Abstract
Diamond-Blackfan anemia (DBA) is a congenital erythroid hypoplasia caused by a functional haploinsufficiency of genes encoding for ribosomal proteins. Among these genes, ribosomal protein S19 (RPS19) is mutated most frequently. Generation of animal models for diseases like DBA is challenging because the phenotype is highly dependent on the level of RPS19 down-regulation. We report the generation of mouse models for RPS19-deficient DBA using transgenic RNA interference that allows an inducible and graded down-regulation of Rps19. Rps19-deficient mice develop a macrocytic anemia together with leukocytopenia and variable platelet count that with time leads to the exhaustion of hematopoietic stem cells and bone marrow failure. Both RPS19 gene transfer and the loss of p53 rescue the DBA phenotype implying the potential of the models for testing novel therapies. This study demonstrates the feasibility of transgenic RNA interference to generate mouse models for human diseases caused by haploinsufficient expression of a gene.
Introduction
Diamond-Blackfan anemia (DBA; Online Mendelian Inheritance in Man [OMIM] no. 105650) is a rare congenital erythroid hypoplasia that presents early in infancy. The classic hematologic profile of DBA consists of macrocytic anemia with selective absence of erythroid precursors in a normocellular bone marrow, normal or slightly decreased neutrophil, and variable platelet count.1 During the course of the disease some patients show decreased bone marrow cellularity that often correlates with neutropenia and thrombocytopenia.2 However, DBA is a developmental disease because ∼ 30%-47% of patients show a broad spectrum of physical abnormalities including craniofacial, heart, and upper limb malformations, and short stature.1,3-5
All known DBA disease genes encode for ribosomal proteins that collectively explain the genetic basis for ∼ 55% of DBA cases.6-11 Twenty-five percent of the patients have mutations in a gene coding for ribosomal protein S19 (RPS19) making it the most common DBA gene.6 The majority of the mutations completely disrupt the expression of the RPS19, whereas the rest are missense mutations interfering with the assembly of RPS19 into 40S ribosomal subunits.12-14 All patients are heterozygous with respect to RPS19 mutations suggesting a functional haploinsufficiency of RPS19 as the basis for disease pathology.
Despite of the recent advances in DBA genetics, the pathophysiology of the disease remains elusive. Cellular studies on patients together with successful marrow transplantation15 have demonstrated the intrinsic nature of the hematopoietic defect. DBA patients have a variable deficit in burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E) progenitors with substantially reduced clonogenic output that correlates with the age of the patient.2,16-19 A similar age-dependent decrease in granulocyte-macrophage progenitor (GMP) numbers has been reported.20 Although present at normal frequencies, the proliferation and differentiation of immature hematopoietic progenitors in DBA have shown to be impaired in vitro.2,18,20,21
Generation of animal models for RPS19-deficient DBA is pivotal to understand the disease mechanisms and to evaluate novel therapies. Matsson et al attempted to create a mouse model with a targeted disruption of Rps19.22 However, although the homozygous loss of Rps19 was lethal before implantation, the mice heterozygous for Rps19 exhibited no reduction in Rps19 expression and a normal hematopoietic phenotype. Recently, a new mouse model for RPS19-deficient DBA was reported that presents a heterozygous missense mutation.23 Similar to the knockout model discussed above, this mutation is lethal if homozygous, whereas the heterozygous mice exhibit a mild anemia and increased apoptosis in bone marrow. Furthermore, a transgenic mouse model with an Rps19 missense mutation shows retarded growth and mild anemia together with reduced numbers of erythroid progenitors.14 In addition, 2 independent groups have generated rps19-deficient zebrafish models that show hematopoietic and developmental abnormalities.24,25
Modeling of haploinsufficient human diseases like DBA in mice is challenging because the phenotype is highly dependent on the level gene down-regulation. As previously mentioned, the complete deletion of essential genes like Rps19 often results in embryonic lethality, whereas the heterozygosity may be compensated efficiently in mice leading only to a mild phenotype. RNA interference (RNAi) provides an alternative approach to down-regulate gene expression. Importantly, RNAi enables a scalable gene silencing that allows a more accurate reproduction of human disease phenotypes. In the current study, we have generated mouse models for RPS19-deficient DBA by taking advantage of transgenic RNAi. These models are engineered to contain a doxycycline-regulatable Rps19-targeting shRNA allowing an inducible and graded down-regulation of Rps19. We demonstrate that the Rps19-deficient mice develop a macrocytic anemia together with leukocytopenia and variable platelet count, and the severity of the phenotype depends on the level of Rps19 down-regulation. We further show that a chronic Rps19 deficiency leads to the exhaustion of hematopoietic stem cells (HSCs) and subsequent bone marrow failure. Finally, both RPS19 gene transfer and the loss of p53 rescue the DBA phenotype implying the potential of the models for testing novel therapies.
Methods
Generation of transgenic mice
Construction of Rps19-targeting shRNAs was done following the guidelines presented by Paddison et al.26 Briefly, a hairpin generation algorithm RNAi Oligo Retriever was used to design miR30-styled shRNAs against mouse Rps19 mRNA (NM_023133). shRNA-D (5′-AGGATGCCCGGAGTTACTGTAA-3′) was newly designed, whereas shRNA-B (5′-CGTCCGGGAAGCTGAAAGTCC-3′) was acquired from a previous experiment.27 shRNAs were cloned into the miR-30 miRNA context of pSH2 retroviral vector (Thermo Scientific). miRNA-styled shRNA cassette was subcloned into the MluI-EcoRI site of pBS31 vector, which was used to target KH2 ES cell line.28 The engineered ES cells were injected into E3.5 C57BL/6 blastocysts to generate chimeric mice. Mice were backcrossed into the C57BL/6 background for at least 5 generations. PCR strategy was used to genotype Rosa26 locus (5′-AAAGTCGCTCTGAGTTGTTAT-3′; 5′-GCGAAGAGTTTGTCCTCAACC-3′; 5′-GGAGCGGGAGAAATGGATATG-3′) and ColA1 locus (5′-TCCCTCACTTCTCATCCAGATATT-3′; 5′-AGTCTTGGATACTCCGTGACCATA-3′; 5′-GGACAGGATAAGTATGACATCATCAA-3′). Rps19 deficiency was induced in vivo by administering 2 mg/mL doxycycline (Sigma-Aldrich) in drinking water supplied with 10 mg/mL sucrose (Sigma-Aldrich). Mice were maintained in the Lund University animal facility and all animal experiments were performed with consent from the Lund University ethical committee.
Blood and histology analysis
Peripheral blood was collected from the tail vain in Microvette tubes (Sarstedt) and cellularity was analyzed using Sysmex XE-5000. Organs for histopathologic analysis were fixed in 4% paraformaldehyde buffered with PBS followed by paraffin embedding and sectioning. Sections were stained with Erlisch eosin for microscopic examination.
Polysome profiles and rRNA analysis
Polysome profiling and rRNA analysis were performed as previously described.29
Flow cytometry
Bone marrow cells were isolated by crushing femur and tibia in PBS containing 2% FCS (GIBCO). Fresh cells were stained with following antibodies: CD44 (BD Biosciences; 553133), CD41 (eBioscience; 12-0411-83), GR1 (Biolegend; 108410), CD11b (Biolegend; 101210), B220 (Biolegend; 103210), CD3 (Biolegend; 100310), Ter119 (eBioscience; 25-5921-82), CD150 (Biolegend; 115910), c-Kit (eBioscience; 47-1171-82), Endoglin (Biolegend; 120404), and Sca-1 (Biolegend; 122520). Streptavidin was purchased from Invitrogen (Q10101MP). Propidium iodide (Invitrogen) was used to exclude dead cells. For cell cycle analysis, cells were stained for surface markers, fixed in 4% paraformaldehyde (Sigma-Aldrich), permeabilized with 0.1% saponin (Sigma-Aldrich), and DNA was stained with DAPI (Sigma-Aldrich). All experiments were performed on FACSCanto II and FACS Aria cell sorters (BD Biosciences) and analyzed with FlowJo Version 9.3.3 software (TreeStar).
Cell cultures
c-Kit expressing cells were enriched using CD117 MicroBeads and MACS separation columns (Miltenyi Biotec). Single preMegE or preCFU-E/CFU-E progenitors were FACS-sorted into Terasaki plates in 20 μL IMDM (Thermo Scientific) supplemented with 20% FCS, 0.1mM β-mercaptoethanol (Sigma-Aldrich), and recombinant growth factors murine stem cell factor (mSCF; 50 ng/mL, PeproTech), murine IL-3 (mIL-3; 10 ng/mL, PeproTech), and erythropoietin (EPO; 2 U/mL, Janssen-Cilag) with or without doxycycline (1 μg/mL). preGM/GMPs were cultured in IMDM containing mSCF, mIL-3 and granulocyte-colony stimulating factor (G-CSF;10 ng/mL, Amgen). Light microscope was used to evaluate preCFU-E/CFU-E, and preMegE and preGM/GMP cultures after 3 or 5 days, respectively. For CFU-C assay, 15 × 103 whole bone marrow cells were seeded into 1.5 mL M3434 methylcellulose (StemCell Technologies) without doxycycline and colonies were scored on day 11.
Quantitative real-time PCR
Total RNA was isolated from FACS-sorted cells using RNAeasy micro kit (QIAGEN) and cDNA transcribed with SuperScript III reverse trancriptase (Invitrogen). Real-time PCR reactions were performed using the Taqman Assay System with the exception of Rps19 that was quantified using the 2XSYBR green master mix (Rps19: 5′-GCAGAGGCTCTAAGAGTGTGG-3′ and 5′-CCAGGTCTCTCTGTCCCTGA-3′; Actb: 5′-CCACAGCTGAGAGGCAAATC-3′ and 5′-CTTCTCCAGGGAGGAAGAGG-3) according to the manufacturer's instructions (Applied Biosystems).
TUNEL assay
TUNEL assay was performed using an Apoptosis Detection Kit following the manufacturer's instructions (GenScript).
RPS19 overexpression
c-Kit enriched bone marrow cells were prestimulated in serum-free medium (StemSpanSFEM; StemCell Technologies) supplemented with penicillin/streptomycin (GIBCO), mSCF (50 ng/mL), human thrombopoietin (hTPO; 50 ng/mL, PeproTech), mIL-3 (20 ng/mL), and human IL-6 (hIL-6; 50 ng/mL, PeproTech) in 6-well plates (non–tissue culture–treated; BD Biosciences) for 24 hours (0.5 × 106 cells per mL). Retronectin-coated (Takara) 6-well plates were preloaded with SFFV-RPS19 or SFFV-GFP vector (100 μL per well corresponding to MOI = 5). One million cells were seeded into each well in 3 mL prestimulation medium and incubated for 2 days. For liquid cultures, 0.5 × 106 transduced cells were seeded into 3 mL SFEM supplemented with mSCF (50 ng/mL), mIL-3 (10 ng/mL), EPO (2 U/mL), and doxycycline (0.5 μg/mL). For BFU-E colony-forming assay, 2 × 104 transduced cells were seeded into 1.5 mL M3436 methylcellulose (StemCell Technologies) with doxycycline (1 μg/mL). Colonies were scored on day 7.
Transplantation assays
Noncompetitive transplantations were performed injecting 5 million unfractionated bone marrow cells in 500 μL PBS into tail vein of irradiated (900 cGy) wild-type recipients. In competitive transplantations 2.5 million unfractionated bone marrow cells from transgenic donors (CD45.2) or wild-type competitor (CD45.1) were mixed and transplanted into irradiated wild-type recipient mice (CD45.1/CD45.2). On peripheral blood reconstitution analysis, red blood cells were lysed using ammonium chloride and samples were stained with the following antibodies (Biolegend): CD45.1 (110730), CD45.2 (109806), B220 (103208), B220 (103212), CD3 (100312), CD11b (101208), and Gr1 (108408).
Statistical analyses
Student t test was used to determine statistical significance, and 2-tailed P values are shown. Error bars represent SD.
Results
Generation of inducible Rps19 knockdown mouse models
We and others have previously demonstrated that RNAi can be used to generate cellular models for DBA.27,30,31 Therefore, we used a similar approach to create mouse models for RPS19-deficient DBA. We took advantage of the KH2 embryonic stem (ES) cell line that is engineered to continuously express M2 reverse tetracycline transactivator (M2-rtTA) at the Rosa26 locus.28 An Rps19-targeting shRNA under a doxycycline-responsive element was targeted into the downstream area of the collagen A1 (ColA1) locus using FLP/frt site-specific recombination (Figure 1A). We created ES cell clones for 2 different miR30-styled Rps19-targeting shRNAs (shRNA-B and shRNA-D) and confirmed the correct recombination using Southern analysis (data not shown). To adjust the level of Rps19 knockdown the mice were bred either heterozygous or homozygous for the shRNA at the ColA1 locus, whereas littermates without shRNAs were used as controls (Figure 1B). All experimental animals were bred homozygous for the M2-rtTA at the Rosa26 locus.
Rps19 knock-down mouse models show a reduction in Rps19 expression on doxycycline administration. (A) Overview of modified loci. (B) Breeding strategy to adjust the level of Rps19 down-regulation. (C) Quantitative real-time PCR analysis of Rps19 mRNA levels in erythroid (preCFU-E/CFU-E), myeloid (preGM/GMP), and multipotent (LSK) hematopoietic progenitors from adult mice that were given doxycycline for 3 days. (n = 4-7 per genotype). (D) Representative polysome profiles of cells derived from the liver after 10 days of doxycycline administration. (E) Pre-rRNA analysis of heterozygous shRNA-B embryonic fibroblasts (EF) cultured for 7 days with or without doxycycline (n = 4), or of cells derived from the liver after 10 days of doxycycline administration. (n = 6-10). Error bars represent SD. SA indicates splice acceptor; and pA, polyadenylation signal. Black arrowheads in panel A indicate transcriptional start sites.
Rps19 knock-down mouse models show a reduction in Rps19 expression on doxycycline administration. (A) Overview of modified loci. (B) Breeding strategy to adjust the level of Rps19 down-regulation. (C) Quantitative real-time PCR analysis of Rps19 mRNA levels in erythroid (preCFU-E/CFU-E), myeloid (preGM/GMP), and multipotent (LSK) hematopoietic progenitors from adult mice that were given doxycycline for 3 days. (n = 4-7 per genotype). (D) Representative polysome profiles of cells derived from the liver after 10 days of doxycycline administration. (E) Pre-rRNA analysis of heterozygous shRNA-B embryonic fibroblasts (EF) cultured for 7 days with or without doxycycline (n = 4), or of cells derived from the liver after 10 days of doxycycline administration. (n = 6-10). Error bars represent SD. SA indicates splice acceptor; and pA, polyadenylation signal. Black arrowheads in panel A indicate transcriptional start sites.
To evaluate the Rps19 knockdown efficiency in vivo, we sorted erythroid (preCFU-E/CFU-E), myeloid (preGM/GMP), and multipotent (Lineage− Sca-1+ c-Kit+; LSK) hematopoietic progenitors from adult mice that were given doxycycline in the drinking water for 3 days. Rps19 mRNA was down-regulated in each cell population and the level of knockdown appeared generally greater in shRNA-D mice (Figure 1C). We also observed a trend toward more efficient knockdown in mice homozygous for a given shRNA compared with heterozygous mice. Furthermore, a similar analysis on erythroid and myeloid progenitors from heterozygous mice induced for 10 days confirmed the persistency of Rps19 knockdown (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Numerous studies have shown that RPS19 is required for the biogenesis of 40S ribosomal subunits.29,32 Polysome profiling of cells derived from the liver of heterozygous shRNA-B and shRNA-D mice after 10 days of doxycycline administration indicated a decrease in the free 40S to 60S subunit ratio relative to control mice (Figure 1D). The 40S subunit deficit in RPS19-deficient cells has been shown to be accompanied by an increased ratio of 21S to 18SE pre-rRNA, which can be used to monitor RPS19 function.29,32 Pre-rRNA processing studies in embryonic fibroblasts as well as livers from induced heterozygous shRNA-B and shRNA-D mice revealed an increased ratio of 21S to 18SE pre-rRNA relative to controls, consistent with a functional deficit of Rps19 (Figure 1E).
Rps19 deficiency results in macrocytic anemia, leukocytopenia, and variable platelet count
To assess the effects of Rps19 down-regulation in vivo, adult mice were given doxycycline in the drinking water. After 10 days of doxycycline administration, the homozygous shRNA-B and shRNA-D mice gradually started to appear moribund and only a small fraction of these mice survived up to 2 months after induction, whereas no mortality was observed in heterozygous mice when followed up to 3 months. Both mouse models showed retarded growth compared with littermate controls, and this was intermediate in the heterozygous and more pronounced in the homozygous mice (supplemental Figure 2). Histopathologic examination of the liver, spleen, kidneys, pancreas, and small intestines revealed no differences between Rps19-deficient and control mice. However, shRNA-B mice had occasional dilated glands in ventricle, large intestine, and colon (data not shown).
Two weeks after induction Rps19-deficient mice showed a reduced number of erythrocytes and reticulocytes (Figure 2A, supplemental Figure 3). Furthermore, at this time-point Rps19-deficient mice had an elevated platelet count, reduced number of lymphocytes, and a variable number of neutrophils (Figure 2A). The severe reduction in lymphocytes was persistent on prolonged Rps19 deficiency and with time Rps19-deficient mice also developed neutropenia and thrombocytopenia. The reduction in erythrocyte numbers was relatively mild, but all Rps19-deficient mice developed macrocytosis.
Induction of Rps19 deficiency leads to severe peripheral blood phenotype that progresses over time. (A) Erythrocyte number, mean corpuscular volume (MCV), neutrophil number, lymphocyte number, and platelet number (n = 12-19 per genotype at week 2; n = 6-16 per genotype at week 7). (B) Erythrocyte number after 15 days of doxycycline administration in lethally irradiated wild-type recipient mice that were transplanted with whole bone marrow cells from transgenic mice. (n = 7-9 recipients per genotype). Error bars represent SD.
Induction of Rps19 deficiency leads to severe peripheral blood phenotype that progresses over time. (A) Erythrocyte number, mean corpuscular volume (MCV), neutrophil number, lymphocyte number, and platelet number (n = 12-19 per genotype at week 2; n = 6-16 per genotype at week 7). (B) Erythrocyte number after 15 days of doxycycline administration in lethally irradiated wild-type recipient mice that were transplanted with whole bone marrow cells from transgenic mice. (n = 7-9 recipients per genotype). Error bars represent SD.
To confirm that the observed phenotype was autonomous to the blood system, unfractionated bone marrow cells from transgenic mice were transplanted into lethally irradiated wild-type recipients followed by doxycycline administration starting 4 weeks after transplantation. On day 15 after induction, the recipient mice reconstituted with shRNA-B or shRNA-D bone marrow had dramatically reduced number of erythrocytes, whereas doxycycline administration had no effect on recipient mice transplanted with control bone marrow (Figure 2B).
Chronic Rps19 deficiency results in bone marrow failure with irreversible exhaustion of HSCs
To gain further insights on the nature of hematopoietic defect in Rps19-deficient mice we applied a FACS strategy that allows a fractionation of myeloerythroid progenitors and erythroid precursors33,34 (supplemental Figure 4A). We decided to use the shRNA-B model as a primary model in the following experiments, because the onset of Rps19 deficiency in shRNA-D mice resulted a severe phenotype that prevented their meaningful analysis.
On day 10 after induction the shRNA-B mice had a decrease in bone marrow cellularity that correlated with the level of Rps19 down-regulation: on average 31% in heterozygous and 48% in homozygous shRNA-B mice (Figure 3A). Simultaneously, both heterozygous and homozygous shRNA-B mice had normal or increased frequencies of hematopoietic stem and progenitor cells compared with controls. However, at this time-point there was a decrease in the frequency of proerythroblasts and more mature erythroid precursors (Figure 3B).
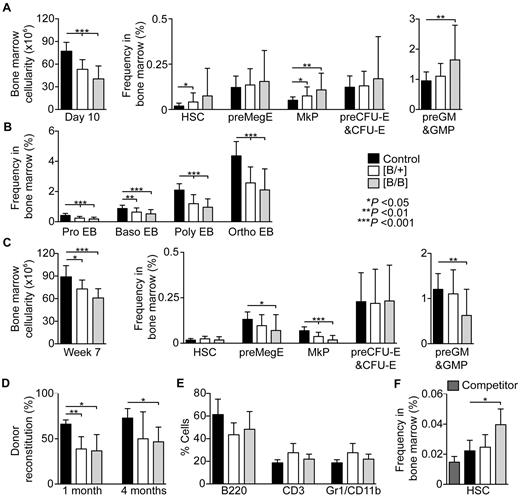
Chronic Rps19 deficiency results in bone marrow failure. (A) Bone marrow cellularity (n = 9-14 per genotype) and hematopoietic stem and progenitor cell frequencies in transgenic mice after 10 days of doxycycline administration (n = 18-31 per genotype). (B) Erythroblast frequencies after 10 days of doxycycline administration (n = 11-18 per genotype). (C) Bone marrow cellularity (n = 10-12 per genotype) and hematopoietic stem and progenitor frequencies in transgenic mice after 7 weeks of doxycycline administration (n = 10-17 per genotype). (D-E) shRNA-B mice (CD45.2; n = 3-5 per genotype) were induced for 30 days followed by 15 days without doxycycline, and unfractionated bone marrow cells were transplanted into lethally irradiated wild-type recipients (CD45.1/CD45.2; n = 3-5 per donor) together with an equal number of wild-type competitor bone marrow cells (CD45.1). (D) Average donor reconstitution 1 and 4 months after transplantation. (E) Lineage distribution of donor-derived cells in peripheral blood 4 months after transplantation. (F) Donor HSC frequencies in bone marrow at the time of transplantation. Error bars represent SD.
Chronic Rps19 deficiency results in bone marrow failure. (A) Bone marrow cellularity (n = 9-14 per genotype) and hematopoietic stem and progenitor cell frequencies in transgenic mice after 10 days of doxycycline administration (n = 18-31 per genotype). (B) Erythroblast frequencies after 10 days of doxycycline administration (n = 11-18 per genotype). (C) Bone marrow cellularity (n = 10-12 per genotype) and hematopoietic stem and progenitor frequencies in transgenic mice after 7 weeks of doxycycline administration (n = 10-17 per genotype). (D-E) shRNA-B mice (CD45.2; n = 3-5 per genotype) were induced for 30 days followed by 15 days without doxycycline, and unfractionated bone marrow cells were transplanted into lethally irradiated wild-type recipients (CD45.1/CD45.2; n = 3-5 per donor) together with an equal number of wild-type competitor bone marrow cells (CD45.1). (D) Average donor reconstitution 1 and 4 months after transplantation. (E) Lineage distribution of donor-derived cells in peripheral blood 4 months after transplantation. (F) Donor HSC frequencies in bone marrow at the time of transplantation. Error bars represent SD.
The immunophenotype became more severe over time and after 7 weeks shRNA-B mice had low frequencies of bipotential megakaryocytic-erythroid (preMegE), megakaryocytic (MkP), and preGM/GMP progenitor cells (Figure 3C). Moreover, when correlating the frequencies with bone marrow cellularity (Figure 3C, supplemental Figure 4B), the absolute numbers of all hematopoietic stem and progenitor cells were reduced. Consistently, the bone marrow of homozygous shRNA-B had low frequencies of colony-forming cells (supplemental Figure 4C).
This finding led us to study the effect of Rps19 deficiency on HSC function. We induced shRNA-B mice for 30 days followed by 15 days without doxycycline to restore Rps19 expression. Unfractionated bone marrow cells from transiently induced mice were transplanted into lethally irradiated wild-type recipients together with an equal number of wild-type competitor bone marrow cells. After 4 weeks the bone marrow cells from the transiently induced shRNA-B mice showed lower reconstitution in peripheral blood compared with the control cells, and the same trend persisted up to 4 months demonstrating the irreversible exhaustion of HSCs on transient Rps19 deficiency (Figure 3D). The lineage distribution in peripheral blood was not significantly changed (Figure 3E). Furthermore, the observed reconstitution defect is probably to be an underestimation because the transiently induced shRNA-B mice had on average more immunophenotypic HSCs on transplantation compared with induced control donor mice (Figure 3F).
Rps19 deficiency results in impaired proliferation and apoptosis of hematopoietic progenitors
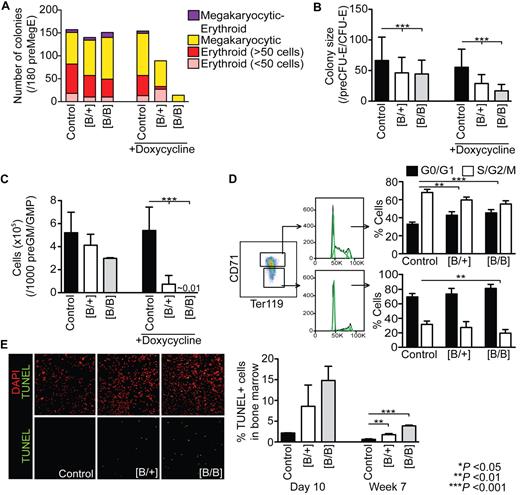
Next we studied the proliferative potential of Rps19-deficient hematopoietic progenitors to gain further understanding on the nature of erythroid defect in DBA. We sorted preMegE and preCFU-E/CFU-E progenitors from induced shRNA-B mice into single-cell liquid cultures with or without doxycycline. preMegEs give rise to megakaryocytic, erythroid, or mixed megakaryocytic/erythroid colonies, whereas preCFU-E/CFU-E progenitors produce only erythroid colonies.33 In the presence of doxycycline, heterozygous shRNA-B preMegEs produced smaller and fewer erythroid colonies compared with controls, whereas no erythroid colonies were observed in homozygous shRNA-B preMegE cultures (Figure 4A). The number of megakaryocytic colonies produced by heterozygous shRNA-B preMegEs was similar to controls, whereas only a few colonies were produced by homozygous shRNA-B preMegEs. Furthermore, Rps19-deficient megakaryocytes appeared smaller in size compared with controls (data not shown). However, the restoration of Rps19 expression on doxycycline removal rescued the colony formation. In contrast to the preMegE cultures, the removal of doxycycline resulted only in a partial rescue of colonies derived from shRNA-B preCFU-E/CFU-E cells and the defect was more pronounced in presence of doxycycline (Figure 4B). Taken together, these data indicate that the most severe erythroid defect is located downstream of preMegE progenitors, probably at the CFU-E–proerythroblast transition, and thus corroborates our FACS data. In addition to the erythroid cultures, the proliferation and survival of shRNA-B preGM/GMPs was dramatically affected in the presence of doxycycline, whereas no significant difference was observed on the restoration of Rps19 expression (Figure 4C).
Rps19 deficiency results in impaired proliferation of hematopoietic progenitors and increased apoptosis. shRNA-B mice were induced for 10 days with doxycycline followed by FACS sorting of (A) preMegE progenitors (n = 180 single-cell cultures), (B) preCFU-E/CFU-E progenitors (n = 240 single-cell cultures) or (C) 1000 preGM/GMP progenitors (n = 3-6 per genotype) into supportive liquid cultures with or without doxycycline. (D) Cell cycle analysis of early (CD71+ Ter119+) and late (CD71low Ter119+) erythroblasts in induced shRNA-B mice (n = 4-6 per genotype). Dean-Jett-Fox model was used to calculate percentages of cells in G1/G0, S, and G2/M phases (shown in green). (E) TUNEL staining of whole bone marrow cells from shRNA-B mice after 10 days (n = 2-3 per genotype) or 7 weeks (n = 3 per genotype) of doxycycline administration. Error bars represent SD.
Rps19 deficiency results in impaired proliferation of hematopoietic progenitors and increased apoptosis. shRNA-B mice were induced for 10 days with doxycycline followed by FACS sorting of (A) preMegE progenitors (n = 180 single-cell cultures), (B) preCFU-E/CFU-E progenitors (n = 240 single-cell cultures) or (C) 1000 preGM/GMP progenitors (n = 3-6 per genotype) into supportive liquid cultures with or without doxycycline. (D) Cell cycle analysis of early (CD71+ Ter119+) and late (CD71low Ter119+) erythroblasts in induced shRNA-B mice (n = 4-6 per genotype). Dean-Jett-Fox model was used to calculate percentages of cells in G1/G0, S, and G2/M phases (shown in green). (E) TUNEL staining of whole bone marrow cells from shRNA-B mice after 10 days (n = 2-3 per genotype) or 7 weeks (n = 3 per genotype) of doxycycline administration. Error bars represent SD.
Consistent with decreased proliferation in vitro, the early erythroid precursors (CD71+ Ter119+) in induced shRNA-B mice showed significant delay in the G1/S transition compared with controls (Figure 4D). Furthermore, Rps19-deficient late erythroid precursors (CD71low Ter119+) had a tendency for earlier cell cycle withdrawal compared with controls. In addition to the proliferative defect, the bone marrow cells of shRNA-B mice showed elevated apoptosis as assessed with TUNEL assay (Figure 4E).
RPS19 overexpression rescues the hematopoietic defect in vitro
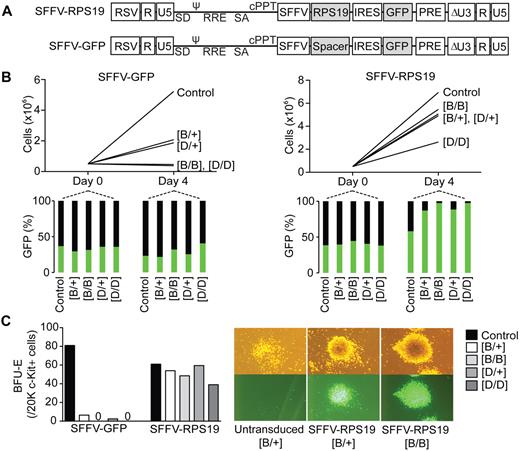
Because shRNAs can mediate off-target gene silencing or alternatively interfere with endogenous miRNAs, we wanted to confirm that the observed hematopoietic phenotype was specifically because of Rps19 deficiency. We designed a lentiviral vector overexpressing a codon-optimized human RPS19, which is not recognized by the Rps19-targeting shRNAs used in generated models (Figure 5A). A similar vector expressing GFP was used as a control. c-Kit–enriched hematopoietic progenitors from uninduced shRNA-B and shRNA-D mice were transduced and seeded into doxycycline-containing liquid cultures. The heterozygous shRNA-B and shRNA-D cells transduced with the control vector proliferated poorly compared with controls, whereas the homozygous shRNA-B and shRNA-D cells showed no proliferation (Figure 5B). However, RPS19 overexpression rescued the proliferative defect in both shRNA-B and shRNA-D cultures. Similarly, RPS19 overexpression rescued the BFU-E colony formation defect (Figure 5C).
RPS19 overexpression rescues the hematopoietic defect in vitro. (A) Design of lentiviral rescue vector overexpressing a codon-optimized human RPS19 and GFP, and the control vector expressing only GFP. (B) c-Kit–enriched hematopoietic progenitors from uninduced mice were transduced, and their proliferation was assayed in liquid cultures in presence of doxycycline. (C) BFU-E colony forming potential of transduced hematopoietic progenitors in presence of doxycycline. Data shown are from a single experiment but are representative of 2 repeat experiments. RSV indicates Rous sarcoma virus; ψ, packaging signal; SD, splice donor; RRE, rev responsive element; SA, splice acceptor; cPPt, polypurine tract; SFFV, spleen focus forming virus promoter; PRE, posttranscriptional regulatory element; IRES, internal ribosome entry site; and GFP, green fluorescent protein.
RPS19 overexpression rescues the hematopoietic defect in vitro. (A) Design of lentiviral rescue vector overexpressing a codon-optimized human RPS19 and GFP, and the control vector expressing only GFP. (B) c-Kit–enriched hematopoietic progenitors from uninduced mice were transduced, and their proliferation was assayed in liquid cultures in presence of doxycycline. (C) BFU-E colony forming potential of transduced hematopoietic progenitors in presence of doxycycline. Data shown are from a single experiment but are representative of 2 repeat experiments. RSV indicates Rous sarcoma virus; ψ, packaging signal; SD, splice donor; RRE, rev responsive element; SA, splice acceptor; cPPt, polypurine tract; SFFV, spleen focus forming virus promoter; PRE, posttranscriptional regulatory element; IRES, internal ribosome entry site; and GFP, green fluorescent protein.
Inactivation of Trp53 rescues the hematopoietic phenotype depending on the level of Rps19 down-regulation
Recent studies have implicated the role of p53 as a sensor for abnormal ribosome biogenesis.23,24,35-37 Consistently, we observed up-regulated expression of multiple p53 transcriptional target genes including Cdkn1a, Zmat3 (Wig1), Phlda3, Ccng1, Ptp4a3, and Bax in Rps19-deficient preCFU-E/CFU-E, preGM/GMP, and LSK progenitors (supplemental Figure 5).
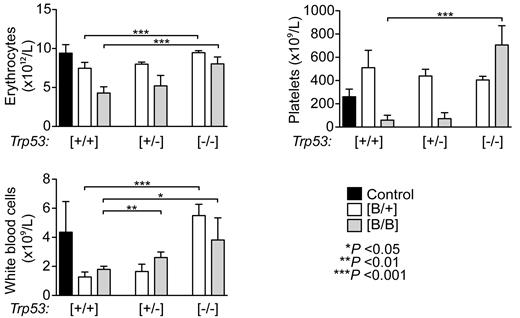
To ask whether the inactivation of Trp53 completely rescues the severe hematopoietic phenotype observed in the Rps19-deficient mice, we crossed shRNA-B mice with Trp53 null mice and transplanted bone marrow cells from these mice into lethally irradiated wild-type recipients. After 2 weeks of doxycycline administration the loss of Trp53 completely rescued the erythrocyte and white blood cell numbers in heterozygous shRNA-B mice, whereas the homozygous shRNA-B mice still showed a mild reduction in erythrocyte number (Figure 6). Furthermore, platelet count was increased in both heterozygous and homozygous shRNA-B mice. Although the deletion of one Trp53 allele in the homozygous shRNA-B mice ameliorated the hematopoietic phenotype, these mice failed to recover from the onset of Rps19 deficiency. Taken together, these data confirm the major role of p53 underlying the Rps19-deficient hematopoiesis.
Inactivation of Trp53 rescues the Rps19-deficient hematopoiesis depending on the level of Rps19 down-regulation. Lethally irradiated wild-type recipient mice were transplanted with 5 × 106 bone marrow cells from transgenic donors (that were either Trp53+/+, Trp53+/−, or Trp53−/−) followed by doxycycline administration to recipients starting 2 weeks after transplantation. Erythrocyte, platelet, and leukocyte numbers in peripheral blood of recipient mice after 2 weeks of doxycycline administration are shown (n = 4-9 recipients per genotype). Error bars represent SD.
Inactivation of Trp53 rescues the Rps19-deficient hematopoiesis depending on the level of Rps19 down-regulation. Lethally irradiated wild-type recipient mice were transplanted with 5 × 106 bone marrow cells from transgenic donors (that were either Trp53+/+, Trp53+/−, or Trp53−/−) followed by doxycycline administration to recipients starting 2 weeks after transplantation. Erythrocyte, platelet, and leukocyte numbers in peripheral blood of recipient mice after 2 weeks of doxycycline administration are shown (n = 4-9 recipients per genotype). Error bars represent SD.
Discussion
In the current study we have generated transgenic mouse models for RPS19-deficient DBA using RNAi as means to down-regulate Rps19 expression. This system allows an inducible and graded down-regulation of Rps19 expression providing an ideal tool to study haploinsufficient conditions like DBA.38 A previous study has demonstrated that CD34+ bone marrow cells in patients with premature termination codons in RPS19 show a 2- to 4-fold reduction in RPS19 mRNA.39 The quantification of Rps19 mRNA in different hematopoietic fractions in the generated mouse models revealed a similar degree of Rps19 down-regulation that was dependent on the used Rps19-targeting shRNA and its copy number.
The onset of Rps19 deficiency resulted in severe anemia, the key feature of DBA, and the severity of the phenotype correlated with the degree of Rps19 down-regulation. The majority of the mice with the most severe Rps19 down-regulation failed to recover from the onset of Rps19 deficiency and developed a bone marrow failure. However, with time the erythroid defect in the recovered mice developed considerably milder than in most patients suggesting a mouse-human species difference in the ability to compensate for Rps19 deficiency in the erythroid lineage. The location of the erythroid failure in DBA has not been fully defined, and studies using erythroid colony-forming assays have revealed a variable deficit of BFU-E and CFU-E progenitors in DBA patients.2,16-19 However, these assays are suboptimal because RPS19 deficiency affects the cloning efficiency and proliferation of erythroid progenitors. Based on FACS analysis and single-cell cultures of prospectively isolated erythroid progenitors we definitely prove that the most severe erythroid defect is located at the CFU-E–proerythroblast transition. Our findings are thus in agreement with previous studies suggesting that the most pronounced erythroid defect is at the onset of EPO-dependent terminal erythropoiesis.18,40 However, despite of slow cell cycle progression the terminal differentiation of Rps19-deficient erythroid precursors appeared normal.
Shortly after the induction of Rps19 deficiency, the mice developed thrombocytosis, a feature that is commonly seen in DBA patients at diagnosis.1 This finding suggests that thrombopoiesis is relatively intact on Rps19 deficiency, and the elevation in platelet count merely reflects the onset of stress erythropoiesis because these lineages originate from a common bipotential progenitor. However, with time the Rps19-deficient mice developed thrombocytopenia and neutropenia that correlated with a decrease in numbers of hematopoietic stem and progenitor cells in bone marrow. Similar progressive phenotype has been reported in DBA patients suggesting that a chronic Rps19 deficiency results in an exhaustion of the HSC compartment.2,19 Indeed, our data show that Rps19 deficiency results in irreversible exhaustion of HSCs that is not rescued by the restoration of Rps19 expression.
Recent studies have implicated the role of p53 as a sensor for abnormal ribosome biogenesis.23,24,35-37 For instance, rps19-deficient zebrafish show p53-dependent abnormalities in development and hematopoiesis.24 Furthermore, in Rps19-deficient Dsk3 mice, the activation of p53 results in melanocytosis together with a mild erythroid defect and increased apoptosis in bone marrow.23 Hematopoietic progenitors in Rps19-deficient mice showed up-regulation of p53 target genes and this was not restricted to the erythroid lineage. This finding is interesting given the recent study demonstrating the selective activation of p53 in RPS19-deficient primary human erythroid progenitors,37 and could partly explain the different hematopoietic manifestation in humans and mice. Consistently, the mice with a hypomorphic Mdm2 allele that results in a constitutive activation of p53 show a highly-similar hematopoietic phenotype compared with the generated Rps19-deficient mice including a severe lymphocytopenia, mild anemia, and neutropenia.41 Similarly to the rps19-deficient zebrafish24 and Dsk3 mice,23 the loss of p53 ameliorated the hematopoietic phenotype. However, our data indicate that the extent of rescue depends on the level of Rps19 down-regulation suggesting that p53-independent pathways may contribute toward the phenotype on severe Rps19 deficiency.
Although the 2 models exhibited some phenotypic differences, RPS19 overexpression rescued the hematopoietic defect in vitro in both models demonstrating that the observed phenotype is mainly because of down-regulation of Rps19. Together with our previous studies,42,43 it also provides a proof of principle for the feasibility of gene therapy as a treatment for RPS19-deficient DBA.
In conclusion, the generated mouse models for Rps19-deficient DBA provide a powerful tool for testing novel therapies for DBA, and the inducible nature of these models enables the collection of high numbers of hematopoietic progenitor cells required for large-scale therapeutic screens. This study demonstrates for the first time the feasibility of transgenic RNAi as means to generate mouse models for a human disease with haploinsufficient expression of a gene.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Konrad Hochedlinger and Rudolph Jaenisch for providing KH2 ES cell line, Ulrike Nuber for the Trp53−/− mice, and Eva Gynnstam for animal care.
This work was supported by the Hemato-Linné grant (Swedish Research Council Linnaeus), the Swedish Cancer Society (S.K.), the Swedish Children's Cancer Society (S.K.), the Swedish Medical Research Council (S.K.), the Tobias Prize awarded by The Royal Swedish Academy of Sciences financed by the Tobias Foundation (S.K.), and the EU project grants CONSERT, STEMEXPAND, and PERSIST. R.Q. was funded by a postdoctoral position from the Swedish Pediatric Cancer Society and J.F. by the Diamond-Blackfan Anemia Foundation. The Lund Stem Cell Center was supported by a Center of Excellence grant in life sciences from the Swedish Foundation for Strategic Research.
Authorship
Contribution: S.K. conceptualized the project and directed the research; S.K., J.F., J.L., K.O., and P.J. designed and generated the mouse models; P.J. analyzed hematopoiesis and performed gene expression analysis; P.J and D.B. performed flow cytometric analyses; R.Q. performed TUNEL analysis; A.H. and S.E. performed polysome and rRNA analyses; A.S. and C.B. generated the lentiviral vectors; M.E. analyzed histopathology; and P.J. and S.K. analyzed the data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefan Karlsson, Molecular Medicine and Gene Therapy, BMC A12, 221 84, Lund, Sweden; e-mail: stefan.karlsson@med.lu.se.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal