | 3.4 | 17.3 | 5.5 | 1.9 | 4.4 |
| Fibrinogen, g/L | 0.9 | 1.5 | 4.2 | 2.1 | 0.9 | 0.6 | 1.2 | 2.3 | 0.6 | Fibrinogen, g/L | 0.5 | 0.6 | 1.2 | 2.4 | 0.6 | 0.7 | < 0.5 | 1.7 | 0.8 | 0.7 | 2.9 | |
| Hemophagocytosis | Yes | Yes | No | No | No | Yes | No | Yes | Yes | Yes | Hemophagocytosis | Yes | No | No | No | No | Yes | ND | No | Yes (CSF) | No | Yes |
| Ferritin, μg/L | 5000 | 6482 | 315 | 2800 | 2399 | 1020 | 1540 | 437 | 4148 | 1858 | Ferritin, μg/L | 2920 | > 15 000 | 5433 | 6597 | 7828 | 2387 | 11 372 | 208 | 8100 | 1147 | 1162 |
| sCD25, U/mL | ND | ND | ND | > 7500 | ND | ND | 20 380 | ND | ND | 30 486 | sCD25, U/mL | 21 976 | ND | 5666 | 14 110 | ND | 11 861 | ND | 8802 | > 7500 | > 7500 | Elevated |
| NK cell activity* | Borderline (11 LU) | ND | Defective | Defective | Defective | Defective | ND | Defective | Borderline (13 LU) | Defective | NK cell activity* | Defective | Defective | Defective | Defective | Defective | Defective | ND | Defective | Defective | Defective | Defective |
| NK cell degranulation | Defective | ND | Defective | Low | Defective | Defective | ND | Defective | Defective | ND | NK cell degranulation | Defective | Defective | Defective | Defective | Defective | ND | ND | ND | Defective | Defective | ND |
| Neurologic manifestations† | Yes | No | No | No | Yes | No | No | No | No | Yes | Neurologic manifestations† | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No |
| Pathologic CSF | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Pathologic CSF | Yes | ND | Yes | Yes | Yes | ND | Yes | Yes | Yes | ND | Yes |
| Treatment active disease | HLH-2004 | HLH-2004 | HSCT | HLH-2004 | HLH-2004 | HLH-2004 | HLH-94 | HLH-2004 | HLH-2004 | HLH-94 | Treatment active disease | HLH-2004 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-94 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-94 |
| Remission at 2 months | Remission, but possible CNS progression | No | NA | No | Remission, but possible CNS progression | Yes | No | No | Yes | No | Remission at 2 months | No | No | No | No | No | No | No | Yes | No | No | Yes |
| Age at HSCT, days | 245 | 191 | 273 | 128 | 297 | 653 | 177 | 373 | 419 | 471 | Age at HSCT | 705 | No | 195 | No | 224 | 371/853 | No | 197 | 150 | 86 | 843 |
| Outcome | Alive | Deceased 226 days | Deceased 289 days | Alive | Deceased 312 days | Alive | Alive | Deceased 460 days | Alive | Alive | Outcome | Alive | Deceased 71 days | Alive | Deceased | Alive | Alive | Deceased 143 days | Alive | Alive | Deceased | Alive |
| . | Patient A . | Patient B:1 . | Patient B:2 . | Patient C . | Patient D . | Patient E . | Patient F . | Patient G . | Patient H . | Patient I . | . | Patient J:1 . | Patient J:2 . | Patient K . | Patient L . | Patient M . | Patient N . | Patient O:1 . | Patient O:2 . | Patient P . | Patient Q . | Patient R . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethnical origin | Croatia | Slovenia | Slovenia | Sweden | Denmark/Ukraine | Croatia | Sweden | Denmark | Sweden | Finland | Ethnical origin | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden | Sweden/USA | Sweden | Norway |
| Familial disease | No | Yes | Yes | No | No | No | No | No | No | Familial disease | Yes | Yes | No | Yes | No | No | Yes | Yes | No | No | No | |
| Parental consanguinity | No | No | No | No | No | No | No | No | No | No | Parental consanguinity | No | No | No | No | No | No | No | No | No | ||
| Sex | Male | Male | Male | Male | Male | Female | Male | Female | Female | Male | Sex | Female | Female | Female | Female | Male | Female | Male | Female | Female | Female | Male |
| Allele 1 | 753 + 1G, splice error | 2642T > C, Leu881Pro | 2642T > C, Leu881Pro | 532delC, Gln178ArgfsX71 | 2346_2349delGGAG, Arg782SerfsX12 | 118-308C > T | Inversion | Inversion | Inversion | Inversion | Allele 1 | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | ||
| Allele 2 | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 118-308C > T | 2346_2349delGGAG, Arg782SerfsX12 | 2176C > T, Gln726X | Allele 2 | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | Inversion | ||
| Age at diagnosis of HLH, days | 55 | 195 | 229 | 57 | 150 | 133 | 87 | 198 | 104 | 148 | Age at diagnosis of HLH, days | 365 | 1 | 79 | 459 | 104 | 127 | 56 | 134 | 73 | 3 | 388 |
| Fever | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Fever | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Splenomegaly | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Splenomegaly | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Hepatomegaly | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Hepatomegaly | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Hb, g/L | 100 | 115 | 105 | 70 | 61 | 81 | 60 | 95 | 46 | 97 | Hb, g/L | 83 | 138 | 62 | 100 | 87 | 100 | 58 | 65 | 82 | 61 | 62 |
| Neutrophils, × 109/L | 0.3 | 0.8 | 1.0 | 1.0 | 0.4 | 0.0 | 0.2 | 0.6 | 0.4 | 0.9 | Neutrophils, × 109/L | 2.9 | 2.9 | 1.4 | 0.8 | 0.7 | 0.4 | 0.1 | 0.7 | 0.2 | 0.3 | 0.8 |
| Platelets, × 109/L | 28 | 143 | 34 | 13 | 22 | 3 | 31 | 72 | 17 | 59 | Platelets, × 109/L | 38 | 15 | 32 | 15 | 12 | 37 | 7 | 76 | 18 | 10 | 33 |
| Triglycerides, mM | 2.8 | 12 | 4.5 | 2.3 | 7.3 | 3.9 | 2.4 | 6.1 | 4.2 | 5.4 | Triglycerides, mM | 3.0 | 2.7 | 11 | 2.6 | 3.4 | 2.7 | 3.4 | 17.3 | 5.5 | 1.9 | 4.4 |
| Fibrinogen, g/L | 0.9 | 1.5 | 4.2 | 2.1 | 0.9 | 0.6 | 1.2 | 2.3 | 0.6 | Fibrinogen, g/L | 0.5 | 0.6 | 1.2 | 2.4 | 0.6 | 0.7 | < 0.5 | 1.7 | 0.8 | 0.7 | 2.9 | |
| Hemophagocytosis | Yes | Yes | No | No | No | Yes | No | Yes | Yes | Yes | Hemophagocytosis | Yes | No | No | No | No | Yes | ND | No | Yes (CSF) | No | Yes |
| Ferritin, μg/L | 5000 | 6482 | 315 | 2800 | 2399 | 1020 | 1540 | 437 | 4148 | 1858 | Ferritin, μg/L | 2920 | > 15 000 | 5433 | 6597 | 7828 | 2387 | 11 372 | 208 | 8100 | 1147 | 1162 |
| sCD25, U/mL | ND | ND | ND | > 7500 | ND | ND | 20 380 | ND | ND | 30 486 | sCD25, U/mL | 21 976 | ND | 5666 | 14 110 | ND | 11 861 | ND | 8802 | > 7500 | > 7500 | Elevated |
| NK cell activity* | Borderline (11 LU) | ND | Defective | Defective | Defective | Defective | ND | Defective | Borderline (13 LU) | Defective | NK cell activity* | Defective | Defective | Defective | Defective | Defective | Defective | ND | Defective | Defective | Defective | Defective |
| NK cell degranulation | Defective | ND | Defective | Low | Defective | Defective | ND | Defective | Defective | ND | NK cell degranulation | Defective | Defective | Defective | Defective | Defective | ND | ND | ND | Defective | Defective | ND |
| Neurologic manifestations† | Yes | No | No | No | Yes | No | No | No | No | Yes | Neurologic manifestations† | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No |
| Pathologic CSF | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Pathologic CSF | Yes | ND | Yes | Yes | Yes | ND | Yes | Yes | Yes | ND | Yes |
| Treatment active disease | HLH-2004 | HLH-2004 | HSCT | HLH-2004 | HLH-2004 | HLH-2004 | HLH-94 | HLH-2004 | HLH-2004 | HLH-94 | Treatment active disease | HLH-2004 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-94 | HLH-2004 | HLH-2004 | HLH-2004 | HLH-94 |
| Remission at 2 months | Remission, but possible CNS progression | No | NA | No | Remission, but possible CNS progression | Yes | No | No | Yes | No | Remission at 2 months | No | No | No | No | No | No | No | Yes | No | No | Yes |
| Age at HSCT, days | 245 | 191 | 273 | 128 | 297 | 653 | 177 | 373 | 419 | 471 | Age at HSCT | 705 | No | 195 | No | 224 | 371/853 | No | 197 | 150 | 86 | 843 |
| Outcome | Alive | Deceased 226 days | Deceased 289 days | Alive | Deceased 312 days | Alive | Alive | Deceased 460 days | Alive | Alive | Outcome | Alive | Deceased 71 days | Alive | Deceased | Alive | Alive | Deceased 143 days | Alive | Alive | Deceased | Alive |
Patient B:1 and B:2 carried a novel missense mutation, c.2642T > C, p.Leu881Pro predicted to be pathogenic using 2 different protein prediction programs (PolyPhen, SIFT30,31 ), whereas the other mutations are previously described.9,25,32
ND indicates no data; NA, not applicable; CSF, cerebrospinal fluid; CNS, central nervous system; and HSCT, hematopoietic stem cell transplantation.
Defective: ≤ 10 lytic units.
Reported at some point during the course of the disease.
UNC13D intron 1 mutation
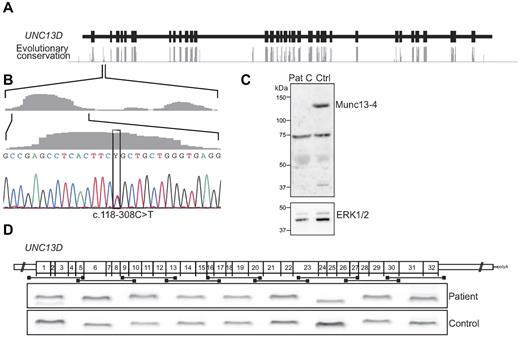
To identify mutations in noncoding regions of UNC13D, evolutionary conservation of nucleotides was evaluated using a phylogenetic hidden Markov model based on sequence from 17 vertebrates, including mammalian, amphibian, bird, and fish species (AlaMut, Interactive Biosoftware34 ; Figure 1A). Aside from the highly conserved coding regions, 4 intronic regions in UNC13D displayed a high degree of evolutionary conservation. In an attempt to identify mutations in the patients carrying a monoallelic UNC13D mutation, these conserved noncoding regions were sequenced. A variant, c.118-308C > T, was identified in a conserved region of intron 1 in 5 of the 7 patients carrying monoallelic UNC13D mutations (4 families, Figure 1B). This variant has previously been reported in a heterozygous state in a single patient diagnosed with FHL3.35 Besides c.118-308C > T, sequencing of the entire intron 1 identified 2 other variants, c.117 + 59C > T and c.118–176G > C. The most upstream variant, c.117 + 59C > T, is a common polymorphism (rs3744010) and thus unlikely to cause disease. The variant c.118–176G > C was found in a homozygous state in 3 and in a heterozygous state in 12 of 96 healthy blood donors. In contrast, the variant c.118-308C > T is located in a conserved region and was not identified in any of the healthy controls, suggesting that this nucleotide variant might represent a disease-causing mutation.
Characterization of a UNC13D intron 1 mutation. (A) Evolutionary nucleotide conservation (gray bars) as predicted by the AlaMut algorithm relative to intronic (black line) and exonic (black boxes) regions of UNC13D. (B) The nucleotide exchange c.118-308C > T is located in a highly conserved region of intron 1. The c.118-308C > T mutation was identified in 7 patients (6 families) in a heterozygous state and in one patient in a homozygous state. (C) PBMCs from patient C and a healthy control were lysed, and protein content was analyzed by SDS-PAGE and Western blotting. Rabbit polyclonal antibodies for detection of Munc13-4 and ERK1 and 2 were used. (D) Nine different primer pairs were used for amplification of overlapping, consecutive UNC13D cDNA fragments. The amplified fragments from patient E, carrying the intron 1 mutation c.118-308C > T in a homozygous state, and a healthy control were separated by gel electrophoresis and are shown relative to their position in the UNC13D transcript.
Characterization of a UNC13D intron 1 mutation. (A) Evolutionary nucleotide conservation (gray bars) as predicted by the AlaMut algorithm relative to intronic (black line) and exonic (black boxes) regions of UNC13D. (B) The nucleotide exchange c.118-308C > T is located in a highly conserved region of intron 1. The c.118-308C > T mutation was identified in 7 patients (6 families) in a heterozygous state and in one patient in a homozygous state. (C) PBMCs from patient C and a healthy control were lysed, and protein content was analyzed by SDS-PAGE and Western blotting. Rabbit polyclonal antibodies for detection of Munc13-4 and ERK1 and 2 were used. (D) Nine different primer pairs were used for amplification of overlapping, consecutive UNC13D cDNA fragments. The amplified fragments from patient E, carrying the intron 1 mutation c.118-308C > T in a homozygous state, and a healthy control were separated by gel electrophoresis and are shown relative to their position in the UNC13D transcript.
In addition to the initial 7 patients with monoallelic UNC13D mutations, 2 infants of Swedish and Danish origin, respectively, with HLH and defective NK cell cytotoxicity were identified carrying the c.118-308C > T mutation in a heterozygous state. Importantly, one infant from Croatia with HLH and defective NK cell cytotoxicity was homozygous for the c.118-308C > T mutation (patient E; Table 1). Western blot analyses of whole cell lysates prepared from PBMCs did not show Munc13-4 expression in patients carrying the c.118-308C > T mutation (Figure 1C), indicating that the conserved intronic sequence may interfere with UNC13D splicing, as hypothesized by Santoro et al,35 or alternatively, regulate UNC13D transcription.
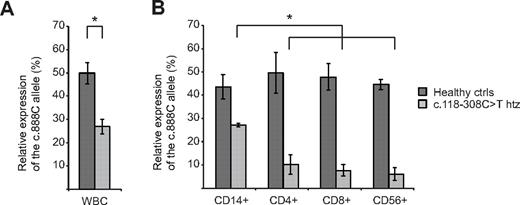
To distinguish between defects in splicing or transcription, UNC13D transcripts were analyzed in patients and heterozygous carriers of the c.118-308C > T mutation in intron 1. First, to determine whether this mutation interfered with UNC13D splicing, overlapping fragments of the UNC13D transcript were amplified in patient E, homozygous for the c.118-308C > T mutation. In this patient, all fragments were readily amplified and displayed the predicted length (Figure 1D). Notably, sequence analysis of the fragments revealed correctly spliced products. Thus, data indicated that the intron 1 mutation did not interfere with UNC13D splicing. To evaluate whether the mutation affected UNC13D transcript levels, allele-specific quantitative real-time PCR was performed on persons heterozygous for the c.118-308C > T mutation that were also heterozygous for a linked, common polymorphism, c.888G > C, in exon 11. For comparison, allele-specific transcription was quantified in healthy controls that did not carry the c.118-308C > T mutation but were heterozygous for the c.888G > C polymorphism. On analysis of allele transcription in WBCs, UNC13D transcripts with the c.888C allele were as frequent as those with the c.888G allele in persons that did not carry the c.118-308C > T mutation (Figure 2A). In contrast, in parents that where heterozygous carriers of the c.118-308C > T mutation, transcription of the allele carrying the linked c.888G > C polymorphism was significantly diminished, representing 27% of total UNC13D transcripts (Figure 2A). Accordingly, a homozygous carrier of the mutation would theoretically have 37% of the normal amount of transcript. In transplanted FHL patients, relapse of HLH is rarely, if ever, observed unless the percentage of donor lymphocytes decreased to < 10%.36,37 Accordingly, such a decrease in UNC13D transcription might not be expected to cause disease.
Allele-specific quantitative real-time PCR in heterozygous carriers of the UNC13D intron 1 mutation. (A) Allele-specific expression analysis of UNC13D was assessed in WBCs from healthy controls and heterozygous carriers of the intron 1 mutation, c.118-308C > T, using real-time PCR (standard curve method). Primers were designed for specific amplification of either the c.888G or the c.888C allele, a single nucleotide polymorphism located in exon 11. Both healthy controls (n = 3) and carriers of the intron 1 mutation (n = 4) carried the SNP, c.888G > C, in a heterozygous state. In all heterozygous carriers of the intron 1 mutation, c.118-308C > T, was located on the c.888C allele. (B) For evaluation of allele-specific transcription in different cell subsets, CD14+, CD4+, CD8+, and CD56+ cells were isolated consecutively by positive magnetic selection. Allele specific real-time PCR was performed in healthy controls (n = 4) and heterozygous carriers of the intron 1 mutation, c.118-308C > T (n = 4). Statistical significance was analyzed using the 2-tailed Mann-Whitney U test. *P < .05.
Allele-specific quantitative real-time PCR in heterozygous carriers of the UNC13D intron 1 mutation. (A) Allele-specific expression analysis of UNC13D was assessed in WBCs from healthy controls and heterozygous carriers of the intron 1 mutation, c.118-308C > T, using real-time PCR (standard curve method). Primers were designed for specific amplification of either the c.888G or the c.888C allele, a single nucleotide polymorphism located in exon 11. Both healthy controls (n = 3) and carriers of the intron 1 mutation (n = 4) carried the SNP, c.888G > C, in a heterozygous state. In all heterozygous carriers of the intron 1 mutation, c.118-308C > T, was located on the c.888C allele. (B) For evaluation of allele-specific transcription in different cell subsets, CD14+, CD4+, CD8+, and CD56+ cells were isolated consecutively by positive magnetic selection. Allele specific real-time PCR was performed in healthy controls (n = 4) and heterozygous carriers of the intron 1 mutation, c.118-308C > T (n = 4). Statistical significance was analyzed using the 2-tailed Mann-Whitney U test. *P < .05.
Thus, we hypothesized that the c.118-308C > T mutation might more severely affect UNC13D transcription in cell types that mediate lymphocyte cytotoxicity, such as CD3+CD8+ T cells and CD3−CD56+ NK cells. To test this, allele-specific transcription was examined in CD14+, CD4+, CD8+, and CD56+ cells isolated sequentially from controls or carriers of the c.118-308C >T mutation that all were heterozygous carriers of the c.888G >C polymorphism. Strikingly, the allele carrying the c.888G >C polymorphism represented < 10% of UNC13D transcripts in CD4+, CD8+, and CD56+ cells from persons carrying the c.118–308C > T mutation, indicating a more than a 9-fold reduction in transcription of the mutated UNC13D allele relative to the wild-type in lymphocytes (Figure 2B). The relative frequency of the transcript from the mutated allele in CD4+, CD8+, and CD56+ cells was significantly less than that observed in CD14+ cells. In persons that did not carry the c.118-308C > T mutation, the frequency of the alleles with or without the c.888G > C polymorphism was similar (Figure 2B). Altogether, the data indicate that the c.118-308C > T mutation specifically impairs UNC13D transcription in lymphocytes.
UNC13D 253-kb inversion
The identification of 2 patients with the monoallelic UNC13D intron 1 mutation, in addition to the 2 previous patients with monoallelic UNC13D mutations that did not harbor the intron 1 mutation, indicated other noncoding UNC13D aberrations. Examination of exonic and intronic single nucleotide polymorphisms throughout UNC13D in these patients revealed a shared haplotype on the second allele in 2 patients of Swedish origin, one of Danish origin, and one of Finnish origin. Remarkably, other nonrelated Scandinavian HLH patients with defective NK cell cytotoxicity were homozygous for this haplotype.
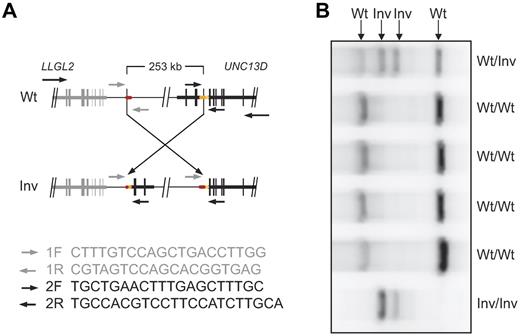
To determine whether these patients carried a common disease-causing mutation affecting UNC13D splicing, overlapping fragments of the UNC13D transcript were amplified in 3 patients homozygous for the second haplotype where RNA was available. Whereas the first 7 fragments were readily amplified in these 3 patients, the last 2 fragments were not detected (Figure 3A). By comparison, all fragments were readily amplified in healthy control samples (Figure 3A). Thus, the data indicated an aberration affecting splicing of the 3′-end of UNC13D. To obtain the 3′-end sequence of the transcript in the patients homozygous for the second haplotype, 3′-rapid amplification of cDNA ends PCR was performed. Three different products were amplified. Cloning and sequencing of the 3 PCR products revealed UNC13D sequence up to the end of exon 30. In the first product, also observed in healthy controls, the sequence of exon 30 was followed by sequence from intron 30. In the 2 other PCR products, specific to the patients, sequence following exon 30 lacked homology to UNC13D. Thus, these 3′-end sequences were blasted against the human genome using the BLAT algorithm.38 The highest homology for both products was located on chromosome 17, 253 kb upstream of UNC13D. Thus, the sequence identity suggested an inversion of approximately 253 kb straddling the UNC13D locus. To confirm the existence of such an inversion in the genomic DNA from the patients, primers were designed in the vicinity of the exon 30 splice site and from the sequence located 253 kb upstream of UNC13D. One primer pair specifically amplified product in the patients, but not in healthy, unrelated controls. PCR products from the patients were isolated, cloned, and sequenced, thereby providing sequence over one of the break points. Similarly, the second break point was amplified with a different set of primers and subsequently cloned and sequenced. In this way, the breakpoints were mapped down to 2 25-bp Alu elements that were identical in sequence (chromosome 17: 73825638-73825663 and 73573065-73573090, human, February 2009, GRCh37/hg19 assembly; Figure 3B). No gain or loss of DNA seemed to have occurred. For rapid detection of the 253-kb inversion, a multiplex PCR assay using 2 different forward primers and 2 different reverse primers was developed (Figure 4). The inversion was detected in a heterozygous state in one of 190 healthy blood donors, in line with this aberration being a cause of FHL in the Scandinavian population.
Characterization of an UNC13D inversion. (A) Nine different primer pairs were used for amplification of overlapping, consecutive UNC13D cDNA fragments. The amplified fragments from a patient (representative of 3 analyzed patients) and a healthy control were separated by gel electrophoresis and are shown relative to their position in the UNC13D transcript. (B) Schematic illustration of a 253-kb inversion identified in patients with FHL. The breakpoints are located in Alu elements containing an identical sequence of 25 bp. Two elements, AluSc8 and AluSx1 (yellow, orange), are located on the reverse strand in intron 30 UNC13D, and an AluY (red) element is located 253 kb upstream of UNC13D on the forward strand. (C) PBMCs from patient O, patient L, and healthy controls were lysed, and protein content was analyzed by SDS-PAGE and Western blotting. Rabbit polyclonal antibodies for detection of Munc13-4 and ERK1 and 2 were used.
Characterization of an UNC13D inversion. (A) Nine different primer pairs were used for amplification of overlapping, consecutive UNC13D cDNA fragments. The amplified fragments from a patient (representative of 3 analyzed patients) and a healthy control were separated by gel electrophoresis and are shown relative to their position in the UNC13D transcript. (B) Schematic illustration of a 253-kb inversion identified in patients with FHL. The breakpoints are located in Alu elements containing an identical sequence of 25 bp. Two elements, AluSc8 and AluSx1 (yellow, orange), are located on the reverse strand in intron 30 UNC13D, and an AluY (red) element is located 253 kb upstream of UNC13D on the forward strand. (C) PBMCs from patient O, patient L, and healthy controls were lysed, and protein content was analyzed by SDS-PAGE and Western blotting. Rabbit polyclonal antibodies for detection of Munc13-4 and ERK1 and 2 were used.
Multiplex PCR assay for detection of the UNC13D inversion. A multiplex PCR assay with 2 different forward and reverse primers was designed for rapid detection of the UNC13D inversion. (A) Primer sequences and schematic representation of primer pair positioning on wild-type (Wt) and inverted (Inv) UNC13D genomic sequence. (B) Gel electrophoresis of multiplex PCR products on genomic DNA from individuals wild-type, heterozygous, or homozygous for the inversion straddling UNC13D. In homozygous carriers of the inversion, 2 products of 725 and 922 bp are amplified. In persons that do not carry the inversion, 2 products of 466 and 1220 bp are amplified. Consequently, in heterozygous carriers of the inversion, all 4 products are amplified.
Multiplex PCR assay for detection of the UNC13D inversion. A multiplex PCR assay with 2 different forward and reverse primers was designed for rapid detection of the UNC13D inversion. (A) Primer sequences and schematic representation of primer pair positioning on wild-type (Wt) and inverted (Inv) UNC13D genomic sequence. (B) Gel electrophoresis of multiplex PCR products on genomic DNA from individuals wild-type, heterozygous, or homozygous for the inversion straddling UNC13D. In homozygous carriers of the inversion, 2 products of 725 and 922 bp are amplified. In persons that do not carry the inversion, 2 products of 466 and 1220 bp are amplified. Consequently, in heterozygous carriers of the inversion, all 4 products are amplified.
Western blot analysis of whole cell lysates prepared from PBMCs did not detect Munc13-4 expression in patients carrying the inversion (Figure 3C), suggesting that the truncated UNC13D transcript generates an unstable protein product.
Effect of mutations on lymphocyte cytotoxic function
Decreased NK cell activity, typically assessed by quantifying lysis of K562 target cells using freshly isolated PBMCs as effector cells, is a diagnostic criterion for HLH.7 A value of ≤ 10 LU is indicative of HLH. With freshly isolated PBMCs, NK cell-mediated lysis of K562 target cells was defective compared with controls and below the limit set as pathologic in most patients tested (Figure 5A). For patients with biallelic mutations in UNC13D and with at least 1 allele of the intron 1 mutation, all but 1 (patient A, 11 lytic units) displayed pathologic NK cell activity (Figure 5A). Similarly, of the patients with biallelic aberrations in UNC13D and with at least 1 allele of the inversion, all but 1 (patient H, 13 LU) displayed pathologic NK cell activity (Figure 5A). This pathologic NK cell activity was evident despite normal or close to normal NK cell frequencies, apart from for 2 patients, who displayed much reduced frequencies of circulating NK cells (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Furthermore, elevated K562 cell lysis coincided with high NK cell frequencies (supplemental Table 1). Degranulation by freshly isolated CD3−CD56+ NK cells in response to K562 cells in FHL2 patients carrying biallelic mutations in PRF1 was comparable with that of healthy adult and infant controls, whereas degranulation was < 3.0% in all patients with the intron 1 mutation and most patients carrying the inversion (Figure 5B). Three patients tested during flares displayed degranulation > 3.0% (patients C, Q, and P; supplemental Table 1). On repeated analysis during HLH-2004 therapy or remission, degranulation in these patients was < 3.0%.
Freshly isolated NK cell cytotoxicity and degranulation in FHL3 patients carrying the intron 1 mutation or the 253-kb inversion. Resting PBMCs (A-B) or PBMCs stimulated with IL-2 for 48 hours (C-D) from healthy adult and infant donors or FHL3 patients were evaluated for cytotoxicity and degranulation toward K562 cells. (A,C) NK cell cytotoxicity was evaluated in a 4-hour 51Cr-release assay. Plots represent lytic units calculated at 25% lysis. (B,D) NK cell degranulation was evaluated in a 2-hour flow cytometric assay. The cells were stained with fluorochrome-conjugated anti–CD3, anti–CD56, and anti–CD107a mAbs. Lymphocytes were gated on forward/side scatter plots, followed by gating on CD3 versus CD56 plots. Plots represent induced CD107a surface expression (ΔCD107a+) on CD3−CD56+ NK cells. Boxes represent 25th, 50th, and 75th percentiles; pluses, mean values; and error bars, 95% CI. For FHL3 patients, each point represents one person. Intron 1 mutant alleles (blue), inversion alleles (red), and alleles with other mutations (gray) are indicated. Bars represent mean and 95% CI.
Freshly isolated NK cell cytotoxicity and degranulation in FHL3 patients carrying the intron 1 mutation or the 253-kb inversion. Resting PBMCs (A-B) or PBMCs stimulated with IL-2 for 48 hours (C-D) from healthy adult and infant donors or FHL3 patients were evaluated for cytotoxicity and degranulation toward K562 cells. (A,C) NK cell cytotoxicity was evaluated in a 4-hour 51Cr-release assay. Plots represent lytic units calculated at 25% lysis. (B,D) NK cell degranulation was evaluated in a 2-hour flow cytometric assay. The cells were stained with fluorochrome-conjugated anti–CD3, anti–CD56, and anti–CD107a mAbs. Lymphocytes were gated on forward/side scatter plots, followed by gating on CD3 versus CD56 plots. Plots represent induced CD107a surface expression (ΔCD107a+) on CD3−CD56+ NK cells. Boxes represent 25th, 50th, and 75th percentiles; pluses, mean values; and error bars, 95% CI. For FHL3 patients, each point represents one person. Intron 1 mutant alleles (blue), inversion alleles (red), and alleles with other mutations (gray) are indicated. Bars represent mean and 95% CI.
On stimulation with IL-2 for 30 to 60 hours, K562 cell lysis and NK cell degranulation increased markedly in healthy controls (Figure 5C-D). By comparison, lysis and degranulation were weak in FHL3 patients. Notably, in patients carrying at least 1 allele of the inversion, degranulation by IL-2–stimulated NK cells was significantly augmented compared with FHL3 patients carrying the intron 1 mutation or with other UNC13D mutations (Figure 5D). Accordingly, in FHL3 patients, mean cytotoxicity of IL-2–stimulated NK cells was higher in patients carrying allele(s) of the inversion relative to patients carrying the intron 1 mutation or with other UNC13D mutations (Figure 5C).
Clinical characteristics, neurologic manifestations, and outcome
Altogether, 21 infants with at least one allele with the intron 1 mutation, c.118-308C > T, or the 253-kb inversion were included in this study (Table 1). All patients fulfilled the diagnostic criteria (n = 19) according to the HLH-2004 guidelines7 or had a familial disease (n = 2). The median age at diagnosis of patients with at least one allele with the intron 1 mutation was 4.7 months (142 days; mean, 138 days; range, 55-229 days, n = 8), and the age at diagnosis of the one patient with the mutation in homozygous was 133 days. The median age at diagnosis in the group of patients with the 253-kb inversion in a homozygous state was 3.4 months (104 days; mean 158 days; range, 1-459 days, n = 12). By comparison, the age at diagnosis is similar to patients with 2 disruptive mutations in UNC13D (3 months).39 Thus, although a slight improvement in degranulation and cytotoxicity was seen in the patients carrying the inversion, no difference was seen in terms of age at onset in this group of patients compared with other FHL3 patients.
Consistent with a severe presentation of HLH, neurologic alterations were reported in 11 of 20 patients (55%) of the patients, whereas 15 of 18 (83%) had abnormal cerebrospinal fluid (cells and/or protein levels) during the course of the disease.
Sixteen of 21 patients were treated according to the HLH-2004 protocol, 4 were treated according to HLH-94,7,40 and one patient went directly to hematopoietic stem cell transplantation. Only 4 of 19 patients (21%) were reported in remission at 2 months after treatment. Thirteen of 21 patients (62%) are alive after hematopoietic stem cell transplantation.
Geographic distribution of new UNC13D aberrations
Further examination of the geographic distribution and frequency of different UNC13D aberrations in FHL3 patients from Scandinavian countries Croatia, and Slovenia revealed a high allele frequency of the intron 1 mutation in Croatia and Slovenia, but the mutation was also found in patients from Denmark and southern Sweden (Figure 6A). Microsatellite markers indicated a common haplotype for the intron 1 mutation (Figure 6B), suggesting a founder mutation that has become widely geographically distributed. In contrast, the 253-kb UNC13D inversion was found only in patients of Scandinavian ancestry. All these affected persons shared a common haplotype (Figure 6C). Moreover, a gradient was observed with particularly high frequency of the inversion in patients from northern Scandinavia, suggesting a founder effect. Thus, the geographic distribution and microsatellite analysis revealed distinct prevalence and origins of the 2 relatively frequent UNC13D disease-causing aberrations.
Geographic distribution and microsatellite marker analyses of noncoding UNC13D mutations. (A) Frequency of distinct UNC13D mutations in families from different European countries. All patients with UNC13D studied from Sweden, Denmark, Norway, Finland, Croatia, and Slovenia are included. Pie charts indicate the relative frequency of the intron 1 mutation (blue), the inversion (red), or other mutations (gray). Numbers represent alleles. (B) Haplotype analyses using microsatellite markers of patients with the c.118-308C > T mutation. (C) Haplotype analyses using microsatellite markers of patients with the inversion. (D) Relative frequency of mutated alleles in infants that fulfilled the diagnostic criteria for HLH, including defective NK cell cytotoxicity in Sweden between December 2005 and January 2011. The pie chart includes 13 patients and 26 mutated alleles.
Geographic distribution and microsatellite marker analyses of noncoding UNC13D mutations. (A) Frequency of distinct UNC13D mutations in families from different European countries. All patients with UNC13D studied from Sweden, Denmark, Norway, Finland, Croatia, and Slovenia are included. Pie charts indicate the relative frequency of the intron 1 mutation (blue), the inversion (red), or other mutations (gray). Numbers represent alleles. (B) Haplotype analyses using microsatellite markers of patients with the c.118-308C > T mutation. (C) Haplotype analyses using microsatellite markers of patients with the inversion. (D) Relative frequency of mutated alleles in infants that fulfilled the diagnostic criteria for HLH, including defective NK cell cytotoxicity in Sweden between December 2005 and January 2011. The pie chart includes 13 patients and 26 mutated alleles.
Discussion
We here describe 2 aberrations outside the coding region of UNC13D that together explain HLH in 21 infants with primary HLH included in this study. These aberrations are not detectable by normal sequencing of the exons and exon/intron boundaries. The findings increase the sensitivity of a molecular diagnosis in patients with suspected FHL, as well as extended prenatal diagnostics and carrier testing for families affected by FHL. Furthermore, the results provide insight into lymphocyte-specific transcriptional regulation of UNC13D.
The first aberration, a point mutation, c.118-308C > T, was identified in 7 patients in a heterozygous state and in one patient in a homozygous state, all with early onset HLH. The mutation was widely distributed in patients originating from Croatia, Slovenia, Denmark, and Sweden. Microsatellite analysis revealed a common haplotype. Together, these data suggest a distant mutation event that has become widely geographically distributed and that may explain a number of HLH cases in whites. The mutation was located in an evolutionary conserved region of intron 1. Defective NK cell degranulation and cytotoxicity, as well as absent Munc13-4 expression, was consistent with a loss-of-function mutation in UNC13D. Interestingly, further experiments revealed that this mutation did not impair splicing, as previously suggested,35 but rather reduced UNC13D transcription specifically in lymphocytes. The conserved intronic sequence in which the c.118-308C > T mutation is located displays homology to STAT4 and Ets-1 consensus binding sites, which are disrupted by the mutation. Thus, this sequence may represent an intronic enhancer. STAT4 and Ets-1 are highly expressed in lymphocytes,41,42 may promote perforin expression,43 but have not previously been implicated in regulation of Munc13-4 expression. Binding of transcription factors to this putative regulatory site remains to be explored experimentally, but it is well documented that such intronic enhancers can control cell-specific transcription, as exemplified by the B cell-specific expression of the immunoglobulin locus.44,45
The second aberration, a 253-kb inversion with one of the breakpoints located within intron 30 of UNC13D, causes a disruption of the gene. Defective NK cell degranulation and cytotoxicity, as well as absent Munc13-4 expression, was consistent with a loss-of-function mutation in UNC13D. Notable, this aberration appears to be the most common cause of FHL in Swedish infants, constituting ∼ 50% of the mutated alleles. The inversion was also identified in HLH patients from neighboring countries. The incidence and geographic distribution of patients carrying this allele suggest a founder effect in Northern Scandinavia. Most likely, the genomic rearrangement occurred by pairing of the inverted Alu repeats, as described in the X-linked recessive hemophilia A and Alport syndrome.46,47 To our knowledge, this is the first paracentric inversion alone that causes an autosomal recessive disease in humans. Functionally, IL-2 stimulation slightly increased degranulation by NK cells carrying the UNC13D inversion relative to NK cells from patients with other UNC13D mutations. UNC13D missense mutations have been associated with milder impairments in NK cell degranulation and cytotoxicity, as well as later disease onset.48,49 Moreover, mutations in STX11 and STXBP2 are associated with partial gain of degranulation after IL-2 stimulation as well as later disease onset relative to patients with nonsense mutations in PRF1 or UNC13D.12,13,28,50 In contrast, we did not find evidence for any later onset of disease in our group of patients with the UNC13D inversion. Thus, the disease relevance of these functional observations is not clear.
Remarkably, with the discovery of the 2 noncoding UNC13D aberrations, a molecular diagnosis could be provided for all 13 infants from Sweden that presented with HLH and defective NK cell cytotoxicity between December 2005 and January 2011 (Figure 6D). The UNC13D c.118-308C > T mutation in intron 1 corresponded to 3.8% of the mutated alleles in Sweden, whereas the UNC13D 253-kb inversion represented 50% of the mutated alleles in this patient cohort. When only considering patients of European ancestry, the c.118-308C > T mutation corresponded to 4.5% of the mutated alleles in Sweden, whereas the 253-kb inversion represented 59.1% of the mutated alleles. Although our analysis suggests a relatively confined geographic distribution of the UNC13D inversion to Northern Sweden, this aberration may also exist in the Northern American population with Scandinavian ancestry. The newly developed multiplex PCR assay for detection of the UNC13D inversion can facilitate a rapid molecular diagnosis of patients carrying this aberration and can be implemented for future Guthrie-based neonatal screening.
Taken together, identification of the 2 genetic aberrations described here provides a molecular diagnosis for the majority of HLH cases among infants in northern Europe. To our knowledge, the UNC13D inversion represents the first paracentric inversion causing an autosomal recessive disease in humans. Furthermore, with its widespread distribution and abundance across Europe, the UNC13D intron 1 mutation may explain many nonconsanguineous white HLH cases. This mutation identified an intronic sequence required for UNC13D transcription in lymphocytes, advancing our knowledge of the genetic regulation of lymphocyte cytotoxicity. These findings highlight aberrations outside the coding regions as a cause of disease and make a case for sequencing of evolutionary conserved noncoding regions in diagnostics of primary immunodeficiency syndromes.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for participation and Sigrid Sahlén and Mårten Winge for technical assistance.
This work was supported by the Swedish Children's Cancer Foundation, Swedish Research Council, Cancer and Allergy Foundation of Sweden, Swedish Cancer Foundation, Mary Béve Foundation, Märta and Gunnar V Philipson Foundation, David and Astrid Hageléns Foundation, Histiocytosis Association of America, Tobias Foundation, Åke Olsson Foundation for Hematologic Research, Swedish Society for Medical Research, Clas Groschinsky's Memorial Fund, Shizu Matsumara's Donation, the Karolinska Institute Research Foundation, and the Stockholm County Council (ALF project). M.M. was supported by Karolinska Institutet (MD/PhD scholarship and research internship).
Authorship
Contribution: M.M. designed research and performed genetic analysis, analyzed and interpreted data, and drafted the manuscript; S.C.C.C. performed evaluations of NK cell activity and interpreted data; S.M.W. performed evaluations of NK cell activity and Western blot and interpreted data; M.E. performed and interpreted genetic analyses, including microsatellite analyses; H.S. performed FACS analysis; B.B. and E.N. performed genetic analysis; C.B., G.J., J.J., H.H., B.-M.H., L.R., S.P., S.R., M.S., T.T.S., T.S., and J.W. cared for patients and provided clinical data; H.-G.L., B.F., and M.N. designed research and contributed to drafting the manuscript; J.-I.H. initiated the study, designed research, analyzed and interpreted data, and drafted the manuscript; and Y.T.B. designed research, analyzed and interpreted data, and drafted the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marie Meeths, Childhood Cancer Research Unit, CMM L8:02, Department of Women's and Children's Health, Karolinska Institutet, Karolinska University Hospital Solna, SE-17176 Stockholm, Sweden; e-mail: marie.meeths@ki.se; and Yenan T. Bryceson, Center for Infectious Medicine, F59, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, S-141 86 Stockholm, Sweden; e-mail: yenan.bryceson@ki.se.
References
Author notes
S.C.C.C. and S.M.W. contributed equally to this study.
J.-I.H. and Y.T.B. share senior authorship.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal