Abstract
The Ldb1/GATA-1/TAL1/LMO2 complex mediates long-range interaction between the β-globin locus control region (LCR) and gene in adult mouse erythroid cells, but whether this complex mediates chromatin interactions at other developmental stages or in human cells is unknown. We investigated NLI (Ldb1 homolog) complex occupancy and chromatin conformation of the β-globin locus in human erythroid cells. In addition to the LCR, we found robust NLI complex occupancy at a site downstream of the Aγ-globin gene within sequences of BGL3, an intergenic RNA transcript. In cells primarily transcribing β-globin, BGL3 is not transcribed and BGL3 sequences are occupied by NLI core complex members, together with corepressor ETO2 and by γ-globin repressor BCL11A. The LCR and β-globin gene establish proximity in these cells. In contrast, when γ-globin transcription is reactivated in these cells, ETO2 participation in the NLI complex at BGL3 is diminished, as is BCL11A occupancy, and both BGL3 and γ-globin are transcribed. In these cells, proximity between the BGL3/γ-globin region and the LCR is established. We conclude that alternative NLI complexes mediate γ-globin transcription or silencing through long-range LCR interactions involving an intergenic site of noncoding RNA transcription and that ETO2 is critical to this process.
Introduction
The human β-globin locus consists of 5 genes expressed sequentially during development: embryonic ϵ, fetal Gγ and Aγ, and adult δ and β. An enhancer, the locus control region (LCR), lies far upstream of the genes and is necessary for the high level of transcription achieved by these genes in erythroid cells.1 Developmental regulation of globin gene expression is incompletely understood. In particular, repression of fetal (γ) globin expression in adult cells is of great interest because an increase in fetal hemoglobin (HbF) in response to drugs or in patients with hereditary persistence of HbF ameliorates the severity of clinical symptoms associated with sickle cell disease and β-thalassemia. Therefore, reactivation of the fetal γ-globin genes in adult cells is a therapeutic target. Recent genetic and biochemical studies have implicated the zinc finger transcription factor BCL11A as a direct negative regulator of HbF expression2-4 ; however, the mechanism of action of BCL11A is largely unknown.
Achieving a high level of globin gene expression is dependent on a physical interaction between the LCR and globin genes.5,6 Proximity between the LCR and β-globin gene in mouse erythroid cells requires GATA-1 and EKLF, as well as the widely expressed protein NLI/Ldb1 (hereafter NLI).7-9 NLI stabilizes a complex including DNA-binding proteins GATA-1 and SCL/TAL1 (hereafter TAL1) and the bridging protein LMO2 on both the β-globin promoter and LCR, and mediates loop formation between them.9 Although EKLF shares many sites genome-wide with GATA-1 or TAL1, sites shared by all 3 proteins appear to be rare in mouse erythroid cells, leaving it unclear whether EKLF and the NLI complex function independently or together in mediating LCR/β-globin proximity or other long-range interactions.10 In human β-globin–transgenic mice, long-range interactions favor LCR/γ proximity but switch to favor LCR/β proximity during erythroid differentiation, which is consistent with the expression patterns of these genes.11 Furthermore, LCR/β-globin contacts diminish in favor of LCR/γ-globin contacts on a human transgene in BCL11A-null erythroid cells.4 It is unclear what protein(s) mediates these long-range interactions in human cells.
NLI, GATA-1, and TAL1 are required for erythropoiesis, and genome-wide localization studies support the idea that most TAL1- and GATA-1–regulatory functions in mouse erythroid cells are carried out in concert with NLI.12 In addition to the LCR, NLI, GATA-1, and TAL1 co-occupy numerous other sequences with potential enhancer activity.13 Association studies suggest these proteins participate in distinct activating and repressing complexes.12-17 Negative function is thought to be conferred by participation in the complex of the corepressor MTG16/ETO2 (also known as CBFA2T3, hereafter ETO2). For example, when ETO2 is recruited by TAL1, select target genes are repressed.17,18 Likewise, several targets of the NLI complex show increased NLI occupancy and decreased ETO2 occupancy as they become activated during MEL cell differentiation.12 The concept of cofactor exchange to modulate transcriptional outcome adds an important dimension to the regulation of gene expression.19
We investigated NLI complex occupancy in human K562 cells and adult erythroid progenitor cells. In addition to the LCR, ChIP-chip studies identified a novel region of NLI/GATA-1/TAL1/LMO2 occupancy 3′ to the Aγ-globin gene. These intergenic sites are within sequences of BGL3, a noncoding RNA transcript (GenBank AY034471). In progenitor cells, this region was similarly occupied by the NLI complex whether they were differentiated to produce cells with high levels of adult hemoglobin or under conditions in which fetal γ-globin transcription was robustly reactivated. BGL3 transcription was consistently associated with γ-globin transcription. Furthermore, the BGL3 region acquired proximity with the LCR and γ-globin genes when transcription was active. This proximity was precluded in cells with low γ-globin transcription in which the corepressor ETO2 participated in the NLI complex. The BCL11A site downstream of Aγ-globin2 is also located within BGL sequences, but BCL11A occupancy was robust only when γ-globin and BGL3 transcription were low, which is consistent with its role as a γ-globin repressor. These results suggest that diverse NLI complexes mediate the ability of the human fetal γ-globin genes to establish productive long-range interactions with the LCR, and that the BGL3 transcript region may play a role in the regulation of γ-globin expression during development through participating in long-range interactions between the LCR and gene.
Methods
Cell culture
Primary human erythroid cells were collected under National Institutes of Health Institutional Review Board–approved procedures and cultured as described previously.20 Briefly, CD34+ cells from healthy donors were grown for 14 days in Eagle minimum essential medium with erythropoietin (Epo) alone (4 U/mL; Amgen) or with Epo, SCF (50 ng/mL; R&D Systems), and TGF-β (1.25 ng/mL; R&D Systems). Nearly identical erythroblast maturation under the 2 regimens yielded cells containing low levels of HbF (<10%) in Epo or high levels of HbF (∼ 40%) with Epo, SCF, and TGF-β. K562 cells were cultured in RPMI 1640 medium containing 10% FBS.
Quantitative RT-PCR
RNA was isolated from approximately 2 × 106 K562 cells or high-HbF and low-HbF cells on day 9 of differentiation with the RNeasy Plus Micro kit (QIAGEN), treated with DNaseI, reverse transcribed, and diluted as described previously.21 Control reactions contained no reverse transcriptase. cDNA was analyzed using iQ SYBR Green Supermix (Bio-Rad) and quantitative RT-PCR was performed in a Prism 7900HT (ABI). Relative enrichment of cDNA sequences was calculated against a genomic DNA standard derived from matched cells using the comparative Ct method.22 Three preparations of RNA for each cell type were analyzed and results normalized to GAPDH. Sequence homology between Gγ- and Aγ-globin precludes reporting results for these genes separately. β-globin locus primers were described previously.21 BGL3 primers are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
ChIP
ChIP was performed essentially as described previously.20 For ETO2 ChIP, cells were first cross-linked using 2mM ethylglycol bis [succinimidylsuccinate] for 30 minutes at 25°C and the reaction was quenched in 20mM Tris, pH 7.5, for 15 minutes at 25°C. Cells were pelleted and resuspended in PBS before formaldehyde cross-linking. Subsequent procedures were unchanged. Chromatin was prepared from 4-12 donor-matched low- and high-HbF cell cultures on day 9 of differentiation. Immunoprecipitated DNA was amplified in 10-μL reactions using iQ SYBR Green Supermix (Bio-Rad). Relative enrichment was calculated against input, and input standard curves were used to control for minor variations in primer efficiencies.23 Between 3 and 12 ChIP samples were analyzed per Ab and results normalized to GAPDH. Abs used included RNA Polymerase II (N-20), Clim2 (NLI/Ldb1), GATA-1, and ETO2 (MTG16), all obtained from Santa Cruz Biotechnology. H3K4me3 Ab was obtained from Millipore and Ctip1 (Bcl11A) Abs were from Abcam. TAL1 Ab was a kind gift of Dr Catherine Porcher (University of Oxford).24
NLI/GATA-1/Pol II ChIP-chip
ChIP DNA was prepared from K562 cells using NLI, GATA-1, and RNA polymerase II Abs and was amplified as recommended (NimbleGen). ChIP-chip analysis was carried out with NimbleGen tiling arrays consisting of 50 mers positioned every 100 bp along nonrepetitive sequences of chr10 and chr11 in the hg18 assembly (chr10:99783524-135370801 and chr11:5000-35341352). We used the ACME algorithm to call peaks.25 The final parameters used were determined for each factor separately by iteratively running ACME with different parameter sets and determining the set resulting in peaks that best fit their documented binding.9 GATA-1: threshold = 0.99, pval = 10−14, window = 750; Pol II: threshold = 0.90, pval = 10−10, window = 750; NLI: threshold = 0.99, pval = 10−14, window = 750. The complete dataset has been deposited in the Gene Expression Omnibus under accession number GSE30047
Chromosome conformation capture
Chromosome conformation capture (3C) was performed as described previously.26 Briefly, K562, low- and high-HbF cell nuclei were fixed in 1% formaldehyde, digested with EcoRI overnight, and ligated for 4 hours with T4 DNA ligase at 16°C. Cross-links were reversed and ligation products extensively purified. Digestion and ligation efficiencies were determined as described previously5 using quantitative PCR, and primer efficiencies were determined using standard curves generated with a control template. Ligation products were amplified using the iQ SYBR Green Supermix (Bio-Rad). Ligation events were confirmed by sequencing quantitative PCR products. Ligation frequencies were calculated using a conserved gene desert region on chromosome 16 (ENCODE region ENr313) to adjust for template differences among samples. Results were normalized to the ligation frequency of the anchor fragment with a fragment outside the globin locus to adjust for observed differences in levels of globin locus-specific background ligation in transformed cells (K562) and normal human erythroid progenitor cells (see supplemental Table 2 for the 3C primer sequences). The interaction frequency reported for the EcoRI fragment containing the Gγ-globin gene represents the value for interaction of both γ-globin genes with the LCR. Only the Aγ-globin gene could be probed uniquely because the 3′ termini of the Aγ and Gγ EcoRI fragments are identical and the 5′ terminus of the Gγ-globin EcoRI fragment is within a repetitive LINE element.
RNAi and overexpression studies
K562 cells were transfected by the Nucleofector technology (Kit V) using program T-016 (Amaxa Biosystems), and cells were harvested at 48 hours after transfection. Transfection efficiency was monitored by EmGFP expression under fluorescence microscopy. The ETO2 (CBFA2T3) RNAi and control vectors were constructed using the BLOCK-iT Pol II miR RNAi expression vector kit (Invitrogen) and the HA-NL1 expression vector was described previously.9 The ETO2 target sequence was: 5′-ATTGACGATCGAGGAGTTTCA-3′.
Western blotting
Western blotting was performed as described previously.9 The blots were immunostained for HA-NLI using an anti-HA mAb (11-867-423001; Roche) at a 1:2000 dilution. ETO2 goat polyclonal Ab (sc-9739; Santa Cruz Biotechnology) was used at a 1:500 dilution. Actin mouse mAb (sc-8432; Santa Cruz Biotechnology) was used at a 1:1000 dilution. GATA-1 Ab (sc-1233; Santa Cruz Biotechnology) was used at a 1:1000 dilution.
Results
The NLI/GATA-1/TAL1/LMO2 complex occupies an intergenic site 3′ to Aγ-globin
In mouse erythroid cells, adult β-globin gene-LCR proximity requires the NLI complex.9 Previously, using ChIP and quantitative PCR, we detected NLI complex members at LCR DNase I–hypersensitive sites in human K562 cells, an erythroleukemia cell line expressing primarily fetal γ-globin.9 However, the complex was not detected at the γ-globin gene promoters but low GATA-1 occupancy was observed, leaving open the question of which proteins mediate long-range LCR contacts of the fetal globin genes.
To begin to address this issue, we performed ChIP-chip analysis in K562 cells with GATA-1 and NLI Abs using a NimbleGen tiling array compromising 70 Mb of nonrepetitive DNA sequences on human chromosomes 10 and 11 (∼ 2% of the human genome) including the β-globin cluster. We included RNA polymerase II (pol II) in the analysis because it occupies the LCR along with the NLI complex and this co-occupancy might be a signature of positive long-range gene interactions mediated by NLI.9,27 To predict sites of NLI and GATA-1 enrichment, we analyzed the data using the ACME algorithm with parameters chosen to maximize peak designation at the LCR sites of NLI complex occupancy previously confirmed by ChIP assay (see “ChIP” for further details).9
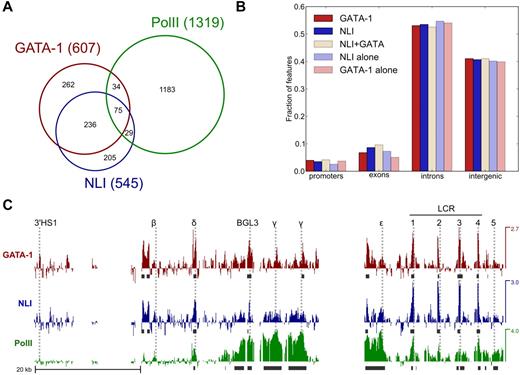
We observed a strong tendency for GATA-1 and NLI co-occupancy with 51% of GATA-1 sites and 57% of NLI sites shared with the other protein, indicating that the most common means by which NLI confers regulatory effects is together with GATA-1 (Figure 1A). Both a limited analysis and a genome-wide study in mouse erythroid cells had found a large fraction of sites occupied by GATA-1, NLI, and TAL1 to be shared by all 3 proteins.12,13 In addition, 24% of the NLI and GATA-1 co-occupied sites were shared with pol II. We interpret this result to indicate that the NLI complex may participate in both activating and possibly repressive interactions with chromatin, as has been suggested previously.13 NLI and GATA-1 occupied primarily intronic and intergenic sites whether they were occupied exclusively or jointly (Figure 1B).
Substantial co-occupancy of genomic sites by NLI and GATA-1 in human cells. Localization of NLI, GATA-1, and RNA pol II in K562 cells was examined by ChIP-chip. (A) Venn diagram showing intersection of genomic binding sites of GATA-1, NLI, and pol II within the region probed by tiling array. Total sites are indicated in parentheses; some pol II sites overlap more than one GATA-1 or NLI site, preventing the sum of interactions in the pol II circle from being equal to the total sites observed. (B) Bar plot of fraction of sites for each factor within promoters, exons, introns, and intergenic space based on RefSeq annotations. Promoter regions are defined as TSS plus 1 kb upstream. Exon and intron categories are not mutually exclusive because of multiple isoforms and overlapping genes. Intergenic space is defined as a lack of annotated transcripts and excludes the 1-kb promoter regions. (C). Tiling array signal (log2 ratios) across the globin locus on chr11. Regulatory elements and globin gene promoters are indicated by dotted lines. Peaks were called by ACME (using the parameters described in “NL1/GATA-1/Pol II ChIP-chip”) and are indicated below each track as dark gray boxes. Results for NLI, GATA-1, and RNA pol II ChIP-chip are shown. Data have been deposited in GEO under accession number GSE30047.
Substantial co-occupancy of genomic sites by NLI and GATA-1 in human cells. Localization of NLI, GATA-1, and RNA pol II in K562 cells was examined by ChIP-chip. (A) Venn diagram showing intersection of genomic binding sites of GATA-1, NLI, and pol II within the region probed by tiling array. Total sites are indicated in parentheses; some pol II sites overlap more than one GATA-1 or NLI site, preventing the sum of interactions in the pol II circle from being equal to the total sites observed. (B) Bar plot of fraction of sites for each factor within promoters, exons, introns, and intergenic space based on RefSeq annotations. Promoter regions are defined as TSS plus 1 kb upstream. Exon and intron categories are not mutually exclusive because of multiple isoforms and overlapping genes. Intergenic space is defined as a lack of annotated transcripts and excludes the 1-kb promoter regions. (C). Tiling array signal (log2 ratios) across the globin locus on chr11. Regulatory elements and globin gene promoters are indicated by dotted lines. Peaks were called by ACME (using the parameters described in “NL1/GATA-1/Pol II ChIP-chip”) and are indicated below each track as dark gray boxes. Results for NLI, GATA-1, and RNA pol II ChIP-chip are shown. Data have been deposited in GEO under accession number GSE30047.
In the β-globin locus, NLI, GATA-1 and pol II co-occupied HS1-HS4 of the LCR (Figure 1C). A peak was called for GATA-1 at the Gγ-globin promoter but not for NLI at either γ-globin promoter, which is consistent with our earlier data,9 although these genes were robustly occupied by pol II, as expected. In addition, several intergenic regions were enriched for NLI, GATA-1, and pol II, including the BGL3 intergenic transcript region 3′ of the Aγ-globin gene. Earlier work had suggested that cis-regulatory elements other than promoters might be involved in long-range chromatin interactions in this locus, but none have been described.28 Our results suggest that the NLI complex may be important for LCR/γ-globin long-range chromatin looping in human erythroid cells and implicate the BGL3 transcript region in the process.
BGL3 transcription is consistently associated with γ-globin transcription and low BCL11A occupancy
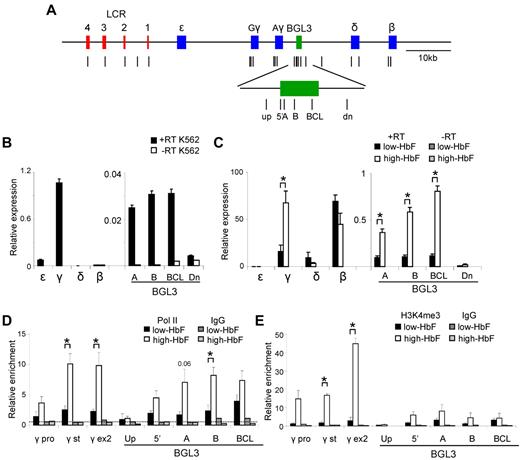
Human erythroid progenitor cells differentiated in the presence of Epo alone or Epo plus SCF and TGFβ produce cells that contain low (< 10%) or high (∼ 40%) levels of HbF (low- or high-HbF cells), respectively.20 The reactivation of γ-globin transcription in these adult erythroid cells is associated with BGL3 transcription (J.L.M., unpublished data). To compare BGL3 transcription in low- and high-HbF cells with K562 cells, in which we had observed the NLI complex binding to BGL3 sequences, expression from the globin genes and intergenic regions was assessed by quantitative PCR (Figure 2A).
BGL3 is preferentially expressed in cells containing high HbF. (A) Map of the human β-globin locus showing the positions of primer pairs used to determine RNA transcript levels and factor occupancy by ChIP and real-time quantitative PCR (vertical lines). The globin genes and LCR DNase I–hypersensitive sites are represented by blue and red rectangles, respectively. The BGL3 region is depicted by a green rectangle. (B) Globin gene and intergenic transcription in K562 cells was evaluated using reverse-transcribed total RNA and quantitative RT-PCR with primers specific to the regions indicated on the x-axis. White bars indicate no reverse transcriptase. Results were normalized to GAPDH and error bars indicate SD. (C) Globin gene and intergenic transcription was evaluated for low-HbF (black bars) and high-HbF cells (white bars) as in panel B. Dark and light gray bars indicate values for reactions with no reverse transcriptase for low- and high-HbF cells, respectively. (D) ChIP was performed using an RNA Polymerase II Ab for low- and high-HbF cells. The locations of primers for PCR are indicated on the x-axis. Results were calculated against an input sample and normalized to GAPDH. Error bars indicate SD; black bars, low-HbF cells; white bars, high-HbF cells. Dark and light gray bars indicate values for an isotype matched control Ab in low- and high-HbF cells, respectively. The dotted line indicates the average of IgG control precipitations across all amplicons. (E) ChIP was performed using an H3K4me3 Ab to determine occupancy in low- and high-HbF cells. All data were calculated and plotted as described in panel D. *P < .05.
BGL3 is preferentially expressed in cells containing high HbF. (A) Map of the human β-globin locus showing the positions of primer pairs used to determine RNA transcript levels and factor occupancy by ChIP and real-time quantitative PCR (vertical lines). The globin genes and LCR DNase I–hypersensitive sites are represented by blue and red rectangles, respectively. The BGL3 region is depicted by a green rectangle. (B) Globin gene and intergenic transcription in K562 cells was evaluated using reverse-transcribed total RNA and quantitative RT-PCR with primers specific to the regions indicated on the x-axis. White bars indicate no reverse transcriptase. Results were normalized to GAPDH and error bars indicate SD. (C) Globin gene and intergenic transcription was evaluated for low-HbF (black bars) and high-HbF cells (white bars) as in panel B. Dark and light gray bars indicate values for reactions with no reverse transcriptase for low- and high-HbF cells, respectively. (D) ChIP was performed using an RNA Polymerase II Ab for low- and high-HbF cells. The locations of primers for PCR are indicated on the x-axis. Results were calculated against an input sample and normalized to GAPDH. Error bars indicate SD; black bars, low-HbF cells; white bars, high-HbF cells. Dark and light gray bars indicate values for an isotype matched control Ab in low- and high-HbF cells, respectively. The dotted line indicates the average of IgG control precipitations across all amplicons. (E) ChIP was performed using an H3K4me3 Ab to determine occupancy in low- and high-HbF cells. All data were calculated and plotted as described in panel D. *P < .05.
In K562 cells, γ-globin was predominantly transcribed (Figure 2B), whereas in low-HbF cells, β-globin transcription predominated and both genes were transcribed in high-HbF cells, as expected (Figure 2C). BGL3 transcript levels represented a similar fraction of γ-globin transcripts in high-HbF and K562 cells (1% and 2%-3%, respectively), although the overall abundance of γ-globin transcripts in K562 cells was only approximately 1%-2% of that in high-HbF cells. The increase of BGL3 and γ-globin transcripts in high-HbF cells compared with low-HbF cells was similar at 4- to 5-fold. Consistent with the correlation of BGL3 and γ-globin transcription, expression of BGL3 was minimal in low-HbF cells. This result is in agreement with studies using a human YAC transgene that showed higher transcript levels through the BGL3 intergenic region during embryonic compared with adult stages of development.29
ChIP experiments indicated that the transcript levels observed in high-HbF cells were reflected by pol II enrichment at γ-globin and BGL3 despite the difference in the overall expression level of the 2 transcripts (Figure 2D). Trimethylation of lysine 4 on histone H3 (H3K4me3), known to be correlated with active transcription, varied across the γ-globin and BGL3 regions in high-HbF cells at levels more reflective of the relative abundance of globin gene and BGL3 transcripts (Figure 2E). These 2 regions were also marked by H3K4me3 and other active histone marks in K562 cells (supplemental Figure 1, The ENCODE Project Consortium 2004). pol II and H3K4me3 were very low in the BGL3 region in low-HbF cells.
The BGL3 transcript region includes a site occupied by BCL11A in primary adult erythroid progenitors.2 To understand whether this interaction occurs in progenitors when γ-globin transcription is reactivated, we investigated BCL11A occupancy in high-HbF and low-HbF cells by ChIP and quantitative PCR (Figure 3). BCL11A occupied LCR HS2 and HS3, as expected.2,4,30 Using several primer sets across the BGL3 transcript region, we found BCL11A occupancy at its reported binding site 3′ to Aγ-globin2 and to a lesser extent at more upstream positions in BGL3 that are near additional BCL11A motifs (supplemental Figure 2). At the primary BCL11A site in BGL3, there was a very strong decrease in BCL11A occupancy in high-HbF compared with low-HBF cells
BCL11A occupancy in BGL3 changes when γ-globin is reactivated. ChIP was performed using a BCL11A Ab to determine occupancy in low- and high-HbF cells. Solid black bars indicate low-HbF cells; white bars, high-HbF cells. Dark and light gray bars indicate values for an isotype control Ab in low- and high-HbF cells, respectively. The dotted line indicates the average of IgG control precipitations across all amplicons. Results were calculated and plotted as described in Figure 2D. *P < .05.
BCL11A occupancy in BGL3 changes when γ-globin is reactivated. ChIP was performed using a BCL11A Ab to determine occupancy in low- and high-HbF cells. Solid black bars indicate low-HbF cells; white bars, high-HbF cells. Dark and light gray bars indicate values for an isotype control Ab in low- and high-HbF cells, respectively. The dotted line indicates the average of IgG control precipitations across all amplicons. Results were calculated and plotted as described in Figure 2D. *P < .05.
These results indicate that γ-globin and BGL3 are consistently expressed together, and that in progenitor cells expressing high levels of γ-globin, BCL11A occupancy within BGL3 is low, supporting the idea that this is an important site through which it influences γ-globin switching.
Alternative NLI complexes bind the LCR and BGL3 region in low- compared with high-HbF cells
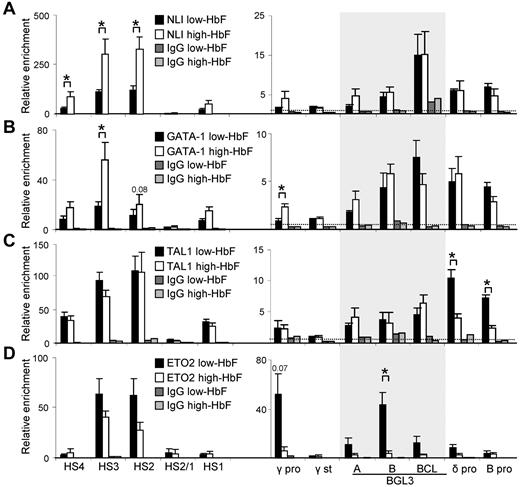
We next assessed the occupancy of NLI, GATA-1, and TAL1 by ChIP and quantitative PCR in low- and high-HbF cells. We included in the analysis ETO2, a corepressor that confers a negative function in association with this complex.12,13,17 Figure 4 shows that all 4 proteins are detectable at the LCR HSs, principally HS2 and HS3, in low-HbF cells. However, when γ-globin is reactivated by cytokines, the composition of the complex at the LCR is altered and there is a substantial increase in NLI and GATA-1 occupancy and a decrease in ETO2, whereas TAL1 is unchanged.
NLI and complex members TAL1, GATA-1, and ETO2 differentially bind to the LCR and globin genes in low- and high-HbF cells. ChIP was performed using Abs specific for NLI (A), GATA-1 (B), TAL1 (C), and ETO2 (D) to determine occupancy in low- and high-HbF cells. Solid black bars indicate low-HbF cells; white bars, high-HbF cells. Dark and light gray bars indicate values for an isotype-matched control Ab in low- and high-HbF cells, respectively. The shaded area shows the 900-bp region encompassed by the 3 primer pairs within BGL3. The dotted line in all panels indicates the average of IgG control precipitations across all amplicons. All data were calculated and plotted as described in Figure 2D. *P < .05.
NLI and complex members TAL1, GATA-1, and ETO2 differentially bind to the LCR and globin genes in low- and high-HbF cells. ChIP was performed using Abs specific for NLI (A), GATA-1 (B), TAL1 (C), and ETO2 (D) to determine occupancy in low- and high-HbF cells. Solid black bars indicate low-HbF cells; white bars, high-HbF cells. Dark and light gray bars indicate values for an isotype-matched control Ab in low- and high-HbF cells, respectively. The shaded area shows the 900-bp region encompassed by the 3 primer pairs within BGL3. The dotted line in all panels indicates the average of IgG control precipitations across all amplicons. All data were calculated and plotted as described in Figure 2D. *P < .05.
Occupancy of NLI, GATA-1, and TAL1 was observed broadly across the BGL3 region where 3 NLI composite E box/GATA-1 motifs are distributed (Figure 4A-C and supplemental Figure 2), with no significant difference in occupancy between low- and high-HbF cells. These results validate detection of NLI and GATA-1 at BGL3 by ChIP-chip in K562 cells (Figure 1). Furthermore, ETO2 was strongly detected within BGL3 in low-HbF cells, whereas occupancy was significantly reduced in high-HbF cells (Figure 4D).
At the γ-globin promoters, ETO2 enrichment was robust in low-HbF cells, and that of NLI, GATA-1, and TAL1 was low but detectable. Similar to the LCR, NLI and GATA-1 enrichment increased at these promoters after γ-globin reactivation and ETO2 occupancy decreased 10-fold. A different pattern was observed at the δ- and β-globin promoters. The NLI complex occupied these promoters in both low- and high-HbF cells, but TAL1 occupancy was lower in high-HbF cells; the significance of this difference is unclear because β-globin is transcribed in both cell types. ETO2 did not significantly occupy these promoters in either condition. Therefore, the corepressor ETO2 participates in the NLI/GATA-1/TAL1 complex occupying LCR/HS2-HS3 and the BGL3 region and occupies the γ-globin promoter in low-HbF cells when γ-globin and BGL3 are not being transcribed, but is greatly reduced at these locations when transcription is activated in high-HbF cells.
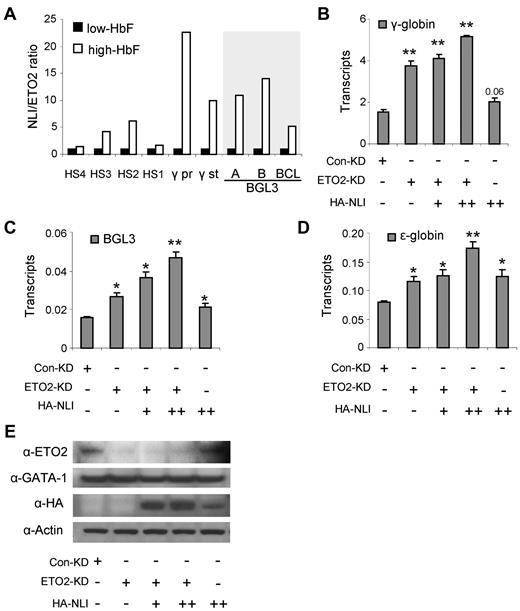
The increase in the ratio of NLI/ETO2 on γ-globin and BGL3 activation (Figure 5A) suggested that ETO2 functions in the NLI complex to repress γ-globin transcription. To test this idea, we reduced ETO2 using RNAi in K562 cells, which resulted in increased γ-globin and BGL3 transcription (Figure 5B-C). Further alteration of the NLI/ETO2 ratio in these cells through overexpression of NLI augmented these effects. Interestingly, ϵ-globin transcription was also augmented by increasing the NLI/ETO2 ratio in K562 cells (Figure 5D), as were the transcript levels of the known NLI targets glycophorin A and P4.2 (supplemental Figure 3). The reduction of ETO2 and the overexpression of NLI was confirmed by Western blot analysis using Abs to these proteins, as shown in Figure 5E, whereas GATA-1 protein levels were unchanged. These results support the idea that γ-globin is a target of the NLI complex and that ETO2 participation in the complex influences the regulatory outcome. Furthermore, factor exchange through the NLI complex may influence transcription of a cohort of erythroid genes.
Increasing the NLI/ETO2 ratio in K562 cells induces γ-globin transcription. (A) The ratio of abundance of NLI/ETO2 at locations across the locus is plotted using the values obtained by ChIP assays shown in Figure 4. (B) The expression of γ-globin was monitored by quantitative RT-PCR after manipulating the NLI/ETO2 ratio in K562 cells. Cells were collected 48 hours after transfection with a control RNAi vector or with an ETO2 RNAi vector alone or together with an NLI-expressing vector (+, 2 μg; ++, 4 μg). Results were normalized to GAPDH and error bars indicate SD. *P < .05; **P < .001. The expression of BGL3 (C) and ϵ-globin (D) were monitored by quantitative RT-PCR after manipulating the NLI/ETO2 ratio in K562 cells exactly as described for panel B. (E) Western blot analysis was performed using Abs to ETO2 and NLI to confirm ETO2 knockdown and NLI overexpression. Actin served as the loading control.
Increasing the NLI/ETO2 ratio in K562 cells induces γ-globin transcription. (A) The ratio of abundance of NLI/ETO2 at locations across the locus is plotted using the values obtained by ChIP assays shown in Figure 4. (B) The expression of γ-globin was monitored by quantitative RT-PCR after manipulating the NLI/ETO2 ratio in K562 cells. Cells were collected 48 hours after transfection with a control RNAi vector or with an ETO2 RNAi vector alone or together with an NLI-expressing vector (+, 2 μg; ++, 4 μg). Results were normalized to GAPDH and error bars indicate SD. *P < .05; **P < .001. The expression of BGL3 (C) and ϵ-globin (D) were monitored by quantitative RT-PCR after manipulating the NLI/ETO2 ratio in K562 cells exactly as described for panel B. (E) Western blot analysis was performed using Abs to ETO2 and NLI to confirm ETO2 knockdown and NLI overexpression. Actin served as the loading control.
The BGL3 region participates in long-range interactions with the LCR and γ-globin genes
Given the distribution of NLI complex members within BGL3 sequences, we hypothesized that the LCR may come into proximity with BGL3 in adult erythroid cells. To determine the proximity of sites in the locus relative to the LCR, we performed 3C analysis interrogating 18 EcoRI fragments spanning the majority of the β-globin locus. The BGL3 5′ region and downstream sequences are present on 2 separate EcoRI fragments, as are the 2 γ-globin genes, with additional small fragments separating these elements, thus providing a high-resolution view of this region.
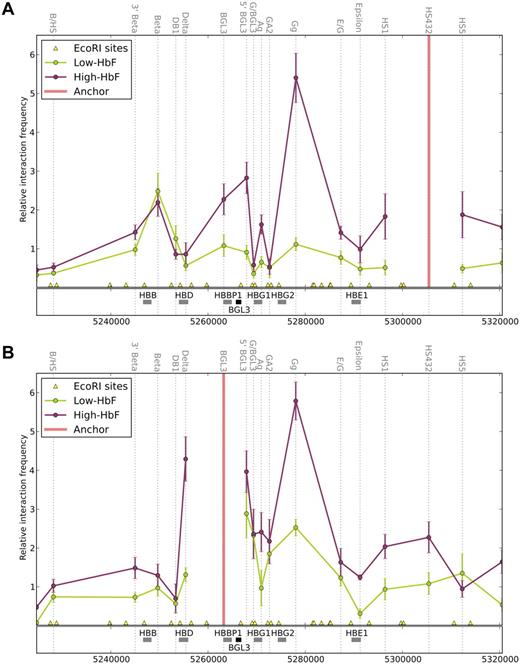
Using the LCR as an anchor fragment, the primary interaction observed in low-HbF cells was between the LCR and β-globin gene, whereas in high-HbF cells, there was an increase in the interaction frequency between the LCR and γ-globin genes (Figure 6A). The higher interaction frequency between the LCR and Gγ-globin compared with Aγ-globin is contributed to significantly by the hybridization of this 3C primer to both γ-globin gene–containing EcoR1 fragments (see “Chromosome conformation capture” for a further explanation), but also may reflect the preponderance of Gγ-globin over Aγ-globin in high-HbF cells.20 The switching of LCR contacts between the γ- and β-globin genes occurs on a transgene in mice as their transcription profiles change during development.11 Nevertheless, in high-HbF cells, the proximity of the β-globin gene and the LCR remained observable, which is consistent with its continued expression in these cells. The δ-globin gene, which is transcribed at a very low level (Figure 2C), was not observed in proximity to the LCR, although NLI complex members occupied the promoter. Therefore, the NLI complex is insufficient, in this case, to establish LCR proximity. Some aspect of the δ-globin promoter may compromise proximity; for example, the absence of an EKLF-binding motif.31,32
Long-range interactions in the β-globin locus include the BGL3 region. 3C analysis was performed for cells expressing high- and low-HbF. Each EcoRI cleavage site is represented on the genome browser by a yellow triangle, and the interaction frequency with the designated anchor fragment is plotted in the middle of each fragment (dotted vertical lines). The globin genes and BGL3 sequences are shown in the middle track as gray and black rectangles, respectively. Colored line graphs represent data from low-HbF cells (green) and high-HbF cells (red). (A) Relative interaction frequency between the LCR2-4 fragment as anchor and other regions of the locus. (B) Relative interaction frequency between the BGL3 region and other parts of the locus. Note that the interaction frequency reported for the EcoRI fragment containing the Gγ-globin gene represents the value for both γ-globin gene interactions with the LCR (see “Chromosome conformation capture”). Error bars indicate SD.
Long-range interactions in the β-globin locus include the BGL3 region. 3C analysis was performed for cells expressing high- and low-HbF. Each EcoRI cleavage site is represented on the genome browser by a yellow triangle, and the interaction frequency with the designated anchor fragment is plotted in the middle of each fragment (dotted vertical lines). The globin genes and BGL3 sequences are shown in the middle track as gray and black rectangles, respectively. Colored line graphs represent data from low-HbF cells (green) and high-HbF cells (red). (A) Relative interaction frequency between the LCR2-4 fragment as anchor and other regions of the locus. (B) Relative interaction frequency between the BGL3 region and other parts of the locus. Note that the interaction frequency reported for the EcoRI fragment containing the Gγ-globin gene represents the value for both γ-globin gene interactions with the LCR (see “Chromosome conformation capture”). Error bars indicate SD.
Interestingly, the BGL3 region, which is present on 2 EcoRI fragments interrogated by BGL3 and 5′BGL3 primers, established close LCR proximity in high-HbF cells (Figure 6A). This interaction was also observed in K562 cells (supplemental Figure 4), but only for the 5′BGL3-containing fragment. The difference may reflect the lower level of transcription of BGL3 and γ-globin in these cells compared with high-HbF cells (Figure 2B-C). In comparison, BGL3/LCR proximity was not elevated in low-HbF cells. To determine the extent of BGL3 interaction with other regions in the β-globin locus directly, the BGL3 fragment was used as an anchor for 3C analysis (Figure 6B). In high-HbF cells, BGL3 interacted strongly with the γ-globin genes; in low-HbF cells, the interaction frequency was diminished. BGL3 proximity with the LCR was also observed in high-HbF cells (Figure 6A), but was reduced in low-HbF cells.
These data establish that the BGL3 region and γ-globin genes make frequent long-range contacts when they are transcribed. At the same time, the regions frequently contact the LCR. These contacts do not occur when ETO2 occupancy is robust at any of these sites. Both low- and high-HbF cells transcribe the β-globin gene, and β-globin/LCR interactions occur in both cell types despite the differing participation of ETO2 in the NLI complex at the LCR. We conclude that the NLI/TAL1/E2A/GATA-1/ETO2 complex specifically disfavors γ-globin and BGL3 transcription by interfering with their long-range LCR interactions.
Discussion
In mouse erythroid cells, the NLI/GATA-1/TAL1 complex mediates long-range interaction between the LCR and β-globin gene. In human erythroid cells, we find that this complex, in addition to occupying the LCR, robustly occupies the BGL3 noncoding transcript region 3 kb downstream of Aγ-globin and that BGL3 participates in long-range LCR interactions. The relative participation in the NLI complex of activating (NLI and GATA-1) or repressing (ETO2) components is a major determinant of whether the γ-globin genes successfully establish proximity to the LCR and transcription activation. These studies highlight ETO2 factor exchange within the NLI complex and the role of the BGL3 region in influencing γ-globin long-range contacts with the LCR and transcription activation.
The BGL3 transcript region and γ-globin transcription
BCL11A is a negative regulator of fetal γ-globin transcription, possibly through interaction with the nucleosome remodeling and histone deacetylase NuRD complex.2,4 Accordingly, in these studies, primary erythroid progenitors were devoid of active histone marks associated with transcription, such as H3K9ac and H3K4me2 and me3 in the region of BCL11A binding.4 We would expect, similar to low-HbF cells in our study, that BGL3 is not being transcribed in these cells. In contrast, in high-HbF cells, H3K4me3 is increased across the BGL3 region and in K562 cells, also transcribing BGL3; ENCODE data show that active histone marks are likewise abundant. Therefore, BCL11A occupancy may antagonize BGL3 transcription and/or long-range interaction of the region with the LCR. It is tempting to speculate that the onset of BGL3 transcription is influential in removing BCL11A from chromatin, allowing changes in histone modifications and long-range LCR looping. However, it remains equally plausible that BGL3 transcription requires removal of BCL11A by other means. Whereas BGL3 and γ-globin transcription appear to be temporally related, whether BGL3 transcription (or its transcript) is consequential for γ-globin expression remains to be clarified.
In earlier studies, a large intergenic deletion between the Aγ- and δ-globin genes (deletion D5) that removes BGL3 sequences did not impair γ-globin transcription from a human transgene, suggesting that BGL3 occupancy by BCL11A (and the ETO2-containing NLI complex, see next paragraph) might be dispensable for γ-globin silencing in the mouse erythroid environment.33 The deletion also removed an addition BCL11A site 1 kb 5′ to δ-globin that we did not study.2 Fetal globin transcription on human transgenes in mouse erythroid cells is silenced prematurely, along with mouse embryonic genes, and BCL11A is present at earlier developmental stages, suggesting that γ-globin repression is divergent between the species.3 Because studies localizing BCL11A (and our ChIP-chip NLI localization) in the β-globin locus revealed other sites of interaction downstream of the LCR in addition to the ones deleted in the transgenic study,4,30 it might be illuminating to determine by ChIP and 3C the factor occupancy and chromatin folding in the D5 transgenic globin locus. Because the large D5 deletion removed putative γ-globin–positive and -negative acting sites, a small deletion limited to BGL3 sequences might be revealing, but it seems likely that definitive information may require that this be carried out in the human erythroid cell environment.
Alternative NLI complexes and γ-globin suppression by corepressor ETO2
The corepressor ETO2 is recruited to the NLI complex by the TAL1/E2A heterodimer, conferring negative function possibly through interaction with histone deacetylases and nuclear receptor corepressor (NCOR).16,17 We found that NLI complex members occupied LCR sites and the γ-globin/BGL3 region in both low- and high-HbF cells; however, when γ-globin transcription was reactivated in high-HbF cells, the relative participation of NLI and GATA-1 in the complex increased, whereas that of ETO2 decreased. How factor exchange occurs in regulatory complexes is unknown, but in this case it might be influenced by BGL3 transcription and/or by chromatin occupancy by BCL11A, which is known to interact with GATA-1.2 The repressive role of ETO2 in γ-globin expression was confirmed by reduction of ETO2 by RNAi, which increased transcription in K562 cells, and overexpression of NLI, which augmented this effect. This result suggests that the relatively low level of γ-globin transcription in these cells compared with erythroid progenitors is at least partially controlled by ETO2 participation in the NLI complex. BCL11A does not seem to be a part of this partial repression because it was not detected in K562 cells.20
ETO2 is down-regulated during erythroid differentiation, suggesting that it may exert a repressive role early in differentiation, whereas its loss at later times allows the expression of genes important for terminal erythroid maturation, such as glycophorin A, band 3 and α-globin.12-14,17,18,34 This contention is supported by activation of erythroid genes in undifferentiated MEL cells after ETO2 knock-down.12 Our observations suggest a novel role for ETO2 in human erythroid cells late in differentiation, when ETO2 occupancy is associated with BGL3 and γ-globin gene repression in low-HbF cells and their reduced interaction with the LCR. ETO2 did not strongly occupy the adult β-globin gene, which is actively transcribed in both high- and low-HbF cells. Overexpressing ETO2 in MEL cells did not prevent full hemoglobinization, although in other studies, a 2-fold increase in β-globin mRNA after ETO2 reduction in G1E cells was observed, leaving open the question of ETO2 influence on β-globin expression.18,35,36
BGL3 and long-range activation of fetal γ-globin genes
3C analysis revealed that the LCR and BGL3 regions establish proximity in high-HbF cells, where BGL3 is transcribed. Similar to our findings, in the first description of γ-globin/LCR interaction by 3C, the BGL3-containing EcoRI fragment was observed in close proximity to the LCR.11 However, a later study did not find BGL3/LCR proximity.4 The reason for this difference is unclear but may reflect the use of different 3C primers. In addition, there is a strong proximity between BGL3 and the γ-globin genes when they are both transcribed. Our data support an important role for the NLI complex in mediating γ-globin activation and long-range LCR contacts through the BGL3 region, however, additional factors may also be critical.
In comparison, BGL3/LCR proximity is not present in low-HbF cells, in which the most prominent interaction is between the LCR and β-globin gene. The NLI complex occupies the β-globin promoter in low- and high-HbF cells, and the gene is transcribed in both. Likewise, EKLF occupies the β-globin promoter, and both NLI and EKLF are required for long-range chromatin interaction between the mouse β-globin LCR and gene7,8 ; whether they act independently or together will be important to explore.
Our experiments raise new questions, but also begin to reveal a plausible scenario for reactivation of fetal γ-globin in human adult erythroid cells, and by inference help to characterize the developmental period when γ-globin transcription is active. BCL11A occupancy is minimal within BGL3 sequences and BGL3 is transcribed. Corepressor ETO2 participation in the NLI complex is very low at binding sites in the LCR, γ-globin promoter, and BGL3 regions. Proximity is established among the BGL3 region, γ-globin genes, and the LCR. Although the temporal relationships among these events remain to be precisely determined, the results shed light on the mechanism underlying BCL11A repression of γ-globin transcription from a position of intergenic occupancy. Based on these results, ETO2, like BCL11A, emerges as a potential therapeutic target to up-regulate γ-globin globin in patients with β-globin disorders.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Sang-Hyun Song and Orapan Sripichai for helpful discussions, Dr Catherine Porcher for TAL1 Abs, and Colleen Byrnes for cell analysis.
This work was supported by the Intramural Program, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD.
National Institutes of Health
Authorship
Contribution: A.D., C.M.K., C.H., and J.L.M. designed the research; C.M.K., J.L., Y.T.L., E.R.M., and C.H. performed the research; C.M.K., J.L., C.H., R.K.D., J.L.M., and A.D. analyzed the data; and C.M.K., J.L., and A.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for C.H. is Department of Biology, Emory University, Atlanta, GA.
Correspondence: Ann Dean, Laboratory of Cellular and Developmental Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bldg 50, Rm 3154, 50 South Dr, MSC 8028, Bethesda, MD 20892; e-mail: anndean@helix.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal