Abstract
A strong clustering of Hodgkin lymphoma in certain families has been long acknowledged. However, the genetic factors in the background of familial Hodgkin lymphoma are largely unknown. We have studied a family of 4 cousins with a rare subtype of the disease, nodular lymphocyte predominant Hodgkin lymphoma. We applied exome sequencing together with genome-wide linkage analysis to this family and identified a truncating germline mutation in nuclear protein, ataxia-telangiectasia locus (NPAT) gene, which segregated in the family. We also studied a large number of samples from other patients with Hodgkin lymphoma, and a germline variation leading to the deletion of serine 724 was found in several cases suggesting an elevated risk for the disease (odds ratio = 4.11; P = .018). NPAT is thus far the first gene implicated in nodular lymphocyte predominant Hodgkin lymphoma predisposition.
Introduction
Nodular lymphocyte predominant Hodgkin lymphoma (NLPHL) is a rare subtype of Hodgkin lymphoma (HL) accounting for approximately 5% of HL. It is distinct from classic Hodgkin lymphoma (cHL), which is the commonest HL subtype. Tumor cells in both NLPHL and cHL originate from germinal center B cells. These cells compose only approximately 1% of the tumor mass the rest consisting mostly of benign cellular infiltrate of reactive lymphocytes and other inflammatory cells.1 In NLPHL, the malignant lymphocyte predominant (previously called lymphocytic and histocytic) CD20-positive cells grow in a nodular, sometimes diffuse, pattern representing transformed germinal centers. They do not usually express CD30 and CD15, which typifies the Hodgkin and Reed-Sternberg cells in cHL. Expression studies on lymphocyte predominant cells have shown down-regulation of several B-cell markers, although not as abundantly as in cHL, as well as active expression of nuclear factor-κB target genes.2
Epidemiologic studies of HL have suggested that certain infections, genetic factors, and deficits in the immune system increase the risk of developing the disease.3,4 However, it is uncertain whether this applies to NLPHL. A hallmark of cHL epidemiology is the bimodal age-specific incidence and the disease in young adults and older adults are probably etiologically different; in particular, there is a low prevalence of Epstein-Barr virus (EBV) in younger cHL cases.3 Evidence for a strong heritable basis to HL is provided by the elevated risk in first-degree relatives and high concordance in monozygotic twins.5,6 The distribution of different HL subtypes seems to be similar in sporadic and familial cases.7 A recent genome-wide association study of HL has confirmed the strong relationship between the major histocompatibility complex region and HL risk and identified 3 additional loci to which common risk variants map.8 Collectively, such loci do not impact significantly on the familial HL risk, and the strong familial clustering in some families suggests the existence of high to moderately penetrant susceptibility. In keeping with this model of susceptibility is the observation that variations of the KLHDC8B gene were found to segregate with cHL in 4 families.9
We have recently reported a family of 4 Finnish cousins with NLPHL.10 All patients had been diagnosed at the ages between 22 and 26 years. We studied KLHDC8B and excluded it as a predisposing gene to NLPHL in this family. In this study, our aim was to further clarify the genetic background of familial NLPHL susceptibility by applying linkage analysis in combination with exome sequencing to this Finnish family. Whole exome sequencing is a new, powerful method that is used to sequence all the protein-coding regions in the genome of a person. It is considered cost-effective and unbiased, and it has been recently used successfully in unraveling genetic defects in familial conditions, including predisposition to cancer.11 In our approach, we combined exome sequencing of one affected family member with genome-wide linkage data and identified a truncating germline deletion of 2 bp in the nuclear protein, ataxia-telangiectasia locus (NPAT) gene, which segregated in the family. This is the first gene implicated in NLPHL predisposition.
Methods
Patient samples and clinical data
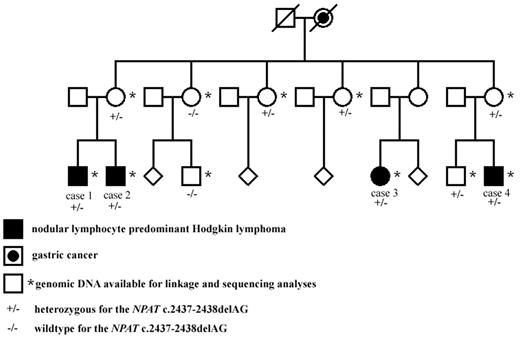
In the Finnish NLPHL family, 4 maternal cousins (3 males and 1 female) had all been affected by early-onset NLPHL (Figure 1). All cases were reexamined by a hematopathologist (K.F.), and the diagnoses were confirmed both in histologic and immunohistochemical studies. Cases 1 to 4 have been diagnosed at the ages of 26, 24, 22, and 23 years with the stages of IIIB, IVB, IIA, and IIIA, respectively (Figure 1). Cases 2 to 4 have achieved remission. Full clinical details of the patients have been previously reported.10 Blood-derived genomic DNA was available from all 4 NLPHL patients and from 2 mothers who are obligate carriers of the putative genetic defect, as well as from 5 healthy family members (Figure 1).
The Finnish NLPHL family. The NLPHL patients as well as those family members whose blood sample was available for linkage analysis and genetic studies are shown. The NPAT deletion status is also shown. The pedigree has been modified for confidentiality.
The Finnish NLPHL family. The NLPHL patients as well as those family members whose blood sample was available for linkage analysis and genetic studies are shown. The NPAT deletion status is also shown. The pedigree has been modified for confidentiality.
A karyotype analysis with the resolution of 400 to 500 bands was performed from the blood sample of one NLPHL patient (case 1, Figure 1). Possible EBV infection was studied using EBV-encoded RNA in situ hybridization, which was performed from paraffin-embedded, NLPHL-affected lymph node tissue received from the 4 NLPHL patients.
Genomic DNA extracted from paraffin-embedded tissue was obtained from 73 Finnish HL patients. Twenty-seven were from families with 1 NLPHL patient and at least 1 close relative with NLPHL, cHL, or non-Hodgkin lymphoma and 38 were < 30 years old at diagnosis. Eight of the cases were from the same geographic region as the original Finnish family of 4 NLPHL patients. The patients were ascertained from a systematic search for related or early-onset NLPHL patients within the Finnish cancer registry. Blood-derived genomic DNA from 282 healthy Finnish blood donors served as a source of controls.
In addition to Finnish samples, we analyzed samples from the United Kingdom. Genomic DNA was obtained from blood samples of 93 patients with HL of whom 26 had a close relative with a lymphoproliferative disorder, and 177 healthy persons with no personal history of malignancy ascertained through the National Study of Colorectal Cancer Genetics study.12 We also screened genomic DNA available from 3 previously reported NLPHL families from United Kingdom/India, France, and Turkey and from an NLPHL-derived cell line, DEV.13-16 A summary of the available samples is shown in Table 1.
NLPHL and HL samples used in NPAT sequencing
| Description of patients . | No. of cases available . | Deletion of c. 2437-2438delAG . | Deletion of serine 724 . |
|---|---|---|---|
| Finland | |||
| Finnish NLPHL family | 4 | 4 | 0 |
| HL patients from other Finnish NLPHL families | 27 | 0 | 0 |
| NLPHL patients with early-onset disease (< 30 years) | 38 | 0 | 1 |
| NLPHL patients from same geographic region as the original family | 8 | 0 | 0 |
| Healthy controls | 282 | 0 | 2 |
| United Kingdom | |||
| HL patients with a family history of a lymphoproliferative disorder | 26 | 0 | 2 |
| Sporadic HL patients | 67 | 0 | 4 |
| Healthy controls | 177 | 0 | 3 |
| Previously reported NLPHL cases | |||
| United Kingdom/India: 2 siblings with NLPHL | 2 | 0 | 0 |
| France: a family of 3 NLPHL patients | 3 | 0 | 0 |
| Turkey: mother and son with NLPHL | 2 | 0 | 0 |
| DEV cell line | 1 | 0 | 0 |
| Description of patients . | No. of cases available . | Deletion of c. 2437-2438delAG . | Deletion of serine 724 . |
|---|---|---|---|
| Finland | |||
| Finnish NLPHL family | 4 | 4 | 0 |
| HL patients from other Finnish NLPHL families | 27 | 0 | 0 |
| NLPHL patients with early-onset disease (< 30 years) | 38 | 0 | 1 |
| NLPHL patients from same geographic region as the original family | 8 | 0 | 0 |
| Healthy controls | 282 | 0 | 2 |
| United Kingdom | |||
| HL patients with a family history of a lymphoproliferative disorder | 26 | 0 | 2 |
| Sporadic HL patients | 67 | 0 | 4 |
| Healthy controls | 177 | 0 | 3 |
| Previously reported NLPHL cases | |||
| United Kingdom/India: 2 siblings with NLPHL | 2 | 0 | 0 |
| France: a family of 3 NLPHL patients | 3 | 0 | 0 |
| Turkey: mother and son with NLPHL | 2 | 0 | 0 |
| DEV cell line | 1 | 0 | 0 |
The number of cases with a deletion in NPAT is shown.
Lymphoblast cell lines were established from all 11 Finnish family members, 4 of the French family members, and from 1 patient in the United Kingdom/India NLPHL family.
The study has been approved by the Ethics Committee of the Hospital district of Helsinki and Uusimaa. All samples were derived either after signed informed consent (blood samples) or authorization from Valvira (National Supervisory Authority for Welfare and Health; paraffin blocks) in accordance with the Declaration of Helsinki. United Kingdom samples were obtained with Ethical board approval and written consent.
Linkage analysis and fine mapping
Whole genome genotype data were obtained from the 11 members of the Finnish family using Affymetrix 50K Xba SNP array. Genotyping was performed according to the manufacturer's standard protocol in the Institute for Molecular Medicine Finland (FIMM) Genome and Technology Center Finland. The array data were analyzed with Merlin,17 using an affected only approach in which penetrance is assumed to be 100%, phenocopy rate 0%, and healthy persons are coded with unknown disease status. Both parametric models, recessive and dominant, were analyzed. Linked regions were haplotyped with Merlin “best” haplotyping function and further mapped with microsatellite markers. For this purpose, we used 40 microsatellite markers, which were either known dinucleotide repeats available in the www.Ensembl.org database or repeats found using the RepeatFinder-program.18 The microsatellite markers are available in supplemental Data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Exome sequencing and sequence analysis
The exome of one NLPHL patient (case 1, Figure 1) was sequenced. Exomic regions of the genome were enriched using Agilent SureSelect Human All Exon Kit totaling approximately 38 Mb of the genome (Agilent Technologies). Paired end short read sequencing with a read length of 56 bp was performed with Illumina Genome Analyzer II in FIMM Genome and Technology Center Finland (Illumina). The sequence data were analyzed with NextGENe- analysis software Version 1.94 (Softgenetics) using NCBI36/Hg18 as reference build. Exomic regions as well as sequence within 2 bp of the exon-intron boundary were analyzed. A minimum sequencing coverage of 5 reads was required for variation analysis; variations existing in only one read or < 20% of reads were considered sequencing errors, and variations resulting to a synonymous amino acid were excluded from the results.
The exome and linkage data were integrated, and potential disease causing variations found in the linked regions were verified with Sanger sequencing implemented on ABI3100xl technology (Applied Biosystems). Segregation of the verified variants was studied in additional family members with direct sequencing using the available samples from the Finnish family. Sequencing primers were designed with ExonPrimer and Primer3 programs (www.ihg.gsf.de/ihg/ExonPrimer.html and www.frodo.wi.mit.edu/primer3) using NCBI36/Hg18 as a reference. Primer sequences and polymerase chain reaction (PCR) conditions are available on request. The PCR products were purified using ExoSAP-IT PCR purification kit (USB Corporation), and the sequencing reactions were performed using the Big Dye Terminator, Version 3.1 kit (Applied Biosystems) according to the manufacturer's instructions. Electrophoresis was performed on an ABI3730 Automatic DNA sequencer (Applied Biosystems at FIMM Genome and Technology Center Finland). Sequences were analyzed using Mutation Surveyor (Softgenetics).
NPAT sequencing
All exons and exon-intron boundaries of NPAT were directly sequenced from the genomic DNA extracted from all available HL samples. We also sequenced the entire NPAT cDNA (CCDS41710) from those samples where we had established lymphoblast cell lines. Separate primers were designed for paraffin, cDNA, and blood-derived samples. Primer design, direct sequencing, and sequence analysis were performed as described in “Exome sequencing and sequence analysis.”
Expression analysis
Gene expression was studied from the blood-derived RNAs of the Finnish family members as well as from 10 controls using GeneChip Human Genome U133 Plus2 (Affymetrix; GSE28536). The genome-wide data were robust multichip average normalized in R using custom chip definition files (ENTREZG, Version 11.0.1).19 Two-tailed t test for NPAT (8 mutation carriers vs 12 nonmutation carriers and controls) was performed with the equal variance assumption.
Results
Karyotyping and EBV status
Before genetic studies, a karyotype analysis was performed in 1 NLPHL patient of the family (case 1, Figure 1). The karyotype was normal, suggesting that chromosomal abnormalities are not associated with NLPHL in the family. The EBV status of the tumor cells in lymph node samples of the patients was evaluated, and all were EBV-negative.
Linkage analysis
According to a dominant model, 13 genomic regions (> 112 Mb) were found to segregate with NLPHL. These were further fine mapped with microsatellite markers. Fine mapping confirmed linkage in all 13 regions. The linked regions are shown in Table 2.
The linked regions in the Finnish NLPHL family according to the dominant model of inheritance (reference build: NCBI36/Hg18)
| Chromosome . | Region start dbSNP ID . | Region end dbSNP ID . | Region size, bp . |
|---|---|---|---|
| 1 | rs3007734 | rs950922 | 3077046 |
| 3 | rs10510592 | rs864391 | 18114197 |
| 5 | rs1161343 | rs2311735 | 6016348 |
| 8 | rs3735915 | rs4388475 | 2001845 |
| 10 | rs10508295 | rs2025465 | 1521857 |
| 11 | rs7120017 | rs633619 | 12464624 |
| 12 | rs4133070 | rs1920706 | 12276245 |
| 13 | rs493041 | rs9300629 | 25624357 |
| 16 | rs2347089 | rs9319570 | 2440380 |
| 18 | rs727929 | rs4797211 | 2070050 |
| 20 | rs6140795 | rs2876404 | 6465648 |
| 21 | rs198057 | rs2831528 | 4988897 |
| X | rs830249 | rs768198 | 15833438 |
| Chromosome . | Region start dbSNP ID . | Region end dbSNP ID . | Region size, bp . |
|---|---|---|---|
| 1 | rs3007734 | rs950922 | 3077046 |
| 3 | rs10510592 | rs864391 | 18114197 |
| 5 | rs1161343 | rs2311735 | 6016348 |
| 8 | rs3735915 | rs4388475 | 2001845 |
| 10 | rs10508295 | rs2025465 | 1521857 |
| 11 | rs7120017 | rs633619 | 12464624 |
| 12 | rs4133070 | rs1920706 | 12276245 |
| 13 | rs493041 | rs9300629 | 25624357 |
| 16 | rs2347089 | rs9319570 | 2440380 |
| 18 | rs727929 | rs4797211 | 2070050 |
| 20 | rs6140795 | rs2876404 | 6465648 |
| 21 | rs198057 | rs2831528 | 4988897 |
| X | rs830249 | rs768198 | 15833438 |
Exome sequencing
The exome of 1 NLPHL patient of the Finnish family was sequenced (case 1, Figure 1) and 7737 variations fulfilling our predefined criteria were found. Common known SNPs and variations found in 13 control samples available from other projects were excluded from the study. In the linked regions, 14 variations unique to the NLPHL patient's exome were identified, and these were directly sequenced using samples from all 4 NLPHL patients, the 2 mothers, and a preliminary set of 3 healthy controls. Seven of the sequence changes were verified, and 6 of them segregated. Three of those were found in at least 1 of the 3 controls. The 3 remaining variations were further sequenced from additional healthy Finnish controls (Table 3). Only 1 was not detectable in the controls. This was a heterozygous deletion of 2 bp, AG (c. 2437-2438), resulting to frame shift and premature stop codon at serine 819 in exon 13 of NPAT gene. Three of 5 healthy family members were shown to carry the deletion (Figure 1).
The unique variations found in the linked regions using exome sequencing of one NLPHL patient
| Gene . | Amino acid change . | Validation status . | Segregation in the family . | Found in controls (of 3) . | Found in additional controls . |
|---|---|---|---|---|---|
| DLEC1 | 1305E > EG | False | |||
| KIF5A | 342W > LW | False | |||
| KRT85 | 189D > DN | True | Yes | 1 | |
| MIP | 107V > VI | True | Yes | 2 | |
| NBPF3 | 170Q > QR | False | |||
| NPAT | c. 2437–2438delAG | True | Yes | 0 | 0/239 |
| PAN2 | 247N > NT | False | |||
| PFKFB3 | 72G > WG | False | |||
| PFKFB3 | 440C > SC | False | |||
| POF1B | 315R > C | True | Yes | 0 | 2/251 |
| SLC22A14 | c. 713–716delTGTT | True | Yes | 1 | |
| UGCGL2 | 983Q > HQ | False | |||
| DDIT3 | 157R > RQ | True | No | ||
| DGKA | 296T > TA | True | Yes | ND | 2/110 |
| Gene . | Amino acid change . | Validation status . | Segregation in the family . | Found in controls (of 3) . | Found in additional controls . |
|---|---|---|---|---|---|
| DLEC1 | 1305E > EG | False | |||
| KIF5A | 342W > LW | False | |||
| KRT85 | 189D > DN | True | Yes | 1 | |
| MIP | 107V > VI | True | Yes | 2 | |
| NBPF3 | 170Q > QR | False | |||
| NPAT | c. 2437–2438delAG | True | Yes | 0 | 0/239 |
| PAN2 | 247N > NT | False | |||
| PFKFB3 | 72G > WG | False | |||
| PFKFB3 | 440C > SC | False | |||
| POF1B | 315R > C | True | Yes | 0 | 2/251 |
| SLC22A14 | c. 713–716delTGTT | True | Yes | 1 | |
| UGCGL2 | 983Q > HQ | False | |||
| DDIT3 | 157R > RQ | True | No | ||
| DGKA | 296T > TA | True | Yes | ND | 2/110 |
The results of the verification by direct sequencing and the segregation of the variation in the family are also shown.
ND indicates not determined.
NPAT sequencing
After identification of the truncating deletion in the NPAT gene in the Finnish NLPHL family, we examined whether NPAT mutations were carried by other patients with HL. NPAT contains 18 exons totaling 1427 amino acids. The exons and exon-intron boundaries of NPAT were sequenced from genomic DNA of the 73 Finnish HL patients, 93 HL patients from United Kingdom, 3 previously published NLPHL families, and from an NLPHL-derived cell line DEV.13-16 A deletion of 3 bp resulting to the loss of serine 724 (S724) was detected in 7 patients (Table 1). One of them was a Finnish early-onset NLPHL patient, and 4 were sporadic HL patients from United Kingdom who had all been diagnosed between ages of 14 and 30 years. In addition, the S724 deletion was detected in 2 HL patients from the United Kingdom of whom one had a relative with non-HL and one with HL. The deletion was sequenced from 282 healthy Finnish and 177 United Kingdom persons, and was found in 5 persons.
We estimated the impact of the S724 deletion on HL risk through unconditional logistic regression calculating odds ratios and 95% confidence interval using STATA (Version 10.0, State College). On the basis of frequencies of S724 deletion seen in the United Kingdom and Finnish populations, the odds ratio for HL associated with this sequence change is 4.11 (95% confidence interval, 1.27-13.35, P = .018). The success rate of sequencing was 93% for the HL patients and 97% for the controls at the site of S724 deletion.
We also screened the NPAT cDNAs of the NLPHL patients in the original Finnish family as well as in the French and English families and the DEV cell line for possible mutations, but none was identified. The NPAT deletion c. 2437-2438delAG product was seen with clearly lowered intensity in the cDNA sequences (approximately one-third of the wt allele) of the Finnish mutation carriers.
Expression analysis
Significant decrease in NPAT mRNA level (fold change = 0.80, t test, P = .016) was also observed in the patients and the carriers of the deletion c. 2437-2438delAG compared with the controls and the nonmutation carriers, suggesting that the mutated product is directed to the nonsense mediated decay.
Discussion
Genetic factors have long been recognized to contribute to the risk of developing HL, and familial cases, including all HL subtypes, have been estimated to represent 4.5% of HL.20 Association to human leukocyte antigen and linkage to non-human leukocyte antigen loci have been suggested in familial HL, and the genetic susceptibility seems mainly to be related to the disease in young adults.6,7,21-23 The interaction of genes and environmental determinants, such as viral infections, have also been implicated, but, for example, EBV is detected in the tumor cells of less than one-third of familial HL patients compared with 40% to 50% in sporadic cases.24,25 In a few families, a variant in KLHDC8B gene resulting in a decreased protein expression has been reported to associate with cHL, a finding awaiting confirmation.9 The remarkable majority of the patients in the studied HL families have been diagnosed with cHL, most often with the nodular sclerosis or mixed cellularity subtype, and it is possible that the etiologic factors are different in different lymphoma subtypes. Four NLPHL families from United Kingdom/India, France, Turkey, and Finland have thus far been published.10,13-15 EBV, which is infrequently found in sporadic NLPHL, was not detected in any of the patients' tumors, and screening of KLHDC8B did not reveal disease-associated mutations.10 Consequently, the cause of familial NLPHL has thus far remained unknown.
We have identified a germline deletion c. 2437-2438delAG of 2 bp, resulting in frame shift in the NPAT gene in the Finnish family with NLPHL susceptibility using linkage analysis and exome sequencing, a new, powerful and unbiased tool for large-scale genome-wide detection of genetic variations. The deletion in NPAT was the only segregating variation located in the linked regions that was neither a previously known polymorphism nor detected in healthy controls. The NPAT mRNA expression was significantly decreased in the carriers of the deletion. The deletion segregated with the disease in the family members, and also 3 healthy relatives were found to have the mutation suggesting that the penetrance for NLPHL susceptibility is low or additional genetic or environmental factors modify disease risk.
We screened a large number of additional HL patients and another variation leading to a deletion of a serine 724 of NPAT was found. This was significantly overrepresented in the patients with HL, further indicating a role for NPAT in the genetic background of HL. It is, however, acknowledged that our estimate of the impact of this rare variant on HL is imprecise because it is based on a relatively small dataset and a subset of cases were enriched for genetic susceptibility by virtue of having a family history of a lymphoproliferative disease.
NPAT is located in chromosome 11q22.3 next to ataxia-telangiectasia mutated gene (ATM) with which it shares a putative promoter region. It is expressed in all human tissues.26,27 NPAT is phosphorylated by cyclin E-cyclin-dependent kinase 2 (CDK2) complex, and its expression peaks in accordance with cyclin E-CDK2 complex activity at the G1 to S transition of the cell cycle promoting entry to the S-phase.28 NPAT interacts with histone nuclear factor P (HiFN-P) and mediates the activation of histone gene transcription in a cyclin E-CDK2-regulated manner.29,30 The HiFN-P/NPAT complex also affects the transcription of other target genes, such as NPAT and ATM; and in B-cell chronic lymphocytic leukemia cells, NPAT has been shown to be down-regulated.31,32 These data suggest that NPAT has an important role in the regulation of the cell cycle; and, together with HiFN-P, it contributes to the activation of the promoter of ATM, a gene in which mutations have been found in different lymphoid malignancies.31,33-35 In addition, the ataxia-telangiectasia patients with germline mutations in ATM are predisposed to different cancers, particularly leukemias and lymphomas.36
This is the first study in which a candidate germline mutation has been identified in familial NLPHL, providing potential insights to the molecular basis of the disease. Although our findings are intuitively interesting, replication in an independent material is highly desirable. Considering that NPAT shares the promoter region with ATM, mediates important functions in the cell cycle, and possibly has an effect on ATM transcription, there are various cellular mechanisms to which a defect in NPAT could putatively cause dysregulation and thus cause cancer susceptibility. It is also possible that NPAT-related pathways are involved in the pathogenesis of sporadic NLPHL or other hematologic malignancies. In familial HL cases where mutations of NPAT were not found, it is possible that mutations in other genes of the same pathway are involved.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sini Marttinen, Mairi Kuris, Iina Vuoristo, and Inga-Lill Svedberg for technical assistance, Dr Sibrand Poppema for providing the DEV-cell line, and the patients and their clinicians who participated in this collection (listed in Enciso-Mora et al8 ).
This work was supported by the Academy of Finland (Center of Excellence in Translational Genome-Scale Biology and grant 212901), Sigrid Juselius Foundation, Paulo Foundation, Blood Disease Research Foundation, Finnish Medical Foundation, Nona and Kullervo Väre Foundation, Otto Malm Foundation, and National Health Service funding to the National Institute for Health Research Biomedical Research Center. Sample collection at the Institute of Cancer Research was supported by Breakthrough Breast Cancer and the European Union.
Authorship
Contribution: S.S. was responsible for the study, acquired, analyzed, and interpreted the data, and wrote the manuscript; M.A. performed sequencing and expression analyses and wrote the manuscript; V.L. contributed to the study design and supervised the study; R.L. performed linkage analysis and wrote the respective sections of the manuscript; K.F. performed pathologic evaluation; H.J.L. performed laboratory experiments; E.K. analyzed expression data; K.A. acquired patients and contributed to the study design; P.B. performed laboratory experiments; J.T. performed pathologic evaluation of EBV-encoded RNA in situ hybridization samples; B.J.B., F.B., A.U., A.J.S., R.C., and M.J.M. acquired patients and provided samples; R.H. performed statistical analysis and wrote the manuscript; P.V. contributed to the study design and supervised the study; L.A.A. was the principal investigator of the study; and all authors read and contributed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lauri A. Aaltonen, Tumor Genomics Department, Medical Genetics Biomedicum Helsinki, PO Box 63 (Haartmaninkatu 8) FIN-00014 University of Helsinki, Helsinki, Finland; e-mail: lauri.aaltonen@helsinki.fi.