Abstract
The CX3C chemokine family is composed of only one member, CX3CL1, also known as fractalkine, which in mice is the sole ligand of the G protein-coupled, 7-transmembrane receptor CX3CR1. Unlike classic small peptide chemokines, CX3CL1 is synthesized as a membrane-anchored protein that can promote integrin-independent adhesion. Subsequent cleavage by metalloproteases, either constitutive or induced, can generate shed CX3CL1 entities that potentially have chemoattractive activity. To study the CX3C interface in tissues of live animals, we generated transgenic mice (CX3CL1cherry:CX3CR1gfp), which express red and green fluorescent reporter genes under the respective control of the CX3CL1 and CX3CR1 promoters. Furthermore, we performed a structure/function analysis to differentiate the in vivo functions of membrane-tethered versus shed CX3CL1 moieties by comparing their respective ability to correct established defects in macrophage function and leukocyte survival in CX3CL1-deficient mice. Specifically, expression of CX3CL1105Δ, an obligatory soluble CX3CL1 isoform, reconstituted the formation of transepithelial dendrites by intestinal macrophages but did not rescue circulating Ly6Clo CX3CR1hi blood monocytes in CX3CR1gfp/gfp mice. Instead, monocyte survival required the full-length membrane-anchored CX3CL1, suggesting differential activities of tethered and shed CX3CL1 entities.
Introduction
Chemokine (CX3C motif) ligand 1 (CX3CL1), also known as fractalkine or neurotactin,1,2 and its receptor CX3CR13 have been assigned their own CX3C chemokine family. This classification is based on the 3 amino acid gap between its N-terminal cysteines in CX3CL1, with no spacing in CC chemokines and only one intervening amino acid in CXC chemokines.4 CX3CL1 is furthermore structurally unique in that it is synthesized as a type I transmembrane protein with the CX3C chemokine domain presented on an extended stalk.1,2 Both CX3CL1 and CX3CR1 are widely expressed throughout the organism; but in given tissues, expression is often highly cell type-specific. Taking advantage of mice that harbor a targeted replacement of the CX3CR1 gene by a GFP reporter,5 we could, for instance, show that CX3CR1 expression in the brain is restricted to microglia. CX3CR1 expression in the gut was found limited to lamina propria macrophages and CX3CR1 expression in the blood is largely restricted to monocytes, which are uniform CX3CR1 positive, albeit with discrete expression levels.6 CX3CR1 is furthermore expressed by macrophage/dendritic cell precursors,7 various dendritic cell (DC) progenitors, a nonclassic CD8α+ DC subset,8 and plasmacytoid DCs. Aside from the prominent expression in the mononuclear myeloid compartment, CX3CR1 receptor expression has been reported for an NK cell subset3,9 and certain T-cell populations.3,10,11 The in vivo expression pattern of the ligand CX3CL1 remains less well defined and controversial12 but has been reported for neurons,13 intestinal epithelium,14 and inflamed endothelium.2 Notably, in humans eotaxin-3/CC chemokine ligand 26 was recently reported to be a functional ligand for CX3CR115 ; in mice, the CCL26 gene, however, is a pseudogene.
The analysis of CX3C receptor and ligand knockout mice5,16 has revealed a number of phenotypes resulting from the lack of CX3CR1/CX3CL1 interactions.17-20 However, in-depth knowledge of the physiologic role of the CX3C axis, including mechanistic insights, is missing. A key to understanding the biologic function of the CX3C chemokine family probably lies in the unique structure of the ligand CX3CL1. Whereas classic small peptide chemokines are secreted and form gradients by binding to ECM proteoglycans,21 CX3CL1 is synthesized as a transmembrane protein with the CX3C chemokine domain presented on an extended highly glycosylated mucin-like stalk.1,2 To date, CX3CL1 shares this unique membrane anchorage only with one other chemokine, the CXCR6 ligand CXCL16.22 Expression of the CX3C transmembrane chemokine on endothelial and epithelial cells was shown to mediate tight, pertussis toxin-resistant and integrin-independent interactions with CX3CR1-expressing leukocytes.23,24 Moreover, the adhesive interactions of CX3CR1 and CX3CL1 were found to be sufficient to support the recruitment of leukocytes through the endothelium of inflamed vasculature.25 Proteolytic cleavage by the disintegrin-like metalloproteinase ADAM10 results in constitutive release of different-sized soluble CX3CL1 entities.26 Moreover, under inflammatory conditions, CX3CL1 shedding is also promoted by ADAM17/TACE.27,28 Cleavage could be required for the detachment of cells tethered by the CX3CL1/ CX3CR1 bond. Furthermore, ecto-domain shedding of CX3CL1 generates a potential chemoattractant that could also actively interfere with additional CX3CL1/ CX3CR1 interactions. However, specific contributions of the membrane-tethered versus shed CX3CL1 isoforms to the known in vivo activities of the CX3C chemokine family remain to be established.
To gain further insight into the physiologic role of the CX3C chemokine family, we used bacterial artificial chromosome (BAC) transgenesis to explore the in vivo expression pattern of CX3CL1 and perform a structure/function analysis of shed and membrane-anchored CX3CL1 entities. Here we report CX3CL1cherry:CX3CR1gfp mice that harbor fluorescent reporter genes embedded in the CX3CL1 and CX3CR1 loci and establish differential in vivo activities of tethered and shed CX3CL1 isoforms.
Methods
Mice
This study involved the use of cx3cr1gfp mice5 and cx3cl1−/− mice16 on C57BL/6 background. For generation of BM chimeras, the indicated recipient animals were lethally irradiated with 950 cGy and subsequently reconstituted with 5 × 106 donor BM cells. Chimeric animals were analyzed 8 weeks after transplantation. All mice were maintained under specific pathogen-free conditions and handled under protocols approved by the Weizmann Institute Animal Care Committee according to international guidelines.
BAC transgene generation and typing
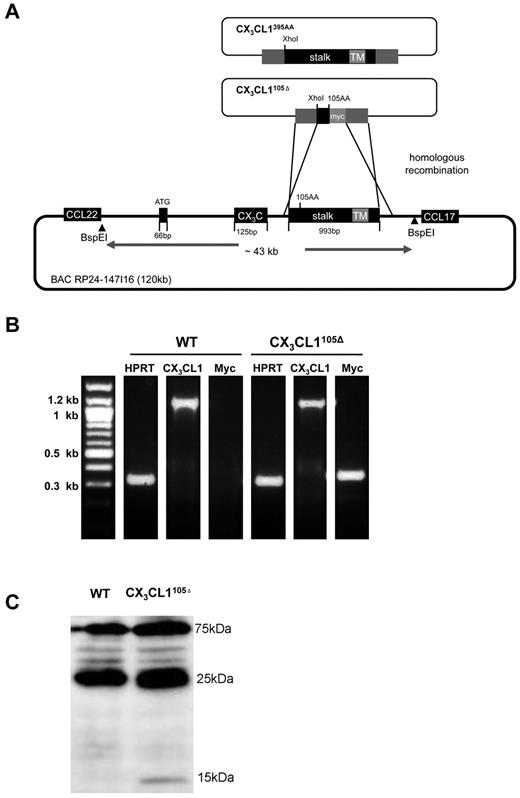
For the generation of the transgenic mice, the RP24–147I16 BAC clone (Roswell Park Cancer Institute) spanning 120 kb, including the murine CX3CL1 locus, was modified as described previously using the pDelsac shuttle vector strategy29 or using Red/ET recombineering.30 The sequences of the CX3CL1105Δ and CX3CL1395AA BAC transgenes can be found in supplemental Figure 4 (see the Supplemental Materials link at the top of the article). Expression of the CX3CL1105Δ transgene can be monitored according to detection of myc-tagged protein. The CX3CL1395AA transgene harbors a silent mutation introducing an Xho1 site. Its expression can therefore be assessed and compared with the expression of endogenous CX3CL1 by quantification of Xho-resistant and -sensitive RT-PCR products. The BAC DNA (1 ng/μL) was injected into the fertilized CB6F1 oocytes; transgenic mice were established and backcrossed to C57BL/6 mice (generations > 8). CX3CL1105Δ mice are typed by PCR using the forward primer 5′-CCAGGCTGGCTATGGTCCAACTG-3′ and the reverse (myc) primer reverse 5′-GAGATGAGTTTTTGTTCGGGCC-3′. CX3CL1395AA mice are typed by PCR using the forward primer 5′-CCAGGCTGGCTATGGTCCAACTG-3′ and the reverse primer 5′-CACTGGCACCAGGACGTATG-3′ that amplify a 1.3-kb fragment from the WT and transgenic CX3CL1 loci; the product of the CX3CL1395AA gene is cleavable by Xho1 into a 300-bp and 1-kb fragment. CX3CL1cherry mice are typed by PCR using the forward primer 5′-CCAGCCGCGAGTGACTAC-3′ and the reverse primer 5′-CCCCAGGGAGATGCAGTC-3′, yielding a 1-kb product from the transgenic locus and a 300-bp product from the endogenous CX3CL1 locus.
Cell preparations
For the isolation of lung cells, lobes were cut and incubated in PBS containing Ca2+ and Mg2+ (Sigma-Aldrich) supplemented with 4 mg/mL of collagenase D (Roche Diagnostics) for 1 hour at 37°C. Kidneys were minced and incubated in RPMI 1640 supplemented with 2 mg/mL of collagenase D and 1 mg/mL of dispase II (Roche Diagnostics) for 1 hour at 37°C. After erythrocyte lysis in ACK buffer (0.15M NH4Cl and 0.01M KHCO3), cells were passed through a 100-mm mesh and centrifuged at 300g for 5 minutes at 4°C. For the flow cytometric analysis, cells were suspended with PBS supplemented with 2mM EDTA, 0.05% sodium azide, and 1% of fetal calf serum.
To obtain intestinal epithelial cells and lamina propria cells, large intestine was longitudinally sectioned and cut into small pieces. Colonic epithelial cells were isolated by incubation in 5 mL HBSS containing 10mM HEPES and 5mM EDTA (pH 8.0) at 37°C on a shaking incubator and collected by passing through a 100-mm mesh. For the isolation of lamina propria cells, remnant colon pieces were digested by 5 mL HBSS containing 10% FBS, 1 mg/mL of dispase II (Roche Diagnostics) and 0.5 mg/mL of collagenase D (Sigma-Aldrich) at 37°C shaking incubator for 1 hour. Completely digested colon was passed through a 100-mm mesh and collected by centrifugation.
Transcription analysis
Total RNA was extracted from the terminal ileum of CX3CL1105Δ BAC transgenic mice using a software-controlled sealed homogenization system (gentleMACS dissociator) and RNA tissue kit (5′). cDNA synthesis was performed with 1 μg of total RNA by Masterscript RT-PCR kit (5′) with exon-spanning primers for HPRT, CX3CL1, and with myc epitope primers for the CX3CL1105Δ transgene.
Protein expression analysis
The 293T cells cultured in serum-free media were transiently transfected with pCDNA3 vector encoding the CX3CL1105Δ protein by polyethylenimine. After 24 hours, cells were lysed and incubated on ice for 20 minutes, and then centrifuged at 20 000g for 30 minutes. Colonic epithelial cells from CX3CL1105Δ mice were lysed and incubated on ice for 20 minutes and then centrifuged at 20 000g for 30 minutes. Protein samples and supernatant were separated on 15% SDS-PAGE and blotted onto a nitrocellulose membrane (GE Healthcare). Detection was performed using mouse anti-cMyc antibody (9E10) and goat anti–mouse HRP (1:10 000, The Jackson Laboratory). Blots were developed with the ECL kit.
Immunohistochemistry
Intestine, kidneys, and lungs were obtained from CX3CL1cherry:CX3CR1gfp mice. Intestine swiss rolls31 and kidney samples were fixed overnight with 2% paraformaldehyde (PFA) at 4°C. Whole lungs were swollen by the injection of 1 mL of 2% PFA into trachea and fixed overnight with 2% PFA at 4°C. Samples were incubated in 30% sucrose at 4°C for 2 to 3 days, embedded in Tissue-Tek optimum cutting temperature compound (Sakura Finetechnical), and frozen in a bath of liquid nitrogen (N2) cold isopentane. Frozen intestine, kidney, and lung tissues were sectioned into 10- to 12-mm sections using a Leica cryo-microtome. Intestinal sections were stained using the periodic acid-Schiff staining protocol32 for goblet cell detection. Brains were fixed overnight with 2.5% fresh PFA at 4°C and soaked in 30% sucrose for 2 to 3 days, frozen, and cut into 25-mm sections using a sliding microtome. Lung sections were stained for Clara cell secretory protein (CCSP) using rabbit anti-CCSP, followed by Dylight 488 conjugated anti–rabbit (The Jackson Laboratory). Brain sections were stained with anti-NeuroN (NeuN) or anti-DCX followed by Cy5-conjugated anti–rabbit Ab or eFluor488 conjugated anti–rabbit Ab. Spinal cord was fixed with 2.5% fresh PFA at 4°C for 2 days and decalcified by 12.5% EDTA (pH7.4) at 4°C for 2 to 3 days. Spinal cord was incubated in 30% sucrose for 2 to 3 days, and lumbar regions (L3-L5) were embedded in optimal cutting temperature (OCT). Frozen L3 to L5 was sectioned into 10-mm sections using a sliding microtome. Sections were stained with biotin anti-calcitonin gene–related peptide (CGRP), followed by streptavidin-conjugated aminomethyl coumarin acetate.
Image acquisition and manipulation
Sections of kidney, lung and brain were imaged from LSM 510 confocal microscopy (Carl Zeiss) equipped with serial tile scanning. Image acquisition was processed with Zeiss LSM Image Browser Software and Adobe Photoshop CS5. Naïve tissue of terminal ileum and intestinal swiss roll sections were also analyzed with LSM 510 confocal microscopy. Spinal cord pictures were taken using a Zeiss Axioskop II fluorescent microscope.
Two-photon microscopy and surgical procedure
Before imaging, mice were anesthetized by intraperitoneal injection of 100 mg ketamine, 15 mg xylazine, and 2.5 mg acepromazine per kilogram. After a midway section through the abdominal wall and peritoneum, the ileum was externalized carefully and immobilized using a custom-built imaging chamber. The lumen was exposed using a cautery (AARON Medical) and carefully flushed with PBS to remove fecal material. During imaging, mice were supplied with oxygen and their core temperature was maintained at 37°C with a warming plate. Imaging was performed using the Ultima 2-photon microscope fitted with a Ti-sapphire Deep-sea Mai Tai pulsed laser with a water immersed 20× objective, the excitation wavelength was set to 970 nm. To create time-lapse sequences, we typically scan 5 to 30 μm of tissue depth, at 5-μm Z-steps every 20 to 40 seconds. The movement of the cells was visualized using Volocity 6.0 software (Improvision Ltd).
Flow cytometry
The following fluorochrome-labeled monoclonal antibodies and staining reagents were used according to the manufacturer's protocols: PE-conjugated anti-CD115 (eBioscience), allophycocyanin-conjugated anti-Gr1 (Ly6C/G, eBioscience), and peridinin chlorophyll protein-conjugated anti-CD11b (eBioscience) were used for blood monocyte staining. Pacific blue-conjugated anti-CD45.2 (eBioscience) was used for intestinal cell staining. CD45-allophycocyanin (eBioscience) was used for kidney and lung staining. Cells were analyzed on a LSR cytometer (BD Biosciences) using FlowJo 7.6 software (TreeStar).
ELISA
For the specific detection of transgene-encoded CX3CL1, it was captured from the serum by rat anti–mouse CX3CL1 (4 μg/mL, R&D Systems). As a secondary antibody, we used biotinylated chicken anti-cMyc (1:5000, Immunology Consultant Laboratories) and streptavidin-HRP. IL-6 and endogenous and transgene-encoded CX3CL1 from the serum were measured by ELISA kit (DuoSet ELISA kit, R&D Systems) following the manufacturer's protocol.
Assessment of TED formation
Indicated mice were challenged by oral gavage with Aspergillus conidia (300 μL of 1-2 × 108 conidia/mL). The following day, the mice were killed and analyzed by microscopic analysis for the presence of transepithelial dendrites (TEDs). Only TEDs that reached the lumen were considered for the quantification.
DSS-induced colitis model and murine colonoscopy
Mice received one cycle (7 days) of dextran sulfate sodium salt (DSS, MP Biomedicals, C-160110) treatment (1.5% in drinking water). For continuous monitoring of colitis, we used a high-resolution murine video endoscopic system (Karl Storz). Endoscopic quantification of colitis was graded as described.33
Statistical analysis
The results were analyzed by 1-way ANOVA followed by Bonferroni Multiple Comparison and are expressed as mean ± SEM or SD.
Results
Generation and analysis of CX3CL1cherry:CX3CR1gfp reporter mice
In-depth understanding of the physiologic role of the CX3C chemokine family requires the definition of the cell types that express the receptor/ligand pair in vivo. Analysis of CX3CL1 expression has so far, however, been restricted to in situ hybridization and immunohistochemistry,13,34,35 which provide limited resolution and preclude the analysis of live tissue. To map the distribution of CX3CL1-expressing cells in tissue context, we hence adopted a reporter gene approach we had previously applied to the CX3CR1 receptor.5 Specifically, we used homologous recombination to replace the first exon of the CX3CL1 gene in a BAC spanning the CX3CL1 locus (RP24–147I16) with a cherry reporter gene36 (Figure 1A). The modified CX3CL1 BAC DNA was then injected into fertilized oocytes of CB6F1 mice to establish transgenic CX3CL1cherry reporter animals, which were found healthy and fertile.
Generation and neuronal expression analysis of CX3CL1 BAC transgenic reporter mice. (A) Schematic of the RedET recombineering method used to replace the first CX3CL1 exon in the BAC RP24–147I16 with a gene encoding monomeric mCherry reporter gene. Note that the BspEI fragment used as transgene lacks the neighboring CCL22 and CCL17 genes. (B-D) Fluorescent microscopic analysis of hippocampus of CX3CL1cherry brain. (B) Red represents CX3CL1/cherry. (C) Blue represents NeuN staining. (D) Merge with mCherry and NeuN. (E) DCX stained hippocampus slice of CX3CL1cherry brain. Red represents CX3CL1/cherry; and green, DCX staining. (F) CGRP-stained section of CX3CL1cherry:CX3CR1gfp spinal cord. (G) Dorsal horn. (H) Dorsal root ganglion. Red represents CX3CL1/cherry; green, CX3CR1/GFP; and blue, CGRP staining. Lumbar regions (L3-L5). Note CX3CL1/cherry+ somata in dorsal horn but absence of CX3CL1/cherry expression from CGRP+ neuronal cell bodies in dorsal root ganglia.
Generation and neuronal expression analysis of CX3CL1 BAC transgenic reporter mice. (A) Schematic of the RedET recombineering method used to replace the first CX3CL1 exon in the BAC RP24–147I16 with a gene encoding monomeric mCherry reporter gene. Note that the BspEI fragment used as transgene lacks the neighboring CCL22 and CCL17 genes. (B-D) Fluorescent microscopic analysis of hippocampus of CX3CL1cherry brain. (B) Red represents CX3CL1/cherry. (C) Blue represents NeuN staining. (D) Merge with mCherry and NeuN. (E) DCX stained hippocampus slice of CX3CL1cherry brain. Red represents CX3CL1/cherry; and green, DCX staining. (F) CGRP-stained section of CX3CL1cherry:CX3CR1gfp spinal cord. (G) Dorsal horn. (H) Dorsal root ganglion. Red represents CX3CL1/cherry; green, CX3CR1/GFP; and blue, CGRP staining. Lumbar regions (L3-L5). Note CX3CL1/cherry+ somata in dorsal horn but absence of CX3CL1/cherry expression from CGRP+ neuronal cell bodies in dorsal root ganglia.
To simultaneously analyze the distribution of CX3CL1- and CX3CR1-expressing cells in tissue context, CX3CL1cherry reporter mice were crossed onto the CX3CR1gfp background.5 Confirming earlier reports based on in situ hybridization,13,34,35 fluorescent microscopy analysis of brain sections of CX3CL1cherry mice revealed prominent CX3CL1 expression in the NeuN+ mature neurons (Figure 1B-D) but not in the DCX+ neuronal precursors (Figure 1E). In steady state, CX3CL1 expression was absent from the brainstem, midbrain, and cerebellum (supplemental Figure 1A) in CX3CL1cherry:CX3CR1gfp mice but restricted to the hippocampus (supplemental Figure 1C), striatum (supplemental Figure 1D), and cortical layer II (supplemental Figure 1E). CX3CR1 was found exclusively expressed by microglial cells. Moreover, CX3CL1 expression was also readily detectable in neurons in the dorsal horn (Figure 1G), but not in CGRP-positive somata in the dorsal root ganglia (Figure 1H).
In the lung, CX3CL1 was found expressed on both alveolar and bronchial epithelial cells, including CCSP+ Clara cells (Figure 2A-B). Flow cytometric analysis of unchallenged lungs revealed CX3CL1/Cherry expression confined to nonhematopoietic (CD45−) cells, whereas CX3CR1/GFP expression was restricted to CD45+ cells (Figure 2C).
Expression of CX3CL1 in lung and kidney of BAC transgenic reporter mice. (A) Confocal imaging analysis of CX3CL1cherry:CX3CR1gfp lung. (B) Immunohistochemical analysis of CX3CL1cherry lung section. Green represents CCSP; red, CX3CL1/cherry; and blue, nuclear Hoechst staining. (C) Flow cytometric analysis of lung tissue of CX3CL1cherry:CX3CR1gfp mouse. (D-E) Confocal imaging analysis of CX3CL1cherry:CX3CR1gfp kidney. (F) Flow cytometric analysis of CX3CL1cherry:CX3CR1gfp kidney. Red represents CX3CL1/cherry; and green, CX3CR1/GFP.
Expression of CX3CL1 in lung and kidney of BAC transgenic reporter mice. (A) Confocal imaging analysis of CX3CL1cherry:CX3CR1gfp lung. (B) Immunohistochemical analysis of CX3CL1cherry lung section. Green represents CCSP; red, CX3CL1/cherry; and blue, nuclear Hoechst staining. (C) Flow cytometric analysis of lung tissue of CX3CL1cherry:CX3CR1gfp mouse. (D-E) Confocal imaging analysis of CX3CL1cherry:CX3CR1gfp kidney. (F) Flow cytometric analysis of CX3CL1cherry:CX3CR1gfp kidney. Red represents CX3CL1/cherry; and green, CX3CR1/GFP.
Analysis of kidneys revealed CX3CL1 expression by tubular epithelial cells and glomeruli, alongside with CX3CR1-expressing DCs37 (Figure 2D-E). Again, flow cytometric analysis of CX3CL1cherry:CX3CR1gfp mice indicated ligand and receptor expression in CD45− and CD45+ cells, respectively (Figure 2F).
In the intestinal lamina propria, CX3CR1 is known to be prominently expressed by a monocyte-derived macrophage population.20,38 Intestinal CX3CL1 expression, on the other hand, was reported for the epithelial cell layer.14 Analysis of CX3CL1cherry:CX3CR1gfp mice confirmed this discrete expression pattern (Figure 3A). Interestingly, however, closer examination of tissue sections revealed a heterogeneous expression of the CX3CL1 reporter in the intestinal epithelia. (Figure 3B-D). Analysis combined with periodic acid-Schiff staining established that the epithelial cells expressing constitutively high CX3CL1 levels were mucus-secreting goblet cells (Figure 3E). Consistent with the changing abundance and location of goblet cells along the proximal and distal gastrointestinal tract, CX3CL1/Cherry-positive cells were found in crypts or superficially in close contact with the gut lumen (supplemental Figures 2 and 3). CX3CL1/Cherry-positive epithelial cells, in particular goblet cells, and CX3CL1/GFP-positive macrophages in the underlying lamina propria were readily detectable by 2-photon microscopy in intravital studies (supplemental Video 1). Taken together, CX3CL1cherry:CX3CR1gfp mice provide a novel powerful tool to investigate the interface of the CX3C chemokine family in tissue context and live animals.
Intestinal epithelium restricted expression of CX3CL1. (A) Flow cytometric analysis of intestinal epithelial cell fraction obtained from WT mice and CX3CL1cherry mice. Cells are gated according to scatter (left). Middle dot plot: WT cells. Right plot: CX3CL1cherry cells. Note the presence of cherryint and cherryhigh cells in the CD45− epithelial cell fraction. (B) Two-photon imaging of CX3CL1cherry:CX3CR1gfp cecum. (C) Confocal imaging of terminal ileum of CX3CL1cherry:CX3CR1gfp mouse. (D) Confocal imaging of colon of CX3CL1cherry:CX3CR1gfp mouse. (E) Ileal section of CX3CL1cherry:CX3CR1gfp mouse shown under fluorescent microscope and after periodic acid-Schiff staining. Note that CX3CL1-positive large granular goblet cells. Red represents CX3CL1/cherry; and green, CX3CR1/GFP.
Intestinal epithelium restricted expression of CX3CL1. (A) Flow cytometric analysis of intestinal epithelial cell fraction obtained from WT mice and CX3CL1cherry mice. Cells are gated according to scatter (left). Middle dot plot: WT cells. Right plot: CX3CL1cherry cells. Note the presence of cherryint and cherryhigh cells in the CD45− epithelial cell fraction. (B) Two-photon imaging of CX3CL1cherry:CX3CR1gfp cecum. (C) Confocal imaging of terminal ileum of CX3CL1cherry:CX3CR1gfp mouse. (D) Confocal imaging of colon of CX3CL1cherry:CX3CR1gfp mouse. (E) Ileal section of CX3CL1cherry:CX3CR1gfp mouse shown under fluorescent microscope and after periodic acid-Schiff staining. Note that CX3CL1-positive large granular goblet cells. Red represents CX3CL1/cherry; and green, CX3CR1/GFP.
Generation and characterization of BAC transgenic mice expressing an obligatory secreted CX3CL1 entity
CX3CL1 is synthesized as a large 373-amino acid type I transmembrane protein with the CX3C chemokine module presented on top of a highly glycosylated stalk.1,2 Cleavage by metalloproteases can subsequently result in the generation of shed CX3CL1 fragments that, depending on the cleavage site, retain or lack the stalk39 (A.L., unpublished observations, August 2010). CX3CL1 can thus potentially act as membrane-anchored adhesion molecule or as classic diffusible chemoattractant; experimental evidence for both activities has been reported.23,24,40 Specific contributions of the membrane-anchored versus shed CX3CL1 isoforms to established in vivo activities of the CX3C chemokine family, however, remain to be shown.
To test for physiologic functions of membrane-tethered and secreted CX3CL1 entities, we decided to perform a structure/function analysis by generating mice that exclusively express a soluble CX3CL1 isoform. Specifically, we used the aforementioned BAC transgenesis approach to introduce into CX3CL1-deficient mice16 a CX3CL1 locus modified to encode an obligatory secreted, truncated CX3CL1 protein lacking the mucin stalk (CX3CL1105Δ). To allow for subsequent detection of the CX3CL1105Δ protein, a C-terminal c-myc epitope was added (Figure 4A; supplemental Figure 4A). In parallel, we reconstituted CX3CL1−/− mice with a BAC transgene encoding a WT CX3CL1 (CX3CL1395AA) harboring a silent mutation for detection (supplemental Figure 4B).
Generation and characterization of BAC transgenes encoding full-length or truncated CX3CL1 isoforms. (A) Schematic of homologous recombination strategy involving pDelSac shuttle vector to replace exon 3 of the CX3CL1 gene in the BAC RP24–147I16 with a fragment harboring a silent mutation and creating a XhoI site (CX3CL1395AA) or a truncated myc-tagged CX3CL1 exon (CX3CL1105Δ). (B) Analysis of transgene expression in intestinal tissue by RT-PCR analysis of ileal tissue of CX3CL1105Δ BAC transgenic mice. (C) Western blot analysis of intestinal epithelial cell fraction of CX3CL1105Δ BAC transgenic mice WT littermates using anti-myc antibody (9E10). Note that the 15-kDa band that is specific for the transgenic tissue-derived sample and probably represents the myc-tagged CX3CL1105Δ protein.
Generation and characterization of BAC transgenes encoding full-length or truncated CX3CL1 isoforms. (A) Schematic of homologous recombination strategy involving pDelSac shuttle vector to replace exon 3 of the CX3CL1 gene in the BAC RP24–147I16 with a fragment harboring a silent mutation and creating a XhoI site (CX3CL1395AA) or a truncated myc-tagged CX3CL1 exon (CX3CL1105Δ). (B) Analysis of transgene expression in intestinal tissue by RT-PCR analysis of ileal tissue of CX3CL1105Δ BAC transgenic mice. (C) Western blot analysis of intestinal epithelial cell fraction of CX3CL1105Δ BAC transgenic mice WT littermates using anti-myc antibody (9E10). Note that the 15-kDa band that is specific for the transgenic tissue-derived sample and probably represents the myc-tagged CX3CL1105Δ protein.
After confirming expression of the respective constructs in vitro (supplemental Figure 5A), we established BAC transgenic mice and backcrossed them to WT C57BL/6 mice. To confirm physiologic transgene expression, we performed RT-PCR analysis of intestinal tissue that revealed the expression of the transgenes in parallel to the endogenous CX3CL1 (Figure 4B). RT-PCR analysis of intestinal tissue of CX3CL1395AA mice revealed comparative expression levels of transgene-encoded CX3CL1 and endogenous CX3CL1, according to similar quantities of Xho cleavable and resistant PCR products (supplemental Figure 5B). Expression of the myc-tagged CX3CL1105Δ protein was furthermore confirmed by Western blot analysis of intestinal epithelia cells (Figure 4C).
Rescue of monocyte survival in CX3CL1-deficient mice requires transmembrane CX3CL1
CX3CR1 is prominently expressed by all circulating blood monocytes, both in human and mice.6 Levels of CX3CR1 expression are, however, notably distinct between the main monocyte subsets (ie, Ly6Clo/CD14dimCD16+ and Ly6Chi/CD14+CD16+/− cells) in the mouse and human.6 We recently showed that engagement of CX3CR1 by its ligand provides a critical survival signal, in whose absence (in CX3CR1gfp/gfp or CX3CL1−/− mice) the number of circulating Ly6Clo CX3CR1hi monocytes is significantly reduced.41 This finding provided a potential mechanistic explanation for the relative protection of CX3CR1 or CX3CL1-deficient animals from atherosclerosis and complications after arterial injury.19,42,43 To probe for a role of the shed CX3CL1 isoform in the control of monocyte survival, we investigated whether introduction of our CX3CL1105Δ BAC transgene would rescue Ly6Clo monocyte survival in CX3CL1−/− mice. To confirm the exposure of leukocytes to the secreted CX3CL1 entity in CX3CL1105Δ:CX3CL1−/− mice, we took advantage of the fact that shed, presumably neuronal-derived CX3CL1 becomes detectable in serum, when CX3CR1 is absent from the peripheral hematopoietic compartment (Figure 5A).44 Accordingly, the myc-tagged CX3CL1 isoform was readily detectable by a myc-epitope directed ELISA in the serum of [CX3CR1gfp/gfp > CX3CL1105Δ:CX3CL1−/−] chimeras (Figure 5B). However, only the CX3CL1395AA transgene encoding full-length CX3CL1 restored Ly6Clo monocyte frequencies to WT levels (Figure 5C). In contrast, the Ly6Clo monocyte compartment remained reduced in CX3CL1105Δ:CX3CL1−/− mice. This finding was corroborated by the analysis of BM chimeras generated by transferring CX3CR1gfp/+ BM into CX3CL1105Δ:CX3CL1−/− and CX3CL1395AA:CX3CL1−/− mice. Again, only the membrane-anchored but not the secreted CX3CL1 isoform resulted in a significant rescue of Ly6Clo monocytes (supplemental Figure 6A).
Restoration of CX3CR1hi Ly6Clo monocyte survival. (A-B) Serum ELISA of indicated BM chimeras for the presence of secreted endogenous and transgene-encoded CX3CL1 isoforms. (A) Detection with anti-CX3CL1 antibody. (B) Detection of CX3CL1105Δ transgene encoded CX3CL1 with anti-myc antibody (9E10). ND indicates not detectable. (C) Flow cytometric analysis of blood monocytes of WT mice, CX3CL1−/− mice, CX3CL1395AA:CX3CL1−/− mice, and CX3CL1105 Δ:CX3CL1−/− mice. (Top) Representative analysis of Ficoll-enriched WT blood cell sample indicating detection of Ly6Clo and Ly6Chi CD115+ monocyte populations. Bar diagram bar represents percentage of Ly6Clo monocytes (R3) of total blood monocyte (R2) for indicated mouse strains. N = 3 or 4 per group in 6 independent experiments. Error bars represent mean ± SEM. ***P < .001 (1-way ANOVA followed by Bonferroni Multiple Comparison).
Restoration of CX3CR1hi Ly6Clo monocyte survival. (A-B) Serum ELISA of indicated BM chimeras for the presence of secreted endogenous and transgene-encoded CX3CL1 isoforms. (A) Detection with anti-CX3CL1 antibody. (B) Detection of CX3CL1105Δ transgene encoded CX3CL1 with anti-myc antibody (9E10). ND indicates not detectable. (C) Flow cytometric analysis of blood monocytes of WT mice, CX3CL1−/− mice, CX3CL1395AA:CX3CL1−/− mice, and CX3CL1105 Δ:CX3CL1−/− mice. (Top) Representative analysis of Ficoll-enriched WT blood cell sample indicating detection of Ly6Clo and Ly6Chi CD115+ monocyte populations. Bar diagram bar represents percentage of Ly6Clo monocytes (R3) of total blood monocyte (R2) for indicated mouse strains. N = 3 or 4 per group in 6 independent experiments. Error bars represent mean ± SEM. ***P < .001 (1-way ANOVA followed by Bonferroni Multiple Comparison).
Collectively, these results suggest that the survival signal for circulating Ly6Clo CX3CR1hi monocytes depends on their encounter of membrane-anchored CX3CL1.
A secreted diffusible CX3CL1 isoform suffices to promote transepithelial dendrite formation by intestinal CX3CR1+ macrophages
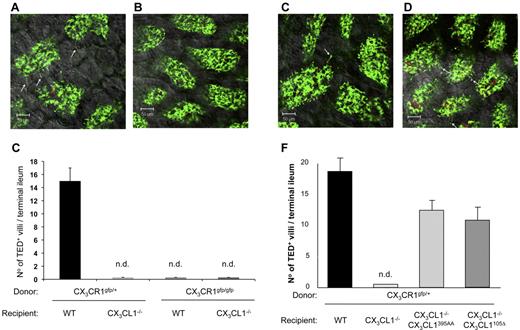
Rescigno et al were the first to report that intestinal mononuclear phagocytes could penetrate epithelial tight junctions to reach the gut lumen for content sensing.45 Taking advantage of CX3CR1gfp mice,5 we subsequently demonstrated these extensions in tissue context and established that formation of TEDs by what are now considered CX3CR1-expressing macrophages46 required the CX3CR1 chemokine receptor.20 In light of a report challenging this notion47 and to confirm the dependence of TED formation on CX3CR1/L1 interaction in ligand mutants, we generated mixed BM chimeras by transferring heterozygote and homozygote mutant CX3CR1gfp BM into lethally irradiated WT and CX3CL1−/− mice. TED formation in the respective chimeras was further boosted by oral gavage with A fumigatus conidia.48 As seen in Figure 6A-C, TEDs were readily formed in the terminal ileum of [CX3CR1gfp/+ > WT] mice but were undetectable in [CX3CR1gfp/+ > CX3CL1−/−] animals, as well as in [CX3CR1gfp/gfp > WT] controls. Collectively, this confirms the requirement of CX3CR1/CX3CL1 interactions for their formation.
Restoration of intestinal transepithelial dendrite formation. (A-B) Representative confocal image of small intestinal villi demonstrating TEDs formed by CX3CR1/GFP+ lamina propria macrophages in panel A (CX3CR1gfp/+ > WT) and panel B (CX3CR1gfp/+ > CX3CL1−/−) BM chimeras after Aspergillus conidia challenge (1-2 × 108 conidia/mL). Green represents lamina propria macrophages; red, conidia; and yellow, merge. Arrows indicate TEDs. (C) Bar diagram summarizing TED quantification in indicated BM chimeras. Note the absence of extension in mice with impaired CX3CR1/L1 interactions. Only TEDs that reached the lumen were considered for the quantification. Results are 1 representative of 3 experiments (N = 4). Error bars represent SD. n.d. indicates not detectable. (D-E) Representative confocal image of small intestinal villi demonstrating TEDs formed by CX3CR1/GFP+ lamina propria macrophages in panel D (CX3CR1gfp/+ > CX3CL1395AA:CX3CL1−/−) and panel E (CX3CR1gfp/+ > CX3CL1105Δ:CX3CL1−/−) BM chimeras after Aspergillus conidia challenge. Arrows indicate TEDs. (F) Bar diagram summarizing TED quantification in indicated BM chimeras. Note the restoration of extensions in CX3CL1−/− mice harboring the CX3CL1395AA and CX3CL1105 Δ transgenes. Results are one representative of 3 independent experiments (N = 3-5). Error bars represent SD.
Restoration of intestinal transepithelial dendrite formation. (A-B) Representative confocal image of small intestinal villi demonstrating TEDs formed by CX3CR1/GFP+ lamina propria macrophages in panel A (CX3CR1gfp/+ > WT) and panel B (CX3CR1gfp/+ > CX3CL1−/−) BM chimeras after Aspergillus conidia challenge (1-2 × 108 conidia/mL). Green represents lamina propria macrophages; red, conidia; and yellow, merge. Arrows indicate TEDs. (C) Bar diagram summarizing TED quantification in indicated BM chimeras. Note the absence of extension in mice with impaired CX3CR1/L1 interactions. Only TEDs that reached the lumen were considered for the quantification. Results are 1 representative of 3 experiments (N = 4). Error bars represent SD. n.d. indicates not detectable. (D-E) Representative confocal image of small intestinal villi demonstrating TEDs formed by CX3CR1/GFP+ lamina propria macrophages in panel D (CX3CR1gfp/+ > CX3CL1395AA:CX3CL1−/−) and panel E (CX3CR1gfp/+ > CX3CL1105Δ:CX3CL1−/−) BM chimeras after Aspergillus conidia challenge. Arrows indicate TEDs. (F) Bar diagram summarizing TED quantification in indicated BM chimeras. Note the restoration of extensions in CX3CL1−/− mice harboring the CX3CL1395AA and CX3CL1105 Δ transgenes. Results are one representative of 3 independent experiments (N = 3-5). Error bars represent SD.
Having established the absence of TEDs in CX3CL1-deficient mice, we next asked whether their formation could be restored by introduction of the obligatory secreted CX3CL1 isoform. To this end, CX3CR1gfp/+ BM was transferred into irradiated BAC transgenic CX3CL1395AA:CX3CL1−/− mice and CX3CL1105Δ:CX3CL1−/− mice, as well as nontransgenic CX3CL1-deficient animals. Seven weeks after BM transfer, the animals were gavaged with A. fumigatus conidia and analyzed for the presence of TEDs in their terminal ileum. As expected, TEDs were absent from CX3CL1−/− recipient mice. Introduction of the BAC transgene encoding the WT CX3CL1 restored TED formation (Figure 6D), albeit at slightly reduced numbers (Figure 6F), thus validating our approach. More importantly, however, TEDs were also readily observed in (CX3CR1gfp/+ > CX3CL1105Δ:CX3CL1−/−) chimeras (Figure 6E-F), establishing that epithelial expression of a secreted CX3CL1 entity was sufficient to allow lamina propria CX3CR1-expressing macrophages to extend protrusions toward the lumen.
Collectively, these results establish that TED formation can be promoted by a secreted, potentially diffusible CX3CL1 entity lacking the glycosylated stalk and is hence independent of signaling by the transmembrane CX3CL1 ligand toward the intestinal epithelial cells.
Restoration of transepithelial dendrites affects the robustness of CX3CL1-deficient mice to withstand a colitis challenge
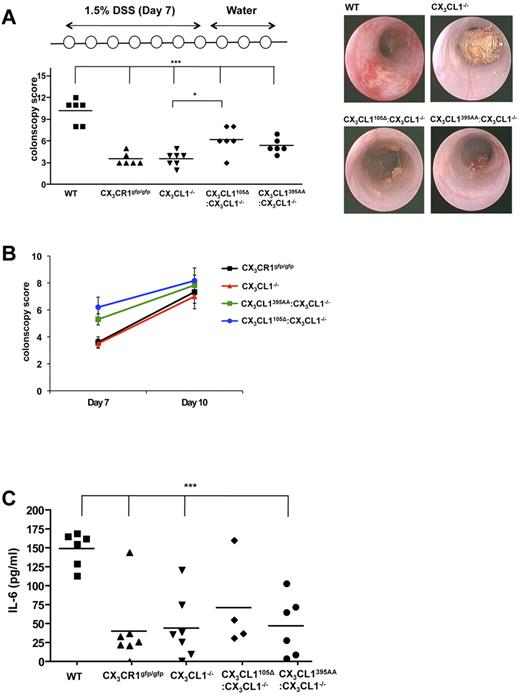
CX3CR1-deficient mice have been reported to display altered sensitivity to protocols inducing gut inflammation.49-51 However, in none of the cases, it has been tested whether the observed phenotype was specifically because of impaired TED formation by CX3CR1+ intestinal macrophages. To investigate this issue, we challenged WT mice, CX3CR1gfp/gfp mice, CX3CL1−/− mice, and CX3CL1−/− mice harboring the CX3CL1395AA or CX3CL1105Δ BAC transgenes by oral administration of DSS. This established colitis model is characterized by ulceration and submucosal inflammation provoked by disruption of the epithelial barrier and exposure to luminal microbiota.52 As previously reported,48 CX3CR1−/− mice displayed a relative protection toward the DSS challenge compared with littermate controls, as did CX3CL1−/− mice judged by endoscopic scores and their serum IL-6 levels (Figure 7). Interestingly, however, when TEDs where restored in CX3CL1−/− mice by introduction of the CX3CL1395AA and CX3CL1105Δ transgenes, the animals developed more severe colitis, as judged by endoscopy and histologic grading (Figure 7A-B; and data not shown). Notably, repeated endoscopy showed that the effect was more pronounced in early stages of colitis development (day 7) compared with a later time point (day 10; Figure 7C). Given the fact that the secreted CX3CL1 failed to reconstitute monocyte survival, these data suggest a direct link of the ability of CX3CR1 macrophages to reach the gut lumen (ie, their formation of TEDs) and the propensity to develop DSS-induced colitis.
Susceptibility to DSS-induced colitis. (A) Colonoscopy scores of WT mice, CX3CR1−/− mice, CX3CL1−/− mice, CX3CL1395AA:CX3CL1−/− mice, and CX3CL1105Δ:CX3CL1−/− mice after indicated DSS challenge (day 7). (B) Representative colonoscopy images taken from mice at day 7. (C) Change of colonoscopy scores between day 7 and day 10. (D) ELISA of serum of WT mice, CX3CR1−/− mice, CX3CL1−/− mice, CX3CL1395AA:CX3CL1−/− mice, and CX3CL1105Δ:CX3CL1−/− mice after indicated DSS challenge for IL-6 (day 10). Results are one representative of 2 independent experiments (N = 5-7). *P < .05 (1-way ANOVA followed by Bonferroni Multiple Comparison). ***P < .001 (1-way ANOVA followed by Bonferroni Multiple Comparison).
Susceptibility to DSS-induced colitis. (A) Colonoscopy scores of WT mice, CX3CR1−/− mice, CX3CL1−/− mice, CX3CL1395AA:CX3CL1−/− mice, and CX3CL1105Δ:CX3CL1−/− mice after indicated DSS challenge (day 7). (B) Representative colonoscopy images taken from mice at day 7. (C) Change of colonoscopy scores between day 7 and day 10. (D) ELISA of serum of WT mice, CX3CR1−/− mice, CX3CL1−/− mice, CX3CL1395AA:CX3CL1−/− mice, and CX3CL1105Δ:CX3CL1−/− mice after indicated DSS challenge for IL-6 (day 10). Results are one representative of 2 independent experiments (N = 5-7). *P < .05 (1-way ANOVA followed by Bonferroni Multiple Comparison). ***P < .001 (1-way ANOVA followed by Bonferroni Multiple Comparison).
Discussion
Here we used a BAC transgenesis approach to study the in vivo expression pattern and structure/function aspects of the unique CX3C chemokine CX3CL1, known as fractalkine. Specifically, we generated reporter animals that harbor genes encoding red and green fluorescent reporter proteins under the control of the CX3CR1 and CX3CL1 promoter control, respectively. Furthermore, we probed for distinct activities of membrane-tethered and shed entities of the CX3CL1 ligand by testing their potential to reconstitute known phenotypes resulting from CX3CL1 deficiencies.
CX3CL1cherry:CX3CR1gfp mice provide a unique tool to study the CX3C interface under various physiologic conditions, including steady state, the main focus of our present study, as well as inflammatory and pathologic conditions. Using a combination of flow cytometric, immunohistochemical, and intravital microscopic approaches, we show that, in all tissues analyzed under homeostasis, CX3CR1 and CX3CL1 expression is confined to hematopoietic and nonhematopoietic compartments, respectively.
In the healthy brain, CX3CL1 expression was found in hippocampal neuronal somata, as well as neurons in the striatum and specific cortical layers. CX3CL1/Cherry expression was absent from neuronal bodies in the brain stem, hypothalamus, midbrain, and cerebellum. CX3CL1/Cherry expression was detected in NeuN+ mature neurons, but absent from DCX+ neuronal precursors. In the spinal cord, CX3CL1 expression was observed in neurons in the dorsal horn, but not the dorsal root ganglia. Expression of the receptor CX3CR1 is restricted to microglial cells throughout the brain and spinal cord, as previously reported.5 No prominent colocalization of CX3CR1- and CX3CL1-expressing cells was observed. Of note, however, expression of the cherry and gfp transgenes in CX3CL1cherry:CX3CR1gfp mice reports on the respective promoter activities but not the actual presence of the receptor/ ligands, as would be the case with fusion proteins. This fact could be particularly relevant with respect to axon and dendrite-bearing neurons and the highly ramified microglia,53 which might well interact through CX3CL1 and CX3CR1 expressed on extensions distant from the actual cell body.
CX3CL1/R1-based neuronal-microglia communication has been proposed to affect the severity of neurodegenerative diseases. Thus, several research groups showed a neuroprotective function of CX3CL1 in vitro.54-56 Furthermore, CX3CR1 deficiency was reported to cause microglial neurotoxicity57 and to promote τ-associated neurodegenerative diseases (tauopathies).58 CX3CR1-deficient mice seem also less sensitive to neuropathic pain.59 Supporting a role of the CX3C axis in pain sensation, we found CX3CL1/Cherry expressed by dorsal horn interneurons. However, in contrast to a previous report,60 CX3CL1 expression was in steady state absent from the dorsal root ganglia sensory neurons that project CGRP fibers to these interneurons. CX3CL1cherry:CX3CR1gfp mice should be instrumental in future studies to define the functional roles of the CX3C chemokine family under pathologic conditions in the central nerve system.
In the healthy lung, we found CX3CL1 expressed by bronchial and alveolar epithelial cells, whereas CX3CR1 expression was confined to resident lung myeloid cells. In humans, CX3CL1 was reported to be induced on bronchial epithelial cells and smooth muscle cells by inflammatory cytokines.61 CX3CR1 is expressed by alveolar DCs that arise under inflammatory conditions from blood monocytes.62 Furthermore, CX3CR1 expression and curiously also CX3CL1 expression can reportedly be triggered on alveolar macrophages by exposure to cigarette smoke.63,64 Moreover, a recent study suggested that under inflammatory conditions CX3CL1 acts as survival factor for CX3CR1+ TH2 cells affecting lung pathology.11
CX3CR1 is expressed by renal DCs and macrophages that form a contiguous network throughout the entire kidney.37 Analysis of the CX3CL1cherry:CX3CR1gfp reporter mice revealed constitutive CX3CL1 expression by glomerular endothelial cells and renal tubular epithelial cells. Notably, CX3CL1 expression was reported to be up-regulated in renal failure and a lupus nephritis model.65,66 Moreover, CX3CR1-deficient mice were reported to be relatively protected in a renal ischemic-reperfusion injury model because of reduced myeloid inflammatory cell infiltrates and reduced fibrosis.67,68 Collectively, CX3C interactions seem to play a critical role in inflammations both affecting leukocyte recruitment and local intercellular communication and CX3CL1cherry:CX3CR1gfp mice should prove useful to decipher the specific role of the CX3C chemokine family in these processes.
Analysis of CX3CL1cherry:CX3CR1gfp double reporter mice corroborated earlier reports of CX3CL1 expression by intestinal columnar epithelial cells in terminal ileum, colon, and cecum. Moreover, our immunohistochemical and intravital microscopic analysis revealed that CX3CL1 expression was especially prominent in mucus-secreting goblet cells in ileum and colon. The physiologic significance of this CX3CL1 expression by goblet cells and its relevance in their potential cross-talk with CX3CR1-expressing intestinal macrophages remain to be studied.
CX3CL1 is a unique chemokine in that it is synthesized as type I membrane protein and thus membrane-anchored with the CX3C chemokine domain displayed on an extended mucin-like stalk.1,2 Subsequent cleavage by metalloproteases, such as ADAM 10 and ADAM 17, can however result in the release of a shed CX3CL1 isoform.26-28 To probe for distinct functions of these membrane-tethered and soluble CX3CL1 entities, we generated BAC transgenic mice expressing a WT or an obligatory secreted chemokine variant and introduced them onto a CX3CL1-deficient genetic background.16 We then asked whether these transgenes would be able to reconstitute known deficiencies caused by the absence of CX3CL1/R1 interactions.
We had previously shown that human and mouse monocytes depend on CX3CR1 engagement as a survival signal.41 In that study, recombinant soluble CX3CL1 was able to protect human CD14++CD16− monocytes from death induced by serum deprivation in vitro.41 In contrast, in the present study, only the membrane anchored full-length CX3CL1 was able to rescue the Ly6Clo CX3CR1hi blood monocytes in [CX3CR1gfp/+ > CX3CL1395AA:CX3CL1−/−] BM chimeras. However, interactions of CX3CR1 and soluble CX3CL1 were insufficient to restore monocyte survival in CX3CL1105Δ:CX3CL1−/− mice. In vivo monocytes seem hence to require the encounter of membrane-anchored CX3CL1, rather than shed CX3CL1 for their survival. Notably, there is no general CX3CL1/Cherry expression detectable on vascular endothelium of CX3CL1cherry:CX3CR1gfp mice in steady state. We hence speculate that circulating monocytes encounter CX3CL1 on their transient passage through specific CX3CL1-decorated vascular compartments, such as the choroid plexus, the lung capillaries, or the kidney glomeruli.
Intestinal lamina propria resident CX3CR1+ macrophages have been reported to extend TEDs to sense and potentially sample luminal pathogens in the lamina propria. We had previously shown that TED formation was dependent on CX3CR1.20 However, Chieppa et al subsequently demonstrated that certain, albeit morphologic distinct, extensions might be formed in the absence of CX3CR1 but rather relied on MyD88-dependent TLR signaling by epithelial cells.47 Here we show, by establishing the absence of TEDs in both CX3CR1- and CX3CL1-deficient mice, that their formation indeed depends on the CX3CL1/R1 axis. Moreover, transgenic expression of both a full-length (CX3CL1395AA) and a truncated, obligatory secreted CX3CL1 entity (CX3CL1105Δ) in CX3CL1-deficient mice restored the ability of CX3CL1+ macrophages to extend dendrites toward the lumen. The physiologic relevance of TEDs is under debate; and, specifically, it remains to be shown how they are related to reported gut-related phenotypes of CX3CR1 KO mice.49-51,69 Interestingly, we establish that TEDs, probably by allowing CX3CR1+ macrophages to sense the gut lumen, can contribute to the robustness of gut homeostasis toward a colitis challenge.
By demonstrating that distinct activities of the CX3C axis, such as the promotion of TED formation and monocyte survival, depend on the presence of shed and membrane-anchored CX3CL1 entities, respectively, we establish, for the first time, unique in vivo activities of the structurally discrete CX3CL1 isoforms. Notably, the anchorage of the CX3C domain might be particularly critical in the circulation, where shed moieties would be expected to be rapidly diluted and might hence not reach critical local concentrations. In the tissue context, however, the soluble CX3C entity might be able to build the required local gradient and hence suffice.
Collectively, the accumulating evidence from our study and the literature suggests CX3CR1/L1 interactions as critical components in the development of inflammatory diseases in diverse organs, including the lung, intestine, and kidney, as well as neurodegenerative diseases. Although much remains to be elucidated, the CX3C family remains an interesting therapeutic target, and the future study on molecular mechanism of the CX3C axis might well provide new avenues for the treatment of inflammatory diseases.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all members of the S.J. laboratory for comments and Dr Chava Rosen for expert advice.
This work was supported by the United States-Israel Binational Science Foundation (S.J.), the Deutsche Forschungsgemeinschaft Research Unit 1336, and the Leona M. and Harry B. Helmsley Charitable Trust (S.J.). S.J. is a Helmsley Scholar at the Crohn's & Colitis Foundation.
Authorship
Contribution: K.-W.K. designed and generated the transgenic mice and performed their characterization; A.V.-E., J.F., and E.S. provided expert technical help; E.Z. helped with the colitis experiment; S.A.L. provided critical KO mice; G.S. and A.L. provided advice; and S.J. designed the study; and S.J. and K.-W.K. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steffen Jung, Department of Immunology, Weizmann Institute of Science, PO Box 26, Rehovot 76100, Israel; e-mail: s.jung@weizmann.ac.il.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal