Abstract
Hematopoiesis is the process whereby BM HSCs renew to maintain their number or to differentiate into committed progenitors to generate all blood cells. One approach to gain mechanistic insight into this complex process is the investigation of quantitative genetic variation in hematopoietic function among inbred mouse strains. We previously showed that TGF-β2 is a genetically determined positive regulator of hematopoiesis. In the presence of unknown nonprotein serum factors TGF-β2, but not TGF-β1 or -β3, enhances progenitor proliferation in vitro, an effect that is subject to mouse strain-dependent variation mapping to a locus on chr.4, Tb2r1. TGF-β2–deficient mice show hematopoietic defects, demonstrating the physiologic role of this cytokine. Here, we show that TGF-β2 specifically and predominantly cell autonomously enhances signaling by FLT3 in vitro and in vivo. A coding polymorphism in Prdm16 (PR-domain-containing 16) underlies Tb2r1 and differentially regulates transcriptional activity of peroxisome proliferator-activated receptor-γ (PPARγ), identifying lipid PPAR ligands as the serum factors required for regulation of FLT3 signaling by TGF-β2. We furthermore show that PPARγ agonists play a FLT3-dependent role in stress responses of progenitor cells. These observations identify a novel regulatory axis that includes PPARs, Prdm16, and TGF-β2 in hematopoiesis.
Introduction
HSCs can self-renew and give rise to all the cells of the blood and the immune system. As they differentiate, HSCs progressively lose their self-renewal capacity and generate lineage-restricted multipotential progenitor (MPP) cells that in turn give rise to mature cells.1,2 Among inbred mouse strains there is extensive genetically determined variation in the hematopoietic stem and progenitor cell (HSPC) compartment. Traits that vary continuously across genetically different persons are determined by the contribution of multiple loci and are called quantitative trait loci (QTLs). Within the hematopoietic system, QTLs have been mapped that affect HSPC number, cycling activity, cytokine responsiveness, mobilization, aging, and gene expression.3-13 Although phenotypic consequences of individual polymorphism may be subtle, the identification of genes underlying QTLs is a powerful approach to show novel regulatory mechanisms.14-16 Furthermore, because many genes or pathways that show quantitative genetic variation in the mouse model also do so in humans,16 this approach allows insight into human genetic variation in a more targeted fashion than is currently possible in genome-wide association studies. Although multiple suggestive “hematopoietic” QTLs have been mapped, only 1 of the underlying genes been identified thus far, latexin, which is involved in the negative regulation of stem cell pool size.13,14
We have shown that signaling by one isoform of TGF-β, TGF-β2, is subject to quantitative genetic variation in HSPCs.17 Although TGF-βs either inhibit HSPC growth in vitro or have no effect on them in vivo,18 we have previously shown that TGF-β2 is a positive regulator of HSPCs. Studies in Tgfb2-deficient mice showed lower frequency, cycling activity, and in vitro proliferative capacity of HSPCs and showed decreased serial repopulating capacity of HSCs compared with wild-type (wt) littermates. In vitro, TGF-β2 has a biphasic dose response on the proliferation of purified HSPCs, defined as lineage−Sca1+Kit+ or LSK cells. Its effect is stimulatory at concentrations in the picogram range, whereas at higher concentrations inhibition of proliferation occurs.17 This proliferative effect requires nonprotein low molecular weight (MW) factors19 and is a quantitative trait, as the magnitude of this effect depended on genetic background.17 We mapped a QTL for this trait with a suggestive level of significance to the telomeric region of chr.4.17 Congenic mice, in which the telomeric 20cM of chr.4 was introgressed from the DBA/2 into the C57BL/6 background (B6.D2-chr.4 mice), had decreased TGF-β2 responsiveness compared with the parental C57BL/6 mice, unequivocally confirming the location of this QTL, Tb2r1, on chr.4.20 Here, we report that TGF-β2 specifically enhances responsiveness of HSPCs to the hematopoietic cytokine, FLT3 ligand (FLT3L), in a predominantly cell autonomous fashion and that, through fine-mapping, we identified Prdm16 (PR-domain-containing 16) as the gene underlying Tb2r1. A coding polymorphism in Prdm16, a gene previously shown by us and by others to be required for the maintenance of HSCs,21,22 affects its capacity to enhance the agonist-induced transcriptional activity of the ligand-activated transcription factor, peroxisome proliferator activated receptor γ (PPARγ).23 Synthetic ligands of PPARγ increase proliferation of HSPCs induced by FLT3L, an effect that requires TGF-β signaling, indicating that the low MW serum factors required for the proliferative effects of TGF-β2 in HSPCs include natural ligands of PPARγ. These data show that QTL analysis can reveal novel regulatory mechanisms and can generate physiologically relevant insights into gene function and into the subfunctionalization of genes that complement those obtained from knockout approaches.
Methods
Mice
C57BL/6J, Tgfb2tm1doe/J,24 B6.129-Ppargtm2Rev/J,25 and B6.Cg-Tg(Mx1-cre)1Cgn/J26 mice were purchased from The Jackson Laboratories. All were backcrossed onto the C57BL/6 background for ≥ 10 generations. FLT3−/− mice27 were obtained from Dr I. Lemischka (Mount Sinai School of Medicine). Animals were housed in a specific pathogen-free facility. Experiments and animal care were performed with approval from the Mount Sinai Institutional Animal Care and Use Committee.
Antibodies and cytokines
FITC-conjugated anti-CD2, -CD3ϵ, -CD8α, -CD4, -CD19, -B220, -Gr1, -Mac1, -CD48; PE-conjugated anti-FLT3, anti-pAKT(pT308), IgG2a, IgG1; PECy7-conjugated streptavidin; and APC-Alexa Fluor 750–conjugated anti–c-kit were purchased from eBiosciences. PE-conjugated anti-Sca1, -CD34, PerCP-Cy5.5–conjugated anti-Mac1 and streptavidin, APC-conjugated anti–c-kit, -IgM, and goat anti–rat Ab, PerCP-conjugated streptavidin, PECy7-conjugated anti-CD19 and -CD4, APC-Cy7–conjugated streptavidin, anti-CD19 and -CD8, Pacific Blue–conjugated anti-B220, and Alexa Fluor 647–conjugated anti-pSTAT5(pY694), -pERK1/2(pT202/pY204), and IgG1-κ were purchased from BD PharMingen. PE-, APC-, and PECy7-conjugated anti-CD150 and Pacific Blue–conjugated anti-Sca1 were purchased from BioLegend. Recombinant mouse Kit ligand, thrombopoietin, IL-6, IL-3, IL-1α, TGF-β1, and TGF-β2, and polyclonal anti-FLT3L neutralizing and goat IgG isotype control Abs, as well as rmFLT3L for in vivo use, were purchased from PeproTech. Human IL-11 was a gift from Dr G. Keller (Ontario Cancer Institute). SB-431542 and GW9662 were purchased from Tocris Biosciences, and rosiglitazone was purchased from Cayman Chemical Company.
Poly(I:C) injections and Ppargfl/fl mouse genotyping PCR
Ppargfl/fl.Mx1-Cre+/− and littermate Ppargfl/fl.Mx1-Cre−/− control mice received 625 μg of poly(I:C) (Sigma-Aldrich) intraperitoneally every other day (4 injections). To test for Pparg deletion, RNA was isolated from BM with the use of TRIzol (Invitrogen) and RNeasy Micro Kit (QIAGEN), and reverse transcribed with SuperScript III (Invitrogen). Sense 5′-GTCACGTTCTGACAGGACTGTGTGAC-3′ and antisense 5′-TATCACTGGAGATCTCCGCCAACAGC-3′ primers anneal to regions in exons A1 and 4 of PPARγ, respectively, distinguishing full-length (700-bp) and recombined (300-bp) transcripts. PCR genotyping was performed with the following primers: for the loxP site, 5′-TGGCTTCCAGTGCATAAGTT-3′ and 5′-TGTAATGGAAGGGCAAAAGG-3′; and for the Cre transgene, 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and 5′-GTGAAACAGCATTGCTGTCACTT-3′. Genomic DNA was amplified 40 cycles at 94°C for 30 seconds, 60°C for 60 seconds, and 72°C for 60 seconds.
Cell sorting, flow cytometry, and culture of LSK cells
Femora and tibia of mice were flushed with cold DMEM (Cellgro; Mediatech) containing 2% FBS (Hyclone) and penicillin/streptomycin (Cellgro). Low-density BM cells were stained with Abs for lineage Ags (CD2, CD3ϵ, CD8α, CD4, CD19, B220, Ter119, Gr1, Mac1), Sca1, and c-Kit and were isolated by cell sorting with the use of FACSVantage SE, Influx (BD Biosciences), or MoFlo (CytoMation) sorters, to obtain LSK cells. Flow cytometric analysis was performed on a Special Order LSRII with DiVa software (BD Biosciences). Data were analyzed with FlowJo software.
LSK cells were cultured in triplicate at 20-60 cells per well in flat bottom 96-well plates in StemPro34 medium (Invitrogen), 10% FCS (Hyclone or PAA), penicillin/streptomycin, in the presence of various cytokines or Abs as mentioned for each experiment. One to 2 hours after plating, the exact number of cells per well was determined by visually counting at 200× magnification. After 5 days of liquid culture in a humidified incubator with 5% CO2 at 37°C the cells were again counted.
Transplantation assays
BM from Ppargfl/fl.Mx1-Cre+/− and littermate Ppargfl/fl.Mx1-Cre−/− control mice was analyzed for deletion of PPARγ 2 weeks after poly(I:C) injection. Pparg−/− or littermate Ppargfl/fl cells (0.5 × 106; C57BL/6 background, CD45.2+, donor) were mixed with 0.5 × 106 CD45.1+ C57BL/6 BM (competitor) and injected into lethally irradiated CD45.1+CD45.2+ (C57BL/6. C57BL/6.SJL-PtprcaPep3b/BoyJ F1) recipient mice. Ten to 12 weeks later, peripheral blood (PB) was stained for CD45.1, CD45.2, CD19, Thy1, Mac1, and Gr1.
Reporter gene assays
COS-7 cells were seeded at 0.5 × 105 cells/well in 12-well plates 1 day before transfection. Cells were transfected with 750 ng of 3 × DR1-luciferase reporter and, when indicated, with 150 ng of pSVSport-PPARγ and pJCXRα or empty vector together with 25 ng of pcDNA3PRDM16(B6) or pcDNA3PRDM16(DBA/2) with the use of FuGENE-6 reagent (Roche Diagnostics). Cells were incubated for 48 hours and, when indicated, were treated for 24 hours before harvest with 1μM rosiglitazone or vehicle (DMSO). Luciferase activity was assayed with the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase reporter gene measurements were normalized to Renilla luciferase activity.
Rosiglitazone diet
C57BL/6 mice received either chow or chow supplemented with rosiglitazone (10 mg/kg; Test Diet). Food was γ-irradiated according to the policy of the institutional animal care and use committee and animal facility at Mount Sinai School of Medicine.
Rosiglitazone and 5-FU treatment
5-Fluorouracil (5-FU; Sigma-Aldrich) was dissolved in PBS at 11.25 mg/mL and filtered through 0.2-μm SFCA filters (Corning). The effect of sublethal doses of 5-FU (150 mg/kg intraperitoneal) was measured by PB counts performed on days 3, 7, 10, and 14 after 5-FU administration with the use of a Beckman Coulter AcTdiff automated hematology machine. Rosiglitazone (15 mg/kg dissolved in DMSO) or DMSO was administered daily on days 0-4 by gavage.
FLT3L administration
rmFLT3L was injected subcutaneously (10 μg/d) in 1 × PBS with mouse serum albumin (MSA; Sigma-Aldrich; 1 μg/mouse/d). Control mice received only 1 μg/d MSA in 1× PBS.
Sequencing of Prdm16 around rs32942538
PCR on genomic DNA was performed with the following primers: forward 5′-TTTGGTTTGGTGGTGAGGTCGAAG-3′ and reverse 5′-CCCTTCCAGTTCCTGCCTAACTTT-3′; amplified by 1 cycle of 94°C for 5 minutes, then 30 cycles of 94°C for 30 seconds, 59°C for 30 seconds, and 72°C for 60 seconds, followed by 1 cycle of 72°C for 7 minutes. PCR product (164 bp) was purified with QIAquick Gel Extraction Kit (QIAGEN) and sequenced with the forward primer.
Statistical analysis
Student 2-tailed t test for paired samples was used, and data represent mean ± SEM. For the analysis of the effect of FLT3L in vivo 3-way ANOVA was used.
Results
TGF-β2 has a relatively modest enhancing effect on cytokine-supported proliferation of LSK cells in the presence of as yet unidentified low MW serum factors in vitro.17,19 The finding that Tgfb2+/− LSK cells proliferate ∼ 50% less in cultures supported by kit ligand (KL), FLT3L and thrombopoietin (TPO) than wt LSK cells suggests an at least partially cell autonomous effect of TGF-β2.17 LSK cells from B6.D2-chr.4 congenic mice, which are hyporesponsive to the proliferative effect of TGF-β2, were specifically hyporesponsive to FLT3L compared with LSK cells from parental C57BL/6 mice.20 Therefore, we hypothesized that TGF-β2 regulates signaling by FLT3L in an at least partially cell autonomous fashion. To test this hypothesis, we cultured Tgfb2+/− (Tgfb2−/− mice die at birth)24 or wt LSK cells (for sort windows see Figure 1A) with increasing concentrations of either FLT3L, KL and TPO or IL-3. Only FLT3L-supported proliferation of Tgfb2+/− LSK cells was significantly reduced compared with that of wt LSK cells (Figure 1B). Interestingly, addition of exogenous TGF-β2 did not rescue the FLT3L responsiveness defect of Tgfb2+/− LSK cells (Figure 1B), suggesting a predominantly cell autonomous effect. Similar to adult Tgfb2+/− LSK cells, the responsiveness of Tgfb2+/− E16 fetal liver LSK cells to FLT3L, but not to the combination KL/TPO, was ∼ 50% lower, whereas that of Tgfb2−/− fetal liver LSK cells was 70% lower compared with wt cells (Figure 1C). To further assess the cell autonomous nature of the TGF-β2 effect, we cultured wt LSK cells in the presence of the specific inhibitor of the type I TGF receptor kinase activity, SB-431542.28 SB-431542 attenuated the response of LSK cells to FLT3, but not to the combination KL/TPO, by 3-fold, an effect that was quantitatively similar to that of homozygous deletion of TGF-β2 (Figure 1D). Because in these experiments no TGF-β2 was added, SB-431542 must block endogenous and presumably autocrinely or paracrinely produced TGF-β. Importantly, FLT3L-induced proliferation of wt and Tgfb2+/− LSK cells was similar in serum-free conditions (Figure 1E), confirming the serum factor requirement for the enhancing effect of TGF-β2 on FLT3L-induced progenitor cell growth. Next, we examined whether TGF-β2 affected the clone size of FLT3-responsive cells or rendered more cells FLT3 responsive. We plated single LSK cells from wt or Tgfb2+/− mice in 96-well plates in the presence of FLT3L and measured the number of growing colonies, as well their clone size. Although the cloning efficiency of Tgfb2+/− LSK cells was similar to that of wt LSK cells, the average clone size was significantly smaller (Figure 1F), indicating that TGF-β2 enhances the growth of FLT3L-responsive cells but does not render FLT3L-resistant cells responsive.
Cell autonomous effect of TGF-β2 on FLT3L responsiveness. (A) Representative example of sort windows used for the isolation of LSK cells. (B) Tgfb2+/− and wt littermate LSK cell proliferation in response to FLT3L, the combination of KL/TPO, or IL3. Experiments in culture supported by FLT3L were also performed in the presence or absence of 1 pg/mL TGF-β2 (n = 3; *P ≤ .05 in the absence of TGF-β2). A 100% response is defined as that of wt LSK cells at 50 ng/mL FLT3L or IL-3 in the absence of TGF-β2. (C) Tgfb2−/− (n = 3), Tgfb2+/− (n = 5), and wt littermate (n = 4) E16 fetal liver LSK cell proliferation in the presence of 10% serum in response to FLT3L (left; *P < .005) or to KL plus TPO (right; P > .4). A 100% response is defined as that of wt LSK cells at 10 ng/mL FLT3L. (D) C57BL/6 LSK cell proliferation in response to FLT3L (left) or KL and TPO (right) in the presence of 10% serum, with or without SB-431542 (10μM in DMSO; n = 3; *P = .002). (E) Tgfb2+/− and wt littermate LSK cell proliferation in serum-free cultures in response to FLT3L (n = 3; P > .1). A 100% response is defined as that of wt LSK cells at 50 ng/mL FLT3L. (F) Clone size (left; n = 80 for wt, n = 83 for Tgfb2+/−; *P = .02 by Student's unpaired t test) and cloning efficiency (right; n = 4; P = .736) of single Tgfb2+/− or wt LSK cells in liquid cultures containing 50 ng/mL FLT3L and 10% serum. Data collected after 5 days of incubation. Error bars represent SEM.
Cell autonomous effect of TGF-β2 on FLT3L responsiveness. (A) Representative example of sort windows used for the isolation of LSK cells. (B) Tgfb2+/− and wt littermate LSK cell proliferation in response to FLT3L, the combination of KL/TPO, or IL3. Experiments in culture supported by FLT3L were also performed in the presence or absence of 1 pg/mL TGF-β2 (n = 3; *P ≤ .05 in the absence of TGF-β2). A 100% response is defined as that of wt LSK cells at 50 ng/mL FLT3L or IL-3 in the absence of TGF-β2. (C) Tgfb2−/− (n = 3), Tgfb2+/− (n = 5), and wt littermate (n = 4) E16 fetal liver LSK cell proliferation in the presence of 10% serum in response to FLT3L (left; *P < .005) or to KL plus TPO (right; P > .4). A 100% response is defined as that of wt LSK cells at 10 ng/mL FLT3L. (D) C57BL/6 LSK cell proliferation in response to FLT3L (left) or KL and TPO (right) in the presence of 10% serum, with or without SB-431542 (10μM in DMSO; n = 3; *P = .002). (E) Tgfb2+/− and wt littermate LSK cell proliferation in serum-free cultures in response to FLT3L (n = 3; P > .1). A 100% response is defined as that of wt LSK cells at 50 ng/mL FLT3L. (F) Clone size (left; n = 80 for wt, n = 83 for Tgfb2+/−; *P = .02 by Student's unpaired t test) and cloning efficiency (right; n = 4; P = .736) of single Tgfb2+/− or wt LSK cells in liquid cultures containing 50 ng/mL FLT3L and 10% serum. Data collected after 5 days of incubation. Error bars represent SEM.
Although the effect of TGF-β2 appears predominantly cell autonomous, exogenously added TGF-β2 enhances the proliferation of HSPCs at low concentrations but is inhibitory at higher concentrations, leading to a biphasic dose response.17,19 A biphasic dose response was observed only when FLT3L was present (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and not when FLT3−/− LSK cells were used or in the presence of neutralizing anti-FLT3L Abs (supplemental Figure 1C-D). HSCs are also defined by the CD150+CD48−CD41− phenotype.29 Consistent with the fact that in the mouse model FLT3 signaling is dispensable for repopulating HSCs30 and with the higher enrichment in repopulating HSC in this population compared with the LSK population,29 FLT3 expression on CD150+CD48−CD41− cells was nearly undetectable (supplemental Figure 1E). TGF-β2 had a similar effect on the FLT3L/KL/TPO-supported proliferation of CD150+CD48−CD41−, LSKCD150+CD48−CD41−, and LSKCD150−CD48+CD41− cells, namely a moderate stimulatory effect at low concentrations and an inhibitory effect at higher concentrations (supplemental Figure 1F). In addition, similar to LSK cells, the enhancing effect of TGF-β2 disappeared when FLT3L was removed from the cytokine cocktail. In contrast to LSK cells, FLT3L alone was insufficient for the growth of CD150+CD48−CD41− cells however, even in the presence of TGF-β2 (not shown). Together, these observations show that TGF-β2 enhances FLT3L responsiveness of HSPCs in a predominantly, although not exclusively, cell autonomous and gene dosage–dependent fashion.
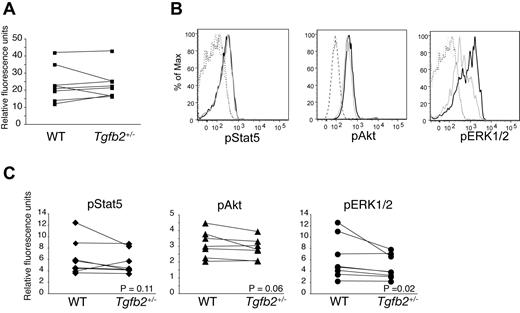
FLT3 protein (Figure 2) and mRNA (not shown) expression were similar in Tgfb2+/− and wt LSK cells. Binding of FLT3L to FLT3 results in the activation multiple signaling pathways, including PI3K/AKT and Ras/ERK cascades. Furthermore, constitutively active, mutated FLT3, but not wt FLT3, also activates STAT5.31 As measured by phosphoflow, pERK1/2 was significantly lower in Tgfb2+/− than in wt LSK cells, whereas the difference in pAKT levels was subtle and just short of statistical significance (Figure 2B-C). These data suggest that TGF-β2 deficiency affects predominantly the Ras/ERK pathways downstream of FLT3.
Effect of TGF-β2 deficiency on STAT5, AKT, and ERK1/2 phosphorylation. (A) FLT3 expression on LSK cells from Tgfb2+/− mice and wt littermates, as measured by flow cytometry (n = 8; P = .93; left). Data are presented as the ratio of FLT3-PE and isotype control IgG2a-PE geometric mean fluorescence. (B) Representative example of flow cytometric analysis of phosphorylation of STAT5, AKT, and ERK1/2 in LSK cells from Tgfb2+/− (gray) and wt littermate (black) mice. IgG controls are shown in dashed lines. (C) Intracellular phosphorylated STAT5 (pSTAT5; n = 6; P = .11), AKT (pAKT; n = 7; P = .06), and ERK1/2 (pERK1/2; n = 6; P = .02) levels in wt and Tgfb2+/− LSK cells. Data are presented as the ratio of pAKT, pSTAT5, or pERK1/2 and isotype control geometric mean fluorescence. Lines connect data from Tgfb2+/− and wt mice within the same litter and experiment. P values were calculated with Wilcoxon signed rank test for paired samples, not normally distributed.
Effect of TGF-β2 deficiency on STAT5, AKT, and ERK1/2 phosphorylation. (A) FLT3 expression on LSK cells from Tgfb2+/− mice and wt littermates, as measured by flow cytometry (n = 8; P = .93; left). Data are presented as the ratio of FLT3-PE and isotype control IgG2a-PE geometric mean fluorescence. (B) Representative example of flow cytometric analysis of phosphorylation of STAT5, AKT, and ERK1/2 in LSK cells from Tgfb2+/− (gray) and wt littermate (black) mice. IgG controls are shown in dashed lines. (C) Intracellular phosphorylated STAT5 (pSTAT5; n = 6; P = .11), AKT (pAKT; n = 7; P = .06), and ERK1/2 (pERK1/2; n = 6; P = .02) levels in wt and Tgfb2+/− LSK cells. Data are presented as the ratio of pAKT, pSTAT5, or pERK1/2 and isotype control geometric mean fluorescence. Lines connect data from Tgfb2+/− and wt mice within the same litter and experiment. P values were calculated with Wilcoxon signed rank test for paired samples, not normally distributed.
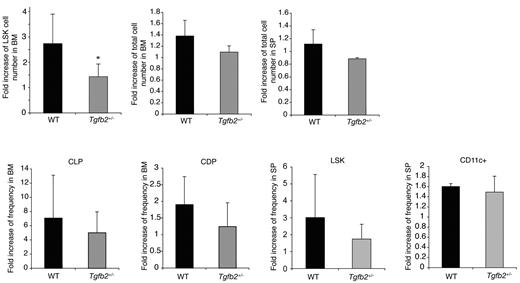
Finally, we examined whether Tgfb2+/− mice were hyporesponsive to FLT3L in vivo. Administration of FLT3L results in expansion of progenitor cells in the BM, mobilization of progenitor cells to the spleen, and expansion of dendritic cells (DCs).32,33 In wt mice, the LSK compartment expanded ∼ 2-fold more than in Tgfb2+/− than in wt mice (Figure 3) after administration of rmFLT3L (10 μg/mouse subcutaneously, once a day for 10 days). The frequency of DCs and of LSK cells in the spleen and of common lymphoid and DC progenitors,34 in the BM, although always tending to be lower in Tgfb2+/− than in wt mice, was similar in both groups however (Figure 3). These data indicate that TGF-β2 regulates the responsiveness of LSK cells to FLT3L to a similar degree in vivo and in vitro. Furthermore, this interaction is limited to the BM LSK cell compartment.
The HSPC compartment of Tgfb2+/− mice is hyporesponsive to FLT3L in vivo. Fold increase in LSK cell number, BM and spleen cellularity, common lymphoid progenitors (CLPs; lineage−IL7Rα+KitloSca1lo), common dendritic progenitors (CDPs, lineage−KitloFLT3+CD115+) in the BM, and LSK cells and DCs (CD11c+) in the spleen in Tgfb2+/− mice and wt littermates after 10 days of FLT3L treatment in vivo. Data are presented as fold expansion in FLT3L-treated mice compared with controls treated with MSA only, from 3 independent experiments, each including 2-4 mice per group and per genotype, with 3 different batches of FLT3L.
The HSPC compartment of Tgfb2+/− mice is hyporesponsive to FLT3L in vivo. Fold increase in LSK cell number, BM and spleen cellularity, common lymphoid progenitors (CLPs; lineage−IL7Rα+KitloSca1lo), common dendritic progenitors (CDPs, lineage−KitloFLT3+CD115+) in the BM, and LSK cells and DCs (CD11c+) in the spleen in Tgfb2+/− mice and wt littermates after 10 days of FLT3L treatment in vivo. Data are presented as fold expansion in FLT3L-treated mice compared with controls treated with MSA only, from 3 independent experiments, each including 2-4 mice per group and per genotype, with 3 different batches of FLT3L.
Fine-mapping of Tb2r1 suggests Prdm16 as the underlying quantitative trait gene
The responsiveness to the serum-dependent proliferative effect of TGF-β2 is genetically determined among inbred mouse strains and maps to a locus on the telomeric region of chr.4, Tb2r1. We unequivocally confirmed the location of this QTL with the use of congenic B6.D2-chr.4 mice.20 To better understand the mechanism underlying regulation of FLT3 and TGF-β2 signaling by serum factors, we fine-mapped Tb2r1. We generated subinterval-specific congenic strains (SISCSs) by backcrossing B6.D2-chr.4 mice onto the C57BL/6 background and selecting F2 offspring in which ≥ 1 crossovers had occurred in the telomeric region of chr.4 (Figure 4A).35,36 More than 200 F2 mice were genotyped for 9 single nucleotide polymorphisms (SNPs) in the introgressed region of chr.4. Mice with informative crossovers were again crossed with C57BL/6 and were interbred to obtain mice in which a segment of the DBA/2-derived introgressed region in B6.D2-chr.4 mice was of C57BL/6 origin (Figure 4A). This strategy established 9 distinct lines that were propagated (Figure 4B). We identified 3 lines, lines 9, 11, and 13, in which the serum-dependent stimulatory effect of low concentrations of TGF-β2 on LSK proliferation, absent in the parental B6.D2-chr.4 mice, was restored (Figure 4C). We focused our efforts on lines 9 and 13, because they were the only responding lines that shared a C57BL/6-derived segment in the otherwise DBA/2-derived introgressed region of chr.4. Further analysis of more narrowly spaced SNPs in the overlapping C57BL/6-derived region of both strains (Figure 4D) showed a much more complex genomic make-up than was suggested by the genotyping of more widely spaced SNPs used to monitor the generation of these lines (Figure 4B), indicating multiple crossovers in this region. Lines 9 and 13 have one segment of C57BL/6-derived genome between 152 055 934 bp and 153 715 384 bp in common (coordinates according to NCBI Build 37.1; Figure 4D). Twelve genes, 13 pseudogenes and processes transcripts (ie, without open reading frame), 1 predicted gene that may encode a protein, and 4 ESTs map to this area, bringing the total of potential candidates to 30, of which 17 encode or may encode a protein. Of these 17, 9 were expressed in LSK cells by quantitative PCR (using spleen, testis, or BM as positive controls; supplemental Table 1). Among these 9 genes, only Prdm16 contains a nonsynonymous coding SNP (ie, a nucleotide substitution that changes the amino acid sequence of corresponding protein). This C/T polymorphism changes amino acid 760 (exon 9, nucleotide 2281) from glutamine in DBA/2 to arginine in C57BL/6 (rs32942538; http://www.informatics.jax.org; Figure 4E). We next confirmed this SNP by sequencing genomic DNA in C57BL/6, DBA/2, and a number of congenic lines (Figure 4F). As expected from the SNP genotyping shown in Figure 4B, C57BL/6 and lines 9 and 13, which responded to TGF-β2, carried the C57BL/6 (C) allele of rs32942538, whereas DBA/2, B6.D2-chr.4, and lines 12 and 16, which did not respond to TGF-β2, carried the DBA/2 (T) allele. LSK cells from line 11 did respond to the enhancing effect of TGF-β2, although according to the SNP analysis shown in Figure 4B, this line did not share C57BL/6 genome with another responding line, line 13. Interestingly, however, line 11 does carry the C57BL/6 allele of Prdm16, showing that at least a small additional portion of C57BL/6 genome was introgressed, again indicative of multiple crossovers in that region. Similarly, line 14, which according to the genotyping of widely spaced SNPs shown in Figure 4B should carry the C57BL/6 allele of Prdm16 but did not respond to TGF-β2, carries the DBA/2 allele, once more indicating multiple crossovers in that region. Together, these data show that the DBA/2 allele of Prdm16 is associated with a lack of responsiveness of LSK cells to the proliferative effects of TGF-β2, whereas the C57BL/6 allele confers responsiveness to this effect of TGF-β2.
Fine-mapping of Tb2r1. (A) Schematic representation of the generation of SISCSs from congenic mice. (B) Map of telomeric region of chr.4 in B6.D2-chr.4 mice and SISCS. DFPSNP indicates distance from preceding SNP. (C) TGF-β2 response of LSK cells from SISCS in FLT3L, KL, and TPO, 50 ng/mL each, 10% serum (n > 3; P < .05). (D) SNP map of the C57BL/6-derived region shared between SISCS lines 9 and 13. (E) Structure of PRDM16 protein. PR indicates PR domain (SET); ZF, zinc finger domain; and NS-SNP, nonsynonymous SNP at aa 760. (F) DNA sequencing of the region surrounding rs32942538 in exon 9 of Prdm16 in C57BL/6 and DBA/2 mice. The left panel shows the alleles of rs32942538 in C57BL/6, DBA/2, B6.D2-chr.4, and several SISC lines.
Fine-mapping of Tb2r1. (A) Schematic representation of the generation of SISCSs from congenic mice. (B) Map of telomeric region of chr.4 in B6.D2-chr.4 mice and SISCS. DFPSNP indicates distance from preceding SNP. (C) TGF-β2 response of LSK cells from SISCS in FLT3L, KL, and TPO, 50 ng/mL each, 10% serum (n > 3; P < .05). (D) SNP map of the C57BL/6-derived region shared between SISCS lines 9 and 13. (E) Structure of PRDM16 protein. PR indicates PR domain (SET); ZF, zinc finger domain; and NS-SNP, nonsynonymous SNP at aa 760. (F) DNA sequencing of the region surrounding rs32942538 in exon 9 of Prdm16 in C57BL/6 and DBA/2 mice. The left panel shows the alleles of rs32942538 in C57BL/6, DBA/2, B6.D2-chr.4, and several SISC lines.
Coding variation in Prdm16 determines regulation of responsiveness of HSPCs to FLT3L and TGF-β2 by PPARγ ligands
PRDM16 is a 140-kDa zinc finger protein that was discovered as a fusion partner in t(1:3)(p36;q21) in t(1;21)(p36;q22) translocations in acute myeloblastic leukemia (AML).37,38 Elevated PRDM16 expression, because of promoter hypomethylation, is frequently observed in karyotypically normal AML, often accompanied by mutation in nucleophosmin.39 We and others showed that, within the hematopoietic system, Prdm16 is expressed very selectively in HSCs and to a lesser extent in MPPs. Consistent with this expression pattern, Prdm16 is critical for the establishment and maintenance of the HSC pool during development and after transplantation.21,22 Because allelic variation in Prdm16 only appears to affect signaling by FLT3/FLT3L, which has been shown to be dispensable for HSC renewal,30 deletion of Prdm16 does not model the effect of a single amino acid change in PRDM16, as observed in DBA/2 and C57BL/6 mice.
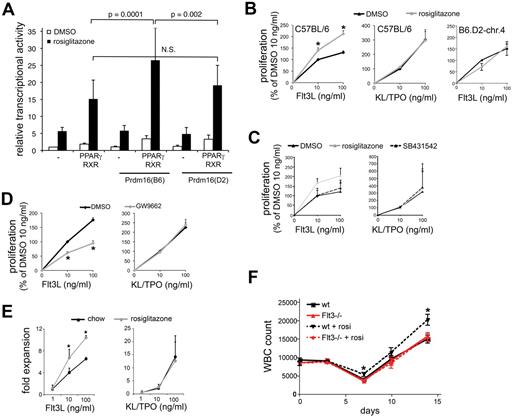
Prdm16 is also expressed in brown adipose tissue, a tissue critical for energy metabolism and thermogenesis. PRDM16 induces brown adipose tissue development from mesodermal precursors, among others, by interacting with and enhancing the transcriptional activity of the ligand-activated transcription factor PPARγ,40 an essential regulator of adipogenesis.23 To establish whether coding variation in Prdm16 affects PPARγ function, we transiently transfected COS-7 cells with a luciferase reporter construct containing 3 tandem repeats of a consensus PPAR response element with the PPARγ/RXRα heterodimer and Prdm16(B6) or Prdm16(D2). Prdm16(B6), but not Prdm16(D2), enhanced the transcriptional activity of PPARγ induced by the synthetic-specific PPARγ agonist, rosiglitazone (Figure 5A). These data directly show that Prdm16 alleles differentially affect PPARγ transcriptional activity. Although the relative physiologic relevance of individual PPARγ ligands is still unclear, its natural ligands are a variety of lipids,23 raising the hypothesis that PPARγ ligands are the nonprotein serum factors required for the regulation of TGF-β2/FLT3 responsiveness. Therefore, we examined whether PPARγ agonists regulate FLT3L and TGF-β2–induced proliferation of LSK cells. The synthetic PPARγ agonist, rosiglitazone, enhanced FLT3L but not KL/TPO-supported proliferation in serum-free conditions in LSK cells from C57BL/6 mice. Moreover, rosiglitazone was unable to enhance the FLT3L-induced proliferation of LSK cells from B6.D2-chr.4 mice, which express the D2 allele of Prdm16 (Figure 5B). Similar data were obtained with ciglitazone (not shown). This effect of rosiglitazone in C57BL/6 LSK cells was absent when endogenous TGF-β signaling was blocked by the addition of SB-431542 (Figure 5C). However, the PPAR inhibitor, GW9662,41 decreased responsiveness of LSK cells to FLT3L but not to KL/TPO when serum was present (Figure 5D). Together, these data indicate that allelic variation in the Prdm16 coding sequence determines the ligand-induced transcriptional activity of PPARγ, that PPAR ligands regulate the responsiveness of HSPCs to FLT3L, and that this effect requires TGF-β signaling. We conclude that the serum factors required for regulation of FLT3L responsiveness of HSPCs by TGF-β2 include natural ligands of PPARγ and that their effect on HSPCs is dictated by allelic variation in Prdm16.
Role of PPARγ. (A) Transcriptional activity of a PPAR-driven reporter gene in COS-7 cells cotransfected with PPARγ/RXRα and with Prdm16(B6) or Prdm16(D2) expression vectors in the presence or absence of rosiglitazone (1μM) (luciferase values relative to renilla luciferase activity; n = 9). (B) Effect of rosiglitazone (1μM) on FLT3L (left) or KL/TPO (middle) supported proliferation of LSK cells from C57BL/6 mice (n = 4; *P < .05), and on FLT3L-supported proliferation of B6.D2-chr.4 congenic mice (right; 1 triplicate experiment). Data were normalized across experiments to proliferation observed at 10 ng/mL FLT3L and DMSO control. (C) Effect of rosiglitazone on FLT3L-induced and KL/TPO-induced proliferation of C57BL/6 LSK cells in the presence and absence of SB431542 (n = 3 triplicate experiments; data expressed as percentage of the proliferation at 10 ng/mL of each cytokine). (D) Effect of GW9662 (1μM) on FLT3L (left) and KL/TPO (right) supported proliferation of C57BL/6 LSK cells in serum-containing cultures (n ≥ 4; *P = .03; data expressed as percentage of the proliferation at 10 ng/mL of each cytokine.). (E) FLT3L- and KL/TPO-supported proliferation of LSK cells isolated from mice fed with chow or rosiglitazone for 3 weeks in the presence of 10% serum (*P < .05; n = 6). (F) PB white blood cell (WBC) counts in FLT3−/− and wt mice gavaged with rosiglitazone (15 mg/kg) or DMSO daily on days 0-4 after administration of 5-FU on day 0 (150 mg/kg intraperitoneally; n = 10; *P < .05 compared with wt DMSO).
Role of PPARγ. (A) Transcriptional activity of a PPAR-driven reporter gene in COS-7 cells cotransfected with PPARγ/RXRα and with Prdm16(B6) or Prdm16(D2) expression vectors in the presence or absence of rosiglitazone (1μM) (luciferase values relative to renilla luciferase activity; n = 9). (B) Effect of rosiglitazone (1μM) on FLT3L (left) or KL/TPO (middle) supported proliferation of LSK cells from C57BL/6 mice (n = 4; *P < .05), and on FLT3L-supported proliferation of B6.D2-chr.4 congenic mice (right; 1 triplicate experiment). Data were normalized across experiments to proliferation observed at 10 ng/mL FLT3L and DMSO control. (C) Effect of rosiglitazone on FLT3L-induced and KL/TPO-induced proliferation of C57BL/6 LSK cells in the presence and absence of SB431542 (n = 3 triplicate experiments; data expressed as percentage of the proliferation at 10 ng/mL of each cytokine). (D) Effect of GW9662 (1μM) on FLT3L (left) and KL/TPO (right) supported proliferation of C57BL/6 LSK cells in serum-containing cultures (n ≥ 4; *P = .03; data expressed as percentage of the proliferation at 10 ng/mL of each cytokine.). (E) FLT3L- and KL/TPO-supported proliferation of LSK cells isolated from mice fed with chow or rosiglitazone for 3 weeks in the presence of 10% serum (*P < .05; n = 6). (F) PB white blood cell (WBC) counts in FLT3−/− and wt mice gavaged with rosiglitazone (15 mg/kg) or DMSO daily on days 0-4 after administration of 5-FU on day 0 (150 mg/kg intraperitoneally; n = 10; *P < .05 compared with wt DMSO).
Next, we examined the effects of PPARγ agonism in vivo. LSK cells from rosiglitazone-fed mice (3 weeks, 10 mg/kg chow) proliferated more in response to FLT3L but not to KL/TPO in vitro compared with LSK cells from control mice fed control chow (Figure 5E), indicating that rosiglitazone also affects FLT3L responsiveness after administration in vivo. Rosiglitazone enhances hematologic recovery from the cytotoxic drug, 5-FU. It was speculated that this effect could be explained by the potentially beneficial metabolic effects of rosiglitazone.42 Because our findings indicate that rosiglitazone specifically enhances the FLT3L responsiveness of HSPCs, we tested the hypothesis that this effect depends on FLT3. We treated wt and Flt3−/− mice with 5-FU and with either rosiglitazone or DMSO for 5 days after 5-FU administration. Rosiglitazone-treated wt but not Flt3−/− mice had more PB leukocytes than DMSO-treated mice on days 7, 10, and 14 after administration of 5-FU (Figure 5F), indicating that, as hypothesized, the rosiglitazone effect on hematologic recovery is FLT3 dependent.
Finally, we investigated the hematopoietic phenotype of conditional Ppargfl/fl mice, because germline Pparg deletion is embryonic lethal.23 In these mice, exons 1 and 2 of PPARγ are flanked by LoxP sites.25 We crossed Ppargfl/fl mice with Mx1-Cre mice, where Cre is expressed under the IFN-α/β– and double-stranded RNA-responsive Mx1 promoter.26 Complete deletion was achieved in the BM after 4 injections of poly(I:C) (625 μg intraperitoneally every 2 days for 8 days; Figure 6A). Ppargfl/fl mice that did not express Cre but received poly(I:C) were used as controls. In LSK cells from conditionally deleted Ppargfl/fl.Mx1-Cre mice treated with poly(I:C) (cPparg−/−), rosiglitazone did not increase FLT3L-induced proliferation in serum-free conditions (Figure 6B), indicating that the effect of rosiglitazone on FLT3L responsiveness depended on PPARγ. However, GW9662 inhibited FLT3L-induced proliferation in the presence of serum to the same extent in Ppargfl/fl and in cPparg−/− LSK cells (Figure 6B). Because GW9662 also inhibits PPARδ and because we have previously shown that PPARδ is also expressed in HSCs,21 these data suggest that the effect of PPAR ligands is not only mediated through PPARγ but also PPARδ. Next, we examined whether PPARγ played a role in the regulation of HSCs. The frequency of LSK cells, LT-HSCs (LSKCD34−FLT3−), ST-HSCs (LSKCD34+FLT3−), and MPPs (LSKCD34+FLT3+)1 in BM (Figure 6C) and spleen (not shown) were similar in cPparg−/− mice and controls (Figure 6C). In competitive repopulation studies, where we injected 0.5 × 106 BM cells from cPparg−/− and control mice (CD45.2+) 2 weeks after treatment with poly(I:C) together with an equal number of BM cells from CD45.1+ congenic C57BL6 mice into lethally irradiated CD45.1+CD45.2+ F1 hosts, the donor CD45.1+ contribution to the B, T, and myeloid lineages in the peripheral blood was similar in recipients of Pparg−/−.Mx1-Cre and Ppargfl/fl donors 10-12 weeks after transplantation (Figure 6B). These data indicate that Pparg is not critical and is probably redundant for hematopoiesis. Because natural ligands of PPARs relatively promiscuously activate all 3 isoforms,23 and because PPARδ expression was detected in HSCs, it is possible that PPARδ compensates for the absence of PPARγ.
Hematopoietic phenotype of Pparg−/− mice. (A) Representative example of genotyping by RT-PCR of Pparg fl/fl.Mx1-Cre or control Pparg fl/fl mice treated with poly(I:C) (625 mg intraperitoneally every 2 days for 8 days). (B) FLT3L-induced proliferation of cPparg−/− cells in the presence of rosiglitazone or GW9662 (n = 4; data expressed relative to DMSO in control LSK cells). (C) Frequency of LSK cells, long-term HSCs (LT-HSCs; LSKCD34−FLT3−), short-term HSCs (ST-HSCs; LSKCD34+FLT3−), and MPP cells (LSKCD34+FLT3+) in the BM of cPparg−/− or control Pparg fl/fl mice treated with poly(I:C) (n = 3). (E) Myeloid (Mac1+Gr1+), B (CD19+) and T (Thy1+) cell contribution of donor-derived PB cells 8-10 weeks after transplantation of 0.5 × 106cPparg−/− or control Ppargfl/fl BM and competitor (CD45.1+ C57BL/6) BM cells into lethally irradiated CD45.1+CD45.2+ C57BL/6 mice (n = 9 for Ppargfl/fl; n = 12 for cPparg−/−).
Hematopoietic phenotype of Pparg−/− mice. (A) Representative example of genotyping by RT-PCR of Pparg fl/fl.Mx1-Cre or control Pparg fl/fl mice treated with poly(I:C) (625 mg intraperitoneally every 2 days for 8 days). (B) FLT3L-induced proliferation of cPparg−/− cells in the presence of rosiglitazone or GW9662 (n = 4; data expressed relative to DMSO in control LSK cells). (C) Frequency of LSK cells, long-term HSCs (LT-HSCs; LSKCD34−FLT3−), short-term HSCs (ST-HSCs; LSKCD34+FLT3−), and MPP cells (LSKCD34+FLT3+) in the BM of cPparg−/− or control Pparg fl/fl mice treated with poly(I:C) (n = 3). (E) Myeloid (Mac1+Gr1+), B (CD19+) and T (Thy1+) cell contribution of donor-derived PB cells 8-10 weeks after transplantation of 0.5 × 106cPparg−/− or control Ppargfl/fl BM and competitor (CD45.1+ C57BL/6) BM cells into lethally irradiated CD45.1+CD45.2+ C57BL/6 mice (n = 9 for Ppargfl/fl; n = 12 for cPparg−/−).
Discussion
With the use of a quantitative genetic approach, we have shown that PPARγ ligands and TGF-β2 cooperatively enhance FLT3L signaling in HSPCs and that allelic coding variation in Prdm16 regulates this effect by differentially affecting the ligand-induced transcriptional activity of PPARγ, thus identifying a novel regulatory axis in hematopoiesis.
We previously reported that the isoform-specific proliferative effects of TGF-β2 on HSPCs16 required low MW, nonprotein serum factors.19 The identification of Prdm16 as the gene underlying Tb2r1 led to the discovery that these low MW serum factors include PPAR ligands. This is the first demonstration of a direct effect of PPARγ agonists on hematopoiesis in vitro and in vivo. Although for each PPAR relatively specific synthetic agonists have been developed, endogenous PPAR ligands encompass a broad range of compounds derived from long-chain fatty acids and include eicosanoids, leukotrienes, and prostaglandins.23 Given the promiscuity of endogenous PPAR ligands in their activation of different PPARs,23 it is not surprising even after deletion of PPARγ that FLT3L responsiveness of LSK cells was normal and could still be inhibited by GW9662, a full antagonist of both PPARγ and PPARδ and a partial antagonist of PPARα.41 Because PPARδ,21 but not PPARα (not shown), is expressed in LSK cells, it is probable that PPARδ compensates for the absence of PPARγ. Probably for the same reason, PPARγ was redundant for HSPC function in vivo. We also determined that the telomeric region of chr.4 contains a QTL that regulates age-associated thymic involution.43 This QTL, Ti1, was confirmed in B6.D2-chr.4 mice, which have delayed thymic involution compared with parental C57BL/6 mice.43 Furthermore, Tgfb2+/− mice have delayed thymic involution.44 Because Prdm16 allelic variation regulates TGF-β2 responsiveness, it is possible that Ti1 and Tb2r1 are in fact the same QTL. These data would fit very well with recent observations that rosiglitazone and PPARγ activation leads to accelerated thymic involution.45
Prdm16, which is required for maintenance of HSCs,21,22 therefore also plays a role in the FLT3L and TGF-β2 responsiveness of early progenitors and potentially in thymic involution through interaction with PPARγ, effects that would not have been discovered through analysis of Prdm16−/− mice. This work shows how analysis of the phenotypic consequences of natural genetic variation has the potential to uncover novel regulatory mechanism and can lead to physiologically relevant insights into gene function and subfunctionalization that complement those obtained from knockout approaches.14-16 Furthermore, QTL identification in a genetically tractable model such as the mouse may allow functional insight into human genetic variation in a more targeted fashion than is currently possible in genome-wide association studies.16 It is interesting to note in this context that the International HapMap project identified several nonsynonymous SNPs in exon 4 (rs12049194) and in exon 8 (rs79653294, rs12031858, rs870124, rs2493292, and rs6175438) of PRDM16 in humans. Although these SNPs do not involve the exact syntenic nucleotide, the exon 8 SNPs are located in the same region of the gene, in between the 2 zinc finger regions. These data raise the hypothesis that allelic variation at the PRDM16 locus in humans may be functionally relevant and play a role in hematopoietic stress responses and thymic involution.
The isoform-specific, serum-dependent enhancing effect of TGF-β2 on FLT3L responsiveness is predominantly cell autonomous. A cell autonomous effect of TGF-β2 on HSPCs is consistent with the phenotype of TGF-β2–deficient mice, in which a progressive decrease in repopulation capacity was observed after serial transplantation into wt hosts.17 It is unclear, however, how the isoform specificity of this effect is explained, how TGF-β2, which is secreted in latent form, is activated, and how TGF-β2 specifically affects FLT3 signaling and not signaling by any other cytokine tested. Because of the low number of LSK cells that can be isolated, together with the absence of a cell line model for this effect of TGF-β2, the precise molecular mechanism of this interaction is currently not accessible. Human FLT3 is frequently mutated in AML, leading to constitutive signaling. One type of mutation, internal tandem duplication (ITD) in the juxtamembrane domain, is associated with a worse prognosis. Expression of a FLT3-ITD in BAF3 or 32D cells leads to growth factor–independent proliferation.31 We found, however, that neither the TGF-RI antagonist SB-431542 nor the PPARγ inhibitor GW9662 affected this proliferation (unpublished observations), indicating that mutated, constitutively active FLT3 is not subject to regulation by the TGF-β2/PPARγ mechanism we identified here. Because signaling by a large array of cytokines activates a relatively limited set of signaling pathways, the specific interaction of TGF-β2 with FLT3L signaling and not with any other cytokine tested suggests that TGF-β2 affects the function of the FLT3 receptor itself. This mechanism does not involve regulation of FLT3 expression, however, but affects predominantly phosphorylation of ERK1/2.
Hyporesponsiveness of Tgfb2+/− to FLT3L administration in vivo was only observed in the LSK compartment. Similarly, although both Tgfb2−/− and FLT3−/− hematopoietic cells display a multilineage defect after transplantation, FLT3−/− cells display a disproportionally profound defect in lymphopoiesis,27 not observed in Tgfb2−/− cells.17 These data suggest that interaction between FLT3L and TGF-β2 signaling is specific to the HSPC compartment or that in more committed progenitor cells this interaction is redundant with other regulatory mechanisms. HSCs in the mouse do not express FLT3 by flow cytometry.46,47 Although initial reports indicated that FLT3−/− HSCs display a multilineage defect after transplantation,26 more recent findings indicate that FLT3 is dispensable for HSCs in mouse.29 Because Tgfb2−/− HSCs do display a defect in renewal after serial transplantation,17 it cannot be excluded that TGF-β2 might affect additional pathways involved in the regulation of HSCs or directly affect HSC function independently of its effect on FLT3 signaling. In humans, however, it has been shown that FLT3 is expressed on HSCs and is required for their survival.48,49 Furthermore, healthy persons have low plasma FLT3L levels, whereas in patients with pancytopenic hematopoietic disorders, such as Fanconi anemia and acquired aplastic anemia, FLT3L levels are elevated.50 Together, these observations suggest that FLT3L, and therefore, given its effect on FLT3 signaling in HSPCs, TGF-β2, may play an important role in HSPC homeostasis, particularly in humans.
In conclusion, coding variation in Prdm16 underlies Tb2r1 and determines the predominantly cell autonomous effect of TGF-β2 on FLT3 signaling through a mechanism involving PPARγ.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr J. Mandeli for advice on statistical analysis of mice treated with FLT3L in vivo.
This work was supported by the National Institutes of Health (grants RO1 AG016327 and HL073760).
National Institutes of Health
Authorship
Contribution: S.A. performed all of the experiments and co-wrote the manuscript; F.A. performed the studies involving cPparg−/− mice and PPARγ transcriptional assays; K.K. performed the 5-FU administration and sequencing studies; and H.-W.S. designed and supervised the experiments and co-wrote the manuscript with S.A.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans-Willem Snoeck, Department of Oncological Sciences, Mount Sinai of School of Medicine, Gustave L. Levy Pl, Box 1496, New York, NY 10029; e-mail: hans.snoeck@mssm.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal