Abstract
In a previous study, we identified somatic mutations of SF3B1, a gene encoding a core component of RNA splicing machinery, in patients with myelodysplastic syndrome (MDS). Here, we define the clinical significance of these mutations in MDS and myelodysplastic/myeloproliferative neoplasms (MDS/MPN). The coding exons of SF3B1 were screened using massively parallel pyrosequencing in patients with MDS, MDS/MPN, or acute myeloid leukemia (AML) evolving from MDS. Somatic mutations of SF3B1 were found in 150 of 533 (28.1%) patients with MDS, 16 of 83 (19.3%) with MDS/MPN, and 2 of 38 (5.3%) with AML. There was a significant association of SF3B1 mutations with the presence of ring sideroblasts (P < .001) and of mutant allele burden with their proportion (P = .002). The mutant gene had a positive predictive value for ring sideroblasts of 97.7% (95% confidence interval, 93.5%-99.5%). In multivariate analysis including established risk factors, SF3B1 mutations were found to be independently associated with better overall survival (hazard ratio = 0.15, P = .025) and lower risk of evolution into AML (hazard ratio = 0.33, P = .049). The close association between SF3B1 mutations and disease phenotype with ring sideroblasts across MDS and MDS/MPN is consistent with a causal relationship. Furthermore, SF3B1 mutations are independent predictors of favorable clinical outcome, and their incorporation into stratification systems might improve risk assessment in MDS.
Introduction
Myelodysplastic syndromes (MDS) are myeloid neoplasms (MPN) characterized by dysplasia in one or more cell lines, ineffective hematopoiesis, and variable risk of progression to acute myeloid leukemia (AML).1 The World Health Organization (WHO) classification criteria for MDS diagnosis require evaluation of peripheral blood and bone marrow morphology combined with cytogenetic analyses, and define the following categories2,3 : refractory cytopenia with unilineage dysplasia, refractory anemia with ring sideroblasts (RARS), refractory cytopenia with multilineage dysplasia (RCMD), refractory anemia with excess blasts (RAEB) type 1 (RAEB-1) and type 2 (RAEB-2), and myelodysplastic syndrome with isolated (del 5q) [MDS del(5q)].
MDSs are heterogeneous disorders ranging from indolent conditions with a near-normal life expectancy to subtypes very close to AML. To improve their prognostication, risk-based stratification systems have been developed. The International Prognostic Scoring System (IPSS) stratifies MDS patients into 4 risk groups by percentage of blasts in the bone marrow, type of cytogenetic abnormality, and number and degree of cytopenias at presentation.4 The WHO classification–based prognostic scoring system (WPSS) takes advantage of the prognostic relevance of the WHO classification,5 is able to classify MDS patients into 5 risk groups showing different survivals and probabilities of leukemic evolution, and can be used not only at diagnosis but also during follow-up.6
Increasing evidence indicates that chromosomal abnormalities play a major role in determining the heterogeneity of MDS.7 This suggests that distinct molecular lesions probably contribute to the variable morphology and clinical outcome of these myeloid neoplasms. Correlating the presence of acquired mutations in MDS with clinical features and outcome will potentially provide a strong molecular basis for future classification and prognostic scoring systems.
Important steps have recently been made in characterizing the molecular basis of MDS. For example, MDS del(5q) appears to derive from haploinsufficiency of genes mapping to chromosome 5q32-q33, in particular from reduced expression of RPS14, miR-145, and miR-146a, and from TP53 overexpression.8-12 Acquired somatic mutations of TET2 have been detected in different myeloid neoplasms and in approximately 25% of MDS patients.13-16 Additional mutant genes, including ASXL1, CBL, ETV6, EZH2, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, and TP53, have been identified in smaller proportions of patients, particularly in those with high risk of leukemic evolution.17 In a recent study, mutations in 5 of these genes have been found to be predictors of poor overall survival (OS), independently of established risk factors.18
Using massively parallel sequencing technology, we recently identified somatically acquired mutations in SF3B1, a gene encoding a core component of RNA splicing machinery, in diverse WHO categories but predominantly in MDS patients with ring sideroblasts.19 This observation and the recent identification of somatic mutations in other key components of the spliceosome20 strongly implicate abnormalities of mRNA splicing in the pathogenesis of MDS. In this work, we performed a comprehensive mutation analysis of SF3B1 in patients with myelodysplastic neoplasms to define the clinical correlates of these mutations.
Methods
Patients
These investigations were approved by the Ethics Committee of the Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Pavia, Italy, and by other local institutional review boards. The procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2000, and samples were obtained after subjects provided informed consent.
We studied 564 patients with MDS, 88 with MDS/MPN, and 40 with AML evolving from MDS (Table 1). The diagnostic criteria of the WHO classification of tumors of hematopoietic and lymphoid tissues were adopted.2 Of 564 patients with MDS, 135 had refractory cytopenia with unilineage dysplasia presenting as refractory anemia (RA), 107 had RARS, 102 had RCMD without ring sideroblasts, 54 had RCMD with ring sideroblasts (RCMD-RS), 87 had RAEB-1, 57 had RAEB-2, and 22 had MDS del(5q). It should be noted that RCMD-RS was a separate MDS category in the 2001 WHO classification of myeloid neoplasms21 but was incorporated into RCMD in the 2008 revision.3 Of 88 patients with MDS/MPN, 67 had chronic myelomonocytic leukemia (CMML),22 18 had RARS associated with marked thrombocytosis (RARS-T),23 and 3 had MDS/MPN unclassifiable (MDS/MPN, U). Forty patients had AML secondary to MDS.2
Patients were studied at diagnosis or during follow-up before any disease-modifying treatment (ie, allogeneic stem cell transplantation, aggressive chemotherapy, or hypomethylating agents). The SF3B1 mutation status of 354 of the aforementioned 564 MDS patients, but not information on mutant allele burden or that on clinical significance of somatic mutation (impact on OS and contribution to the predictive power of IPSS or WPSS), was included in our previous report.19 The SF3B1 mutation status of the 67 patients with CMML, already reported in our previous study,20 was included in this paper to compare CMML and RARS-T within MDS/MPN.
In a subgroup of 325 MDS patients, a quantitative enumeration of ring sideroblasts after Perls staining was performed using recently established consensus criteria24 to study the relationship between SF3B1 mutation status or mutant allele burden and the proportion of bone marrow ring sideroblasts.
Sample collection and cell separation
Mononuclear cells were separated from bone marrow samples by standard density gradient centrifugation, and granulocytes were isolated from peripheral blood as previously described.23 Mononuclear cells were labeled with CD34 MicroBeads, and CD34+ cells were isolated using MACS magnetic cell separation columns (Miltenyi Biotec) according to the manufacturer's recommendations.25 CD34+ cell purity was evaluated with FACS and was > 90%. Genomic DNA was obtained from bone marrow mononuclear cells, CD34+ cells, or peripheral blood granulocytes by following standard protocols for human tissue.
Mutation analysis of SF3B1 and SF3B1 mutant allele burden
The coding exons of SF3B1 were screened using massively parallel pyrosequencing of DNA pools using the genome sequencer FLX system (Roche Diagnostics). Briefly, optimized primers for PCR (shown in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were designed against all coding exons of SF3B1 (CCDS33356, ENST00000335508).19 Where appropriate, overlapping primer sets were used to allow for precise quantification of variant allelic burden estimates. On the 5′ end of each primer, oligonucleotide 8-mer indexes (shown in supplemental Table 2) were incorporated to allow effective separation of sample specific sequence information from the DNA pools.
High throughput sequencing of pooled PCR products was performed on the FLX 454, sequencing reads were mapped to the human genome (Build 37) with Burrows-Wheeler Aligner, and individual sample sequencing information was deconstructed as previously described.19 All novel variants were verified using PCR-based Sanger sequencing.
For each reported SF3B1 variant, the proportion of cells in a DNA sample that carry the variant can be directly estimated by calculating the proportion of sequencing reads reporting the mutant allele. Allelic burden estimates were derived for all variants with minimum sequence coverage of 25 reads and mutant allele representation of at least 10% of the total reads. Base sequence and mapping quality thresholds were both set to 25 (58 in Burrows-Wheeler Aligner).
JAK2 and MPL mutation analysis
Mutation analyses of JAK2 and MPL were performed exclusively on patients with RARS-T.23 A quantitative real-time RT-PCR–based allelic discrimination assay was used for detecting the JAK2 (V617F) mutation and evaluating granulocyte mutant allele burden applying the standard curve method.23 The sensitivity of this assay is approximately 0.2% of mutant alleles. MPL mutation scanning was performed using high resolution melting analysis,26 and mutations were then verified using Sanger sequencing.
Statistical analyses
Numerical variables are summarized by median and range; categorical variable are described with count and relative frequency (percentage) of subjects in each category. Comparison of numerical variables between groups was carried out using a nonparametric approach (Mann-Whitney test or Kruskal-Wallis ANOVA). Comparison of the distribution of categorical variables in different groups was performed with either the Fisher exact test (when computationally feasible) or the χ2 test (larger tables).
Survival analyses were performed with the Kaplan-Meier method. OS was defined as the time (in months) between the date of diagnosis and the date of death (for patients who deceased) or last follow-up (for censored patients). Leukemia-free survival (LFS) was defined as the time (in months) between the date of diagnosis and the date of leukemic transformation (for patients who progressed to AML) or last follow-up (for censored patients). Event-free survival (EFS) was defined as the time (in months) between the date of diagnosis and the date of first event (death or leukemic transformation for cases having at least 1 of these events) or last follow-up (for censored patients). Comparison between survival curves was carried out by the Wilcoxon test. Multivariate survival analyses were performed by Cox proportional hazards regression. All analyses accounted for left censoring of the observations at the time of mutation assessment.
Likelihood ratio tests were carried out to compare nested models with different covariates and parameterizations (eg, categorical vs continuous), and to test for interaction between covariates. To compare regression models with different covariate cut-off points, we used the Akaike information criterion.27 This criterion provides a measure of the relative goodness of fit of a statistical model and a means for comparison among models, a lower Akaike information criterion value indicating a better trade-off between fit and complexity.
Analyses were performed using the Stata SE Version 11.2 software (StataCorp LP; http://www.stata.com).
Results
Somatic mutations of SF3B1 in patients with myeloid neoplasms
Sufficient sequence coverage was obtained for 654/692 patients studied, and the main results of mutation analysis of SF3B1 are reported in Table 1.
Proportion of patients carrying somatic mutations of SF3B1 in the study population
| WHO category . | No. of patients studied . | No. of sequencing failures* . | No. of evaluable patients . | No. (%) of patients carrying SF3B1 mutations . |
|---|---|---|---|---|
| MDS | ||||
| RA | 135 | 13 | 122 | 14 (11.5) |
| RARS | 107 | 2 | 105 | 83 (79.0) |
| RCMD | 102 | 6 | 96 | 6 (6.3) |
| RCMD-RS† | 54 | 2 | 52 | 30 (57.7) |
| RAEB-1 | 87 | 4 | 83 | 7 (8.4) |
| RAEB-2 | 57 | 4 | 53 | 6 (11.3) |
| MDS del(5q) | 22 | 0 | 22 | 4 (18.2) |
| MDS total | 564 | 31 | 533 | 150 (28.1) |
| MDS/MPN | ||||
| CMML | 67 | 5 | 62 | 4 (6.5) |
| RARS-T | 18 | 0 | 18 | 12 (66.7) |
| MDS/MPN, U | 3 | 0 | 3 | 0 |
| AML secondary to MDS | 40 | 2 | 38 | 2 (5.3) |
| All patients studied | 692 | 38 | 654 | 168 (25.7) |
| WHO category . | No. of patients studied . | No. of sequencing failures* . | No. of evaluable patients . | No. (%) of patients carrying SF3B1 mutations . |
|---|---|---|---|---|
| MDS | ||||
| RA | 135 | 13 | 122 | 14 (11.5) |
| RARS | 107 | 2 | 105 | 83 (79.0) |
| RCMD | 102 | 6 | 96 | 6 (6.3) |
| RCMD-RS† | 54 | 2 | 52 | 30 (57.7) |
| RAEB-1 | 87 | 4 | 83 | 7 (8.4) |
| RAEB-2 | 57 | 4 | 53 | 6 (11.3) |
| MDS del(5q) | 22 | 0 | 22 | 4 (18.2) |
| MDS total | 564 | 31 | 533 | 150 (28.1) |
| MDS/MPN | ||||
| CMML | 67 | 5 | 62 | 4 (6.5) |
| RARS-T | 18 | 0 | 18 | 12 (66.7) |
| MDS/MPN, U | 3 | 0 | 3 | 0 |
| AML secondary to MDS | 40 | 2 | 38 | 2 (5.3) |
| All patients studied | 692 | 38 | 654 | 168 (25.7) |
Failure was the result of insufficient sequence coverage.
Overall, 150 of 533 (28.1%) MDS patients had a somatic mutation of SF3B1, and the proportion of positive patients was significantly higher in the RARS and RCMD-RS subgroups than in the remaining WHO categories (72.0% vs 9.8%, P < .001). Within patients with MDS/MPN, 16 of 83 (18.3%) carried a somatic mutation in SF3B1, and the proportion of positive patients was significantly higher in RARS-T than in the other WHO categories (66.7% vs 6.2%, P < .001). Only 2 of 38 patients (5.3%) with AML evolving from MDS carried an SF3B1 mutation.
Clinical and hematologic features of patients with SF3B1 mutation were compared with those of patients without mutation in a subgroup of 325 subjects whose data at the time of molecular evaluation were available. Overall, the prevalence of SF3B1 mutations was higher in females than in males (27% vs 18%, P = .02), and patients with SF3B1 mutation showed higher platelet count (median value, 257 × 109/L vs 104 × 109/L, P < .001), higher proportion of ring sideroblasts (40% vs 0%, P < .001), and marrow erythroblasts (40% vs 27%, P < .001), and lower proportion of bone marrow blasts (2% vs 3%, P < .001; supplemental Figure 1).
SF3B1 mutation types and mutant allele burden
Twenty-three mutations were identified mapping in 15 different codons. As shown in Table 2, the most frequent mutation involved codon 700 (57.7% of all mutated cases), whereas lower frequencies were observed for mutations in codons 666 (10.7%), 662 (10.1%), 625 (6%), and 622 (7.1%). All the mutations identified were missense substitutions. No significant difference was found among different types of mutations as regards WHO subgroup, clinical features, or hematologic parameters.
SF3B1 mutation type and SF3B1 mutant allele burden
| SF3B1 mutations . | No. of patients with mutation . | Proportion of patients with mutation as a percentage of all mutated cases . | Mutant allele burden, %, median (range) . |
|---|---|---|---|
| K700E | 97 | ||
| Codon 700 | 97 | 57.7 | 38.4 (7.5-70.3) |
| K666R | 9 | ||
| K666N | 3 | ||
| K666Q | 3 | ||
| K666T | 2 | ||
| K666M | 1 | ||
| Codon 666 | 18 | 10.7 | 44.6 (17.1-62.7) |
| H662Q | 15 | ||
| H662D | 1 | ||
| H662Y | 1 | ||
| Codon 662 | 17 | 10.1 | 39.1 (7.0-52.1) |
| E622D | 12 | ||
| Codon 622 | 12 | 7.1 | 39.4 (20.2-45.3) |
| R625L | 6 | ||
| R625C | 3 | ||
| R625G | 1 | ||
| Codon 625 | 10 | 6.0 | 37.5 (10.6-45.1) |
| D781G | 3 | ||
| E592K | 2 | ||
| A744P | 2 | ||
| E491G | 1 | ||
| R590K | 1 | ||
| N626D | 1 | ||
| V701F | 1 | ||
| V701I | 1 | ||
| G740R | 1 | ||
| A1188V | 1 | ||
| Miscellaneous codons | 14 | 8.4 | 36.4 (5.4-50.0) |
| SF3B1 mutations . | No. of patients with mutation . | Proportion of patients with mutation as a percentage of all mutated cases . | Mutant allele burden, %, median (range) . |
|---|---|---|---|
| K700E | 97 | ||
| Codon 700 | 97 | 57.7 | 38.4 (7.5-70.3) |
| K666R | 9 | ||
| K666N | 3 | ||
| K666Q | 3 | ||
| K666T | 2 | ||
| K666M | 1 | ||
| Codon 666 | 18 | 10.7 | 44.6 (17.1-62.7) |
| H662Q | 15 | ||
| H662D | 1 | ||
| H662Y | 1 | ||
| Codon 662 | 17 | 10.1 | 39.1 (7.0-52.1) |
| E622D | 12 | ||
| Codon 622 | 12 | 7.1 | 39.4 (20.2-45.3) |
| R625L | 6 | ||
| R625C | 3 | ||
| R625G | 1 | ||
| Codon 625 | 10 | 6.0 | 37.5 (10.6-45.1) |
| D781G | 3 | ||
| E592K | 2 | ||
| A744P | 2 | ||
| E491G | 1 | ||
| R590K | 1 | ||
| N626D | 1 | ||
| V701F | 1 | ||
| V701I | 1 | ||
| G740R | 1 | ||
| A1188V | 1 | ||
| Miscellaneous codons | 14 | 8.4 | 36.4 (5.4-50.0) |
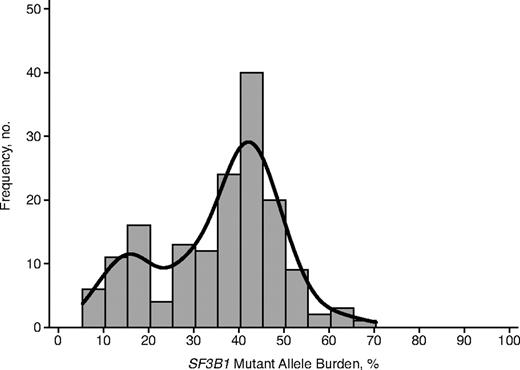
The SF3B1 mutant allele burden could be accurately assessed (as defined under “Mutation analysis of SF3B1 and SF3B1 mutant allele burden”) in 161 of 168 mutated cases. The overall median value was 39.5% (range, 5.4%-70.3%), and frequencies of observations are reported in Figure 1. The histogram of mutant allele burden was compatible with a bimodal distribution that resulted from a mixture of 2 normal distributions with different variance and means. The first normal distribution accounted for 22% of observations with a mean value for mutant allele burden equal to 15%, whereas the second distribution accounted for 78% of observations with a mean value of 41%. Seventeen patients showed a mutant allele burden higher than 50%, but only in 6 cases this value was higher than 60%. In none of these 6 cases, however, was there sufficient coverage that the 95% confidence interval (CI) excluded 50%. These findings are consistent with the presence of a dominant clone with heterozygous SF3B1 mutation in most of the cases, and with the existence of a minority mutant clone in a small fraction of patients.
Histogram of SF3B1 mutant allele burden. The bimodal distribution has been highlighted by adding a Gaussian kernel density plot to the histogram. The first normal distribution (on the left) accounts for 22% of observations, with a mean value for SF3B1 mutant allele burden equal to 15%. The second normal distribution (on the right) accounts for 78% of observations, with a mean value equal to 41%.
Histogram of SF3B1 mutant allele burden. The bimodal distribution has been highlighted by adding a Gaussian kernel density plot to the histogram. The first normal distribution (on the left) accounts for 22% of observations, with a mean value for SF3B1 mutant allele burden equal to 15%. The second normal distribution (on the right) accounts for 78% of observations, with a mean value equal to 41%.
To account for potential bias of DNA source in the evaluation of SF3B1 mutation status and mutant allele burden, we studied potential differences between these subgroups (supplemental Figure 2). Median values for SF3B1 mutant allele burden were 40.6% (24.6%-52.1%) in the CD34+ cell DNA subgroup, 40.9% (7.5%-70.3%) in the granulocyte DNA subgroup, and 35.7% (10.1%-51.1%) in the bone marrow DNA subgroup. Kruskal-Wallis 1-way ANOVA by ranks showed no significant difference in SF3B1 mutant allele burden between these subgroups (P = .077).
No significant relationship was found between mutant allele burden and clinical or hematologic features, including clinical outcome. However, the proportion of WHO categories defined by ring sideroblasts (RARS, RCMD-RS, and RARS-T) was higher within patients with a mutant allele burden ≥ 25% (indicating a dominant clone) than in those with a burden < 25% (indicating a minority clone, P = .022)
Relationship between SF3B1 mutations and ring sideroblasts
An accurate quantitative enumeration of ring sideroblasts using consensus criteria24 was performed in 325 MDS patients. Ring sideroblasts were identified at variable percentages (range, 1%-78%) in 74 of 212 patients assigned to WHO categories that are not defined by this morphologic feature, including MDS del(5q), RA, RCMD, RAEB-1, and RAEB-2. Of these, 44 subjects (10 RA, 22 RCMD, 5 RAEB-1, and 7 RAEB-2) showed a proportion of ring sideroblasts below the diagnostic threshold of 15% for assignment to a sideroblastic subtype (RARS or RCMD-RS), whereas 30 patients had a proportion of ring sideroblasts ≥ 15% [7 MDS del(5q), 23 RAEB].
Of the aforementioned 325 patients, 101 (31%) carried a mutation in SF3B1. A strong association was found between SF3B1 mutation and disease phenotype with ring sideroblasts. Of 101 patients with an SF3B1 mutation, indeed, 91 had more than 15% ring sideroblasts, 7 patients had 1% to 14%, and only 3 patients did not show any ring sideroblasts in the bone marrow (Kendall Tau-b correlation coefficient = 0.54, P < .001). Based on these data, the SF3B1 mutation status had a positive predictive value for disease phenotype with ring sideroblasts of 97.7% (95% CI, 93.5%-99.5%), whereas the absence of ring sideroblasts had a negative predictive value for SF3B1 mutation of 97.8% (95% CI, 93.8%-99.5%).
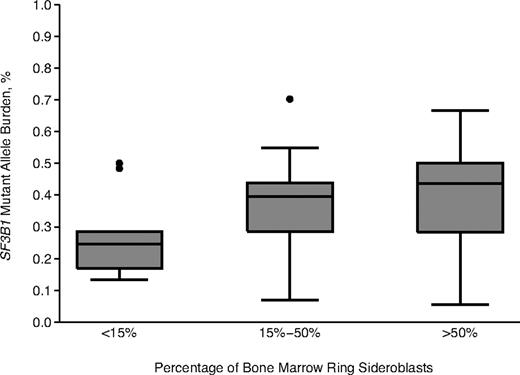
Significant relationships were found between SF3B1 mutant allele burden and percentage of ring sideroblasts (P = .002, Figure 2), or percentage of bone marrow erythroblasts (P = .01).
Relationship between SF3B1 mutant allele burden and proportion of ring sideroblasts. Values for percentage of ring sideroblasts are grouped here in 3 arbitrary categories: < 15% (n = 183), 15% to 50% (n = 85), and > 50% (n = 57). Data are shown in a box plot depicting the smallest and largest observation (lowest and highest horizontal line, respectively), lower and upper quartile with median value (box), and outliers (dots).
Relationship between SF3B1 mutant allele burden and proportion of ring sideroblasts. Values for percentage of ring sideroblasts are grouped here in 3 arbitrary categories: < 15% (n = 183), 15% to 50% (n = 85), and > 50% (n = 57). Data are shown in a box plot depicting the smallest and largest observation (lowest and highest horizontal line, respectively), lower and upper quartile with median value (box), and outliers (dots).
Relationship between SF3B1 mutations and clinical outcome in MDS
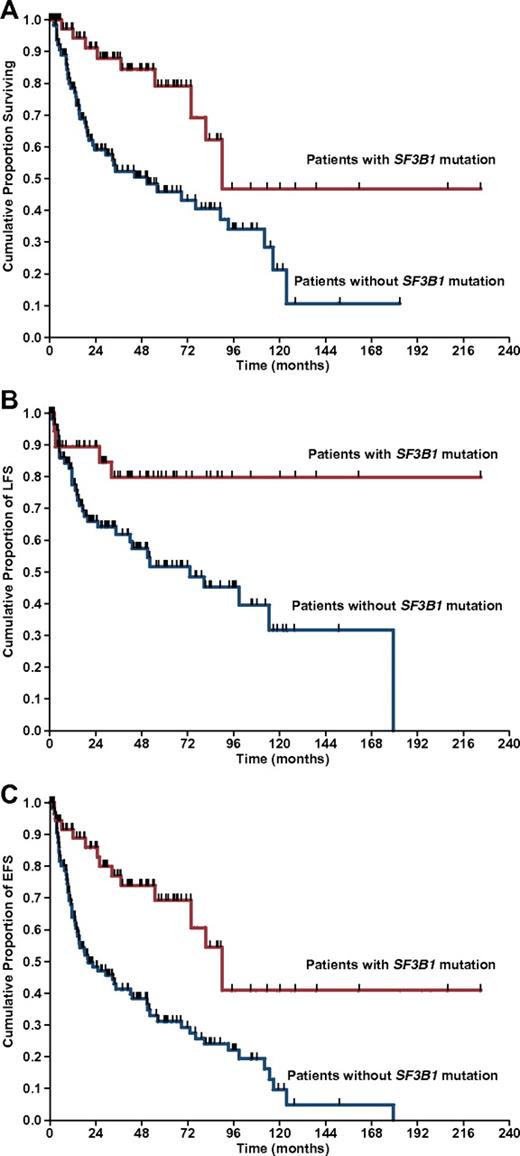
Survival data were available for 323 of the 533 patients with MDS successfully analyzed for SF3B1 mutation. Median observation time from diagnosis was 23 months (range, 1-267 months), whereas median time from diagnosis to mutation analysis was 4.5 months (range, 0-182 months). Patients carrying an SF3B1 mutation showed a significantly better OS compared with those without mutation (median survival, 90 vs 50 months, P = .001; Figure 3A).
Kaplan-Meier analysis of survival in MDS patients stratified according to SF3B1 mutation status. (A) OS. (B) LFS. (C) EFS. Vertical tick-marks indicate right-censored patients.
Kaplan-Meier analysis of survival in MDS patients stratified according to SF3B1 mutation status. (A) OS. (B) LFS. (C) EFS. Vertical tick-marks indicate right-censored patients.
To further define the prognostic relevance of SF3B1 mutations, we performed univariate and multivariate analyses using a Cox proportional hazards regression model. In univariate analysis, SF3B1 mutation positively affected OS (HR = 0.35, 95% CI, 0.17-0.72, P = .009). The significant value of SF3B1 mutation was retained in a multivariate analysis, including age, sex, hemoglobin level, absolute neutrophil count, platelet count, cytogenetic risk, bone marrow blasts, and ring sideroblasts (HR = 0.15, 95% CI, 0.03-0.78, P = .025).
Patients with the SF3B1 mutation showed a significantly better LFS than those without mutation (HR = 0.32, 95% CI, 0.11-0.91, P = .032; Figure 3B). This result was confirmed in a multivariate analysis, including age, sex, hemoglobin level, absolute neutrophil count, platelet count, cytogenetic risk, bone marrow blasts, and ring sideroblasts (HR = 0.33, 95% CI, 0.11-0.99, P = .049).
EFS was calculated by combining the 2 previous outcomes (death or leukemic transformation). Patients with the SF3B1 mutation showed a significantly better EFS compared with those without the SF3B1 mutation in both univariate (HR = 0.33, 95% CI, 0.19-0.62, P < .001) and multivariate analysis, adjusted for demographic and hematologic variables (HR = 0.29, 95% CI, 0.10-0.82, P = .019; Figure 3C).
We then evaluated OS and LFS in the subgroup of patients with RARS and RCMD-RS specifically. Univariate analysis of SF3B1 mutation status showed a significant effect on OS (HR = 0.28, 95% CI, 0.09-0.94, P = .039) but no effect on LFS (HR = 0.62, 95% CI, 0.1-3.78, P = .6) in this subgroup. The protective effect of SF3B1 mutation on OS was lost in multivariate analysis because of significant colinearity among predictor variables.
Relative contribution of SF3B1 mutation status to OS as predicted by IPSS or WPSS
Both IPSS and WPSS are currently used for clinical prognostication of MDS. To establish whether the SF3B1 mutation status can add to the predictive power of IPSS or WPPS, we fitted multivariable Cox models, including age, sex, IPSS or WPSS, and SF3B1 mutation status.
Age was initially modeled as a continuous covariate but was then parameterized as a binary variable with respect to a cut-point. This was set at 70 years (ie, the round value closest to the median age). Its optimal fit compared with other cut-points (ie, 55, 60, 65, and 75) was confirmed by the Akaike information criterion.27 Both IPSS and WPSS were modeled as continuous covariates, as the likelihood ratio test of each model compared with the respective one having a categorical parameterization showed no significant difference between these alternatives.
As shown in Table 3, the IPSS risk group was strongly associated with OS, but the SF3B1 mutation status remained an independent predictor of better survival even after adjustment for IPSS. This was true also in patients with low or intermediate-1 IPSS risk. Indeed, within these risk groups, patients carrying an SF3B1 mutation showed significantly better OS and EFS compared with those without mutation (P = .038 and P = .026; Figure 4).
Relative contribution of SF3B1 mutation status to the ability of IPSS or WPSS to predict OS in MDS patients*
| Risk factor . | Hazard ratio . | P . |
|---|---|---|
| Multivariate analysis including IPSS | ||
| Age (< 70 y vs ≥ 70 y) | 1.52 | .224 |
| Sex (male vs female) | 0.69 | .292 |
| IPSS risk group | 2.16 | < .001 |
| SF3B1 mutation status (mutation present vs absent) | 0.21 | .038 |
| Multivariate analysis including WPSS | ||
| Age (< 70 y vs ≥ 70 y) | 1.39 | .324 |
| Sex (male vs female) | 0.86 | .668 |
| WPSS risk group | 1.96 | < .001 |
| SF3B1 mutation status (mutation present vs absent) | 0.23 | .050 |
| Risk factor . | Hazard ratio . | P . |
|---|---|---|
| Multivariate analysis including IPSS | ||
| Age (< 70 y vs ≥ 70 y) | 1.52 | .224 |
| Sex (male vs female) | 0.69 | .292 |
| IPSS risk group | 2.16 | < .001 |
| SF3B1 mutation status (mutation present vs absent) | 0.21 | .038 |
| Multivariate analysis including WPSS | ||
| Age (< 70 y vs ≥ 70 y) | 1.39 | .324 |
| Sex (male vs female) | 0.86 | .668 |
| WPSS risk group | 1.96 | < .001 |
| SF3B1 mutation status (mutation present vs absent) | 0.23 | .050 |
Multivariate analysis of OS was performed using a Cox proportional hazards regression model that included age, sex, IPSS risk group or WPSS risk group, and SF3B1 mutation status. IPSS and WPSS were modeled as a continuous covariate.
Kaplan-Meier analysis of survival in MDS patients with low or intermediate-1 IPSS risk stratified according to SF3B1 mutation status. (A) OS. (B) EFS. Vertical tick-marks indicate right-censored patients.
Kaplan-Meier analysis of survival in MDS patients with low or intermediate-1 IPSS risk stratified according to SF3B1 mutation status. (A) OS. (B) EFS. Vertical tick-marks indicate right-censored patients.
The WPSS risk group was strongly associated with OS (Table 3), whereas the significance of SF3B1 mutation status as an independent predictor of favorable outcome was borderline (P = .05). The likelihood ratio test performed on the inclusion of SF3B1 mutation status into the model based on WPSS was nonetheless significant (P = .018), indicating that such inclusion added to the model's predictive power. It should be noted that the RARS category is present as a favorable variable in the WPSS6 ; therefore, the close association of RARS with SF3B1 mutations reduces the independent predictive power of the latter in a multivariable Cox model that includes both variables.
Somatic mutations of SF3B1 in patients with MDS/MPN
In RARS-T patients, the mutant allele burden ranged from 17% to 62% (median, 41%), and was not significantly different from that of other WHO categories with ring sideroblasts. No difference was found in clinical and hematologic variables between mutated and unmutated RARS-T patients.
Four RARS-T patients had the JAK2 (V617F) mutation, whereas an additional one had the MPL (W515L) mutation. Two patients with the JAK2 (V617F) mutation also carried an SF3B1 mutation, and the SF3B1 mutant allele burden was greater than that for JAK2 mutation (57% vs 13% in one case, and 17% vs 10% in the other one). The RARS-T patient carrying both an SF3B1 mutation and MPL (W515L) had a fully dominant clone that was heterozygous for both mutations.
Discussion
The findings of this study indicate that there is a close relationship between SF3B1 mutations and ring sideroblasts across a variety of myelodysplastic neoplasms and clearly demonstrate that this molecular lesion is associated with a favorable clinical outcome in these disorders.
Ring sideroblasts are erythroblasts with iron-loaded mitochondria visualized by Prussian blue staining (Perls reaction) as a perinuclear ring of blue granules.28 The iron of ring sideroblasts is stored in mitochondrial ferritin,28 encoded by the FTMT gene, which is typically overexpressed in RARS.29,30
The sideroblastic anemias include both hereditary and acquired conditions, and the most common acquired forms are RARS and RARS-T.31 RARS is characterized by erythroid dysplasia and ineffective erythropoiesis, and its clinical course is stable for years in most cases with a low risk of leukemic evolution.32,33 CD34+ cells from patients with RARS have a particular gene expression profile characterized by overexpression of mitochondria-related genes and, in particular, genes involved in heme synthesis (eg, ALAS2),34 and reduced expression of ABCB7, a gene encoding a protein involved in the transport of iron/sulfur clusters from mitochondria to the cytoplasm.35 RARS-T is a myeloid neoplasm with both myelodysplastic and myeloproliferative features at the molecular and clinical levels23,36 that may develop from RARS through the acquisition of somatic mutations of JAK2, MPL, or other as-yet-unknown genes.23
In this study, 79% of patients with RARS, 57.7% of those with RCMD-RS, and 66.7% of those with RARS-T carried somatic mutations of SF3B1 (Table 1). A recent independent report describes similar proportions of mutated patients: 19 of 23 (82.6%) cases of RARS and 39 of 53 (73.6%) cases of RCMD-RS.20 There was a significant association of SF3B1 mutations with the presence of ring sideroblasts, and the mutant gene had a positive predictive value for this morphologic feature of 97.7% (95% CI, 93.5%-99.5%). These findings strongly support a causal relationship between SF3B1 mutations and ring sideroblasts, and more generally between abnormalities of mRNA splicing, abnormal gene expression profiles, and mitochondrial iron overload.19,34,35 Of note, similar values for SF3B1 mutant allele burden were found in CD34+ cells and circulating granulocytes (40.6% vs 40.9%; supplemental Figure 2), indicating that somatic mutations occur in multipotent hematopoietic stem cells and are then transmitted to their myeloid progeny. It should also be noted that patients with SF3B1 mutation had a higher percentage of bone marrow erythroblasts than those without mutation. This establishes a relationship between SF3B1 mutations and expanded but ineffective erythropoiesis, which represents a distinctive feature of RARS within MDS.32,33,37
The median SF3B1 mutant allele burden was approximately 40% (Figure 1), indicating that in most patients hematopoiesis was sustained by a dominant clone heterozygous for the mutation. This was particularly true for patients with WHO categories defined by ring sideroblasts (RARS, RCMD-RS, and RARS-T), suggesting that somatic mutations of SF3B1 might be an early pathogenetic event in these patients. In none of these mutated patients, however, was the mutant allele burden significantly higher than 50%. This excludes chromosomal defects, such as copy number variation or copy-neutral loss of heterozygosity involving chromosome 2q33.1, where SF3B1 maps. In approximately one-fifth of cases, moreover, the relatively low SF3B1 mutant allele burden was more compatible with a minority clone, suggesting that myelodysplastic transformation had been driven by other molecular mechanisms in these subjects and that the SF3B1 mutation represented a secondary genetic event.
The RCMD-RS category was created in the 2001 WHO classification of myeloid neoplasms21 but was then incorporated into RCMD in the 2008 WHO classification.2 The rationale for this incorporation was that multilineage dysplasia represents a strong negative prognostic factor per se irrespective of the presence or absence of ring sideroblasts.3 Our findings suggest that RCMD and RCMD-RS should be instead considered as separate conditions, or at least that a distinction should be made between patients with wild-type SF3B1 and those with a somatic mutation of this gene.
Within MDS/MPN, a remarkable difference in the incidence of SF3B1 mutations was found between CMML and RARS-T (Table 1). In the last few years, several mutant genes have been detected in CMML patients, including TET2, ASXL1, CBL, RUNX1, and EZH2.38 Rarely, however, ring sideroblasts are observed in these patients, and this is reflected in the low incidence of SF3B1 mutations observed in this study. It should be noted, however, that somatic mutations in genes encoding other key components of the spliceosome (SRSF2, USAF35, ZRSR2, U2AF65, and SF3A1) have been recently reported in 50% of CMML patients, clearly indicating that abnormalities of splicing machinery play a major role in the pathophysiology of this myelodysplastic/myeloproliferative neoplasm.20 Ring sideroblasts represent one of the morphologic hallmarks of RARS-T,39 and in the current study most of these patients indeed carried a somatic mutation of SF3B1. Our observations suggest that RARS-T may result from a combination of SF3B1 and JAK2 (or MPL) mutations, responsible for myelodysplastic and myeloproliferative features,23 respectively.
Approximately one-fourth of patients with ring sideroblasts (RARS, RCMD-RS, and RARS-T subgroups) had a wild-type SF3B1. This suggests that additional mutant gene(s) may be involved in the pathogenesis of ring sideroblasts in myeloid neoplasms. Indeed, somatic mutations of SRSF2 or ZRSR2 have been recently identified in 7% of cases of MDS with ring sideroblasts that had wild-type SF3B1.20 Nonetheless, approximately 15% of these patients did not carry any mutation in genes encoding key components of the spliceosome.
The fact that a small proportion of patients with WHO categories not specifically associated with ring sideroblasts carried an SF3B1 mutation was expected. Indeed, clonal evolution of MDS is probably a multistep process in which several genetic events occur.19 A somatic mutation of SF3B1 may therefore represent a second genetic event: for instance, it may occur in a patient with MDS del(5q) in whom del(5q) was the initiating event. Alternatively, a RARS patient carrying a mutant SF3B1 gene may acquire another somatic mutation that involves excess of blasts and modifies his/her clinical phenotype from RARS to RAEB.18 However, this latter progression appears to be relatively unlikely, and this is particularly true with respect to progression from RARS to AML. Indeed, previous studies clearly indicated that the risk of leukemic evolution is low in patients with RARS,5,33 a notion indirectly confirmed also by our observation that only 2 of 38 (5.3%) patients with AML evolving from MDS carried a somatic mutation of SF3B1.
So far, individual risk assessment in MDS has been based on the use of scoring systems that include clinical, morphologic, and cytogenetic parameters.40 We and others propose that assessment of somatic mutations probably improves both diagnosis and prognostication in patients with MDS,41 although a few simple parameters (eg, the degree of anemia42 ) will continue to be of pivotal importance for risk assessment. In a recent study,18 mutations in 5 genes (TP53, EZH2, ETV6, RUNX1, and ASXL1) were found to be independently associated with decreased OS in MDS. This study, however, was mainly focused on somatic mutations that are associated with unfavorable outcome and disease progression toward AML. On the other hand, somatic mutations of TET2 represent a marker of clonal proliferation in MDS14,43 but are not associated with any WHO category nor have prognostic relevance.16,18
In the current study, we show that SF3B1 is the first mutated gene in MDS to be strongly associated with a specific disease phenotype (ie, ring sideroblasts). From a clinical viewpoint, SF3B1 mutations are independent predictors of favorable clinical outcome in terms of better OS and lower risk of leukemic evolution. As shown in Table 3, incorporation of SF3B1 mutations may add relevant information to the risk stratification systems currently used in clinical practice. Incorporating additional mutant genes, in particular those associated with unfavorable outcome,18 might further refine clinical decision making in MDS.44
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Studies performed at the Department of Hematology Oncology, Fondazione Istituti di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo & University of Pavia, Pavia, Italy were supported by Associazione Italiana per la Ricerca sul Cancro (Special Program Molecular Clinical Oncology 5x1000, project 1005), Fondazione Cariplo, Regione Lombardia, and Italian Ministry of Health. Studies performed at the Sanger Institute were supported by the Wellcome Trust (grant 077012/Z/05/Z) and the Kay Kendall Leukaemia Fund. P.J.C. was supported by a Wellcome Trust Senior Clinical Research Fellowship (grant WT088340MA). D.T.B., M.J.G., S.T., N.K., and A.H. were supported by the Tayside Tissue Bank and Tayside Leukemia Research and Endowment Fund. J.B. and J.S.W. were supported by the Medical Research Council, Leukaemia Lymphoma Research, and the Oxford National Institute for Health Research Biomedical Research Center. E.H.-L. was supported through the Swedish Cancer Society, the Scientific Research Council, and the Cancer Society in Stockholm.
A detailed description of the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative project is available at www.progettoagimm.it.
Wellcome Trust
Authorship
Contribution: M.C., L.M., E.P., D.T.B., J.B., J.S.W., P.A.F., M.R.S., P.J.C., and E.H.-L. designed research; L.M., E.P., M.G.D.P., E.T., M.J.G., A.L.G., I.A., A.G., M.C.D.V., S.C., S.T., N.K., A.H., J.H., and L.J.M. performed research; M.C., L.M., E.P., and C.P. analyzed and interpreted data; M.C., L.M., E.P., and P.J.C. wrote the manuscript; and all authors critically revised the manuscript and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mario Cazzola, Department of Hematology Oncology, Fondazione Istituti di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, University of Pavia, 27100 Pavia, Italy; e-mail: mario.cazzola@unipv.it.
References
Author notes
L.M. and E.P. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal