Abstract
Although the critical requirement for the transcription factor RUNX1/AML1 at the onset of hematopoietic development is well established, little is known about its transcriptional targets at this pivotal stage of blood development. Using microarrays, we identified the uncharacterized gene AI467606 as a gene whose expression level is dramatically reduced in the absence of RUNX1. We further demonstrated by chromatin immunoprecipitation and promoter assay a direct regulation of its transcription by RUNX1. Using a bacterial artificial chromosome transgenic approach, we established that AI467606 is expressed during the development of the hematopoietic system in vivo and in vitro and that its expression is detected within the CD41+ population and marks definitive hematopoietic potential. Similarly, in the adult mouse, all hematopoietic cell lineages, except mature erythrocytes, express AI467606. Taken together, these findings indicate that AI467606 is a novel transcriptional target of RUNX1/AML1 at the onset of hematopoietic development that is extensively expressed within the hematopoietic system.
Introduction
The hemangioblast is a precursor that generates hematopoietic, endothelial, and smooth muscle lineages.1,2 This clonal precursor detected during embryonic stem (ES) cell differentiation and in gastrulating embryos gives rise in culture to blast colonies with both endothelial and hematopoietic potential.3,4 We have recently demonstrated that the generation of definitive hematopoietic cells from the hemangioblast goes through a hemogenic endothelium intermediate and that the transcription factor RUNX1 is essential for this process.4 Several other in vivo and in vitro studies have similarly established this generation of blood cells from a hemogenic endothelium and the requirement for RUNX1 in this process.5-12 However, little is known about the transcriptional targets controlled by RUNX1 at this pivotal stage of blood development. In the present work, we identified AI467606 as a novel transcriptional target of RUNX1 at the onset of hematopoietic development and demonstrated by chromatin immunoprecipitation (ChIP) and promoter assay a direct regulation of its transcription by RUNX1. We next characterized the pattern of expression of this gene in hematopoiesis in vitro using the differentiation system of ES cells as well as in vivo in embryos and adult mice. Our data demonstrate that AI467606 is extensively expressed in hematopoietic cells and that the AI467606-expressing population contains all definitive hematopoietic progenitors.
Methods
ES cell culture, embryoid bodies (EB) differentiation, bacterial artificial chromosome recombineering, and flow cytometry were performed as previously described.13-15 Real-time polymerase chain reaction (PCR) was performed on an ABI 7900 system (Applied Biosystems) using the Roche universal probe library and primer designer (Roche Applied Science). Rabbit IgG (315-005-003 from Jackson ImmunoResearch Laboratories), Pol II (N-20; sc-899; Santa Cruz Biotechnology), and Runx1 (PC284; Calbiochem) antibodies were used for ChIP. The Dual-Luciferase Reporter Assay System (Promega) was used for luciferase assays. Time-lapse photography was performed using the IncuCyteFLR live-cell imaging system (Essen Instruments) equipped with a 20× objective lens. Immunohistochemistry sections were analyzed in a Mirax Histology System (Zeiss). Slides were scanned at 200× magnification with a Zeiss Plan-Apochromat 20×/0.8 NA objective lens.
Results and discussion
Identification of a new transcriptional target of RUNX1
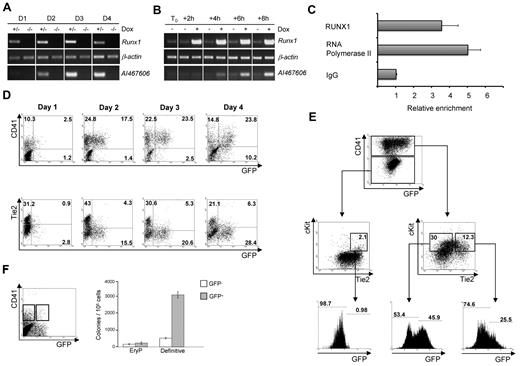
It is now well established that hematopoietic cells are generated from hemogenic endothelium through a RUNX1-dependent process.4-12 To identify transcriptional targets of RUNX1 at this early stage of hematopoiesis, we differentiated Runx1+/− and Runx1−/− ES cells for 3.5 days, sorted hemangioblast-enriched cell populations based on Fetal Liver Kinase-1 expression, and replated them in blast colony conditions. After 24 hours, at which time the hemogenic endothelium population is established, the transcriptome of the 2 cell lines was compared by DNA microarray analysis (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article; http://bioinformatics.picr.man.ac.uk/vice/PublicProjects.vice, accession GE_CRUK/SCBR[2]). Among other genes, we identified AI467606 as a gene with lower expression level in the absence of RUNX1 (supplemental Figure 1B). The AI467606 gene encodes for a protein of 225 amino acids with no known function. To confirm the DNA microarray results, we performed semiquantitative reverse-transcription PCR analysis comparing Runx1+/− and Runx1−/− cells over the 4-day time course of blast colony development. In the presence of RUNX1, the expression of AI467606 was weak at day 1, readily detectable by day 2 of culture, and further increased over time. In contrast, in the absence of RUNX1, AI467606 expression was barely detectable (Figure 1A). We further investigated the influence of RUNX1 on AI467606 transcription using a Runx1−/− ES cell line in which Runx1 expression is induced on the addition of doxycycline.4 After 2 days of blast culture, doxycycline was added to the media and the cells were harvested for RNA analysis every 2 hours. AI467606 transcription was quickly up-regulated within 4 hours after the induction of Runx1 expression (Figure 1B), suggesting that AI467606 is a direct RUNX1 transcriptional target. Subsequent ChIP experiments demonstrated binding of RUNX1 to the AI467606 promoter during blast colony development (Figure 1C). Within a small 84-bp region of the AI467606 promoter, we found 3 RUNX1-binding sites that are conserved in rat, human, chimpanzee, and mouse (supplemental Figure 1C). Both deletion of this 84-bp sequence and specific mutation of the 3 Runx1 binding sites resulted in a net decrease of RUNX1-induced transcription in luciferase assays (supplemental Figure 1D). Overall, these results indicate that AI467606 is a direct transcriptional target of RUNX1 at the onset of hematopoietic development.
Identification of AI467606 as a novel transcriptional target of RUNX1 and its expression pattern at the onset of hematopoietic development. (A) Semiquantitative reverse-transcription PCR analysis of expression of Runx1 and AI467606 during the course of 4 days of Runx1+/− and Runx1−/− blast development. (B) Semiquantitative reverse-transcription PCR analysis of expression of Runx1 and AI467606 after Runx1 induction in Runx1−/− ES cell. (C) ChIP analysis of day 2 blast colony cells. The precipitated chromatin was analyzed using primers specific for AI467606 promoter. All values are relative to the IgG value. (D-E) FACS analysis of AI467606-GFP, CD41, and Tie2 coexpression in blast colonies from day 1 to day 4. Numbers indicate percentages of respective populations. (F) Number of hematopoietic colonies generated by CD41+GFP− (white boxes) and CD41+GFP+ (gray boxes) cell populations. Definitive colony counts include macrophages, macrophages/erythrocytes, granulocytes-macrophages, and mix granulocyte-macrophage-erythroid colonies. EryP indicates primitive erythrocytes. Mean numbers of colonies from 3 dishes of a representative experiment are shown. Error bars represent SEM (n = 3).
Identification of AI467606 as a novel transcriptional target of RUNX1 and its expression pattern at the onset of hematopoietic development. (A) Semiquantitative reverse-transcription PCR analysis of expression of Runx1 and AI467606 during the course of 4 days of Runx1+/− and Runx1−/− blast development. (B) Semiquantitative reverse-transcription PCR analysis of expression of Runx1 and AI467606 after Runx1 induction in Runx1−/− ES cell. (C) ChIP analysis of day 2 blast colony cells. The precipitated chromatin was analyzed using primers specific for AI467606 promoter. All values are relative to the IgG value. (D-E) FACS analysis of AI467606-GFP, CD41, and Tie2 coexpression in blast colonies from day 1 to day 4. Numbers indicate percentages of respective populations. (F) Number of hematopoietic colonies generated by CD41+GFP− (white boxes) and CD41+GFP+ (gray boxes) cell populations. Definitive colony counts include macrophages, macrophages/erythrocytes, granulocytes-macrophages, and mix granulocyte-macrophage-erythroid colonies. EryP indicates primitive erythrocytes. Mean numbers of colonies from 3 dishes of a representative experiment are shown. Error bars represent SEM (n = 3).
AI467606 is expressed in CD41+ cells during blast colony formation
To track and analyze AI467606 expression at the single-cell level, we engineered a bacterial artificial chromosome reporter construct by replacing the coding sequence of AI467606 with a green fluorescent protein (GFP)–neomycin cassette and established transgenic AI467606-GFP ES cell lines. The expression of GFP was detected in a large fraction of these ES cells (supplemental Figure 2A left) and correlated with endogenous AI467606 transcripts (supplemental Figure 2A middle and right). We next investigated AI467606 expression pattern during blast colony development. Consistent with our PCR results (Figure 1A), GFP expression was detected by fluorescence-activated cell sorter (FACS) from day 1 of culture onward (Figure 1D) and was morphologically associated with the appearance of round cells emerging from clusters of adherent cells (supplemental Figure 2B). GFP expression was correlated with the expression of CD41, the first marker expressed by emerging hematopoietic cells,16-19 whereas fewer emerging AI467606-GFP+ cells coexpressed the endothelial marker Tie-220 (Figure 1D). We have recently shown that Tie2+cKit+CD41− hemogenic endothelium cells generate hematopoietic cells through the acquisition of CD41 expression followed by the down-regulated expression of endothelial markers, such as Tie2. The proximal Runx1 promoter is active in the Tie2+cKit+CD41− hemogenic endothelium intermediate population, whereas the distal Runx1 promoter becomes active in cells expressing CD41 and having a low Tie2 expression level.21 When we analyzed the expression of AI467606, we observed that it was barely detectable within the Tie2+cKit+CD41− hemogenic endothelium population (Figure 1E), whereas within the subsequent stages of maturation AI467606-GFP was detected in 25.5% of the Tie2+cKit+CD41+ population and 46% of the Tie2lowcKit+CD41+ cells (Figure 1E). The absence of AI467606 expression in hemogenic endothelium suggests that the activity of the proximal RUNX1 alone is not sufficient to drive AI467606 expression. Altogether, these results indicate that the onset of AI467606 expression is observed within CD41+ cells. As the expression of CD41 is correlated with the acquisition of hematopoietic potential, we next assessed whether the expression of AI467606 within the CD41+ fraction defines a subpopulation of cells with enriched hematopoietic potential. GFP+ and GFP− cells from the CD41+ cell population were sorted from day 5 EBs and tested in clonogenic replating assay for hematopoietic precursors. Primitive erythrocytes and definitive colonies were generated from both populations, but the double-positive population was highly enriched in definitive hematopoietic precursors (Figure 1F). Together, these observations indicate that AI467606 expression by CD41+ cells marks a subpopulation of cells highly enriched in definitive hematopoietic progenitors.
AI467606 expression in vivo
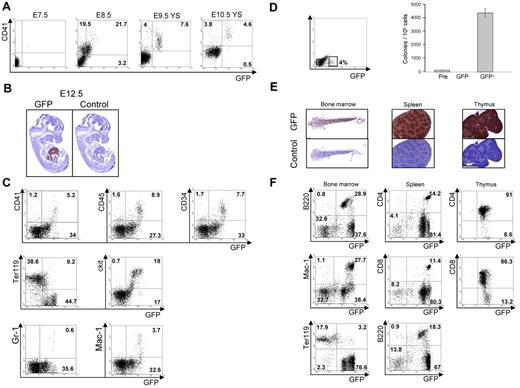
We next analyzed AI467606 expression in vivo by establishing an AI467606-GFP bacterial artificial chromosome transgenic mouse line. GFP+ cells were first detected at embryonic stage E8.5 to E10.5 within the CD41+ population in agreement with the data obtained in vitro (Figure 2A). Immunostaining of sections of whole embryos indicated a strong specific staining in fetal liver at E12.5 and fetal liver and thymus at E14.5 (Figure 2B; supplemental Figure 3). Flow cytometric analysis revealed that all CD41+, CD34+, cKit+, CD45+, Gr-1+, Mac-1+ cells were detected within an AI467606-GFPhigh fraction. In contrast, the AI467606-GFPlow population was mainly negative for these markers, with the exception of cKit and Ter119 (Figure 2C). These results suggest that AI467606 is uniformly expressed in hematopoietic precursors but only weakly expressed in early erythroid cells and not expressed at later stages of erythropoiesis, a pattern reminiscent of Runx1 expression.22-24 To investigate the hematopoietic potential of the AI467606-GFP+ cells, AI467606-GFP− and GFP+ subpopulations isolated from GFP+ embryos at E8.5 were tested in a clonogenic assay. We observed that all the hematopoietic potential resides in the AI467606-GFP+ population (Figure 2D). Similar results were obtained for E11.5 fetal liver and yolk sac and E14.5 fetal liver (data not shown). These results indicate that the onset of expression of AI467606 takes place at the early stages of hematopoietic development in CD41+ cells and that AI467606 expression is sustained in hematopoietic cells in the liver. In adult mice, immunostaining indicated that AI467606 is widely expressed in bone marrow, spleen, and thymus (Figure 2E). In the bone marrow, all cells positive for the B-cell markers B220, IgM, and IgD and the myeloid markers Gr-1 and Mac-1 coexpressed AI467606, whereas in contrast erythroid Ter119+ cells were mostly AI467606-GFP− (Figure 2F; supplemental Figure 4A). Similarly, in spleen and thymus, most of the cells expressed AI467606 (Figure 2F; supplemental Figure 4B-C).
Analysis of the expression of AI467606 in mouse embryos and adult hematopoietic organs. (A) FACS analysis of CD41 and AI467606-GFP coexpression in single embryos at E7.5, E8.5, yolk sac at E9.5 and E10.5. (B) GFP and control antibody immunohistochemistry of paraffin sections of transgenic embryos at E12.5. (C) FACS analysis of AI467606-GFP coexpression with CD41, CD45, CD34, Ter119, cKit, Gr-1, and Mac-1 in E12.5 fetal liver. Numbers indicate percentages of respective populations. (D) GFP FACS profile of E8.5 embryos and FACS sorting gates. Number of colonies generated by each cell population in methylcellulose. Mean numbers of colonies from 3 dishes of a representative experiment are shown. Error bars represent SEM (n = 3), Pre corresponds to the total population (Pre-sort). (E) GFP and control antibody immunohistochemistry of paraffin sections of bone marrow, spleen, and thymus of transgenic embryos. (F) FACS analysis of AI467606-GFP coexpression with B220 B-cell marker, Mac-1 myeloid marker, Ter119 erythroid marker, and CD4 and CD8 T-cell markers in bone marrow (left), spleen (middle), and thymus (right) of AI467606-GFP mice. Numbers indicate percentages of respective populations.
Analysis of the expression of AI467606 in mouse embryos and adult hematopoietic organs. (A) FACS analysis of CD41 and AI467606-GFP coexpression in single embryos at E7.5, E8.5, yolk sac at E9.5 and E10.5. (B) GFP and control antibody immunohistochemistry of paraffin sections of transgenic embryos at E12.5. (C) FACS analysis of AI467606-GFP coexpression with CD41, CD45, CD34, Ter119, cKit, Gr-1, and Mac-1 in E12.5 fetal liver. Numbers indicate percentages of respective populations. (D) GFP FACS profile of E8.5 embryos and FACS sorting gates. Number of colonies generated by each cell population in methylcellulose. Mean numbers of colonies from 3 dishes of a representative experiment are shown. Error bars represent SEM (n = 3), Pre corresponds to the total population (Pre-sort). (E) GFP and control antibody immunohistochemistry of paraffin sections of bone marrow, spleen, and thymus of transgenic embryos. (F) FACS analysis of AI467606-GFP coexpression with B220 B-cell marker, Mac-1 myeloid marker, Ter119 erythroid marker, and CD4 and CD8 T-cell markers in bone marrow (left), spleen (middle), and thymus (right) of AI467606-GFP mice. Numbers indicate percentages of respective populations.
Overall, these findings indicate a pan-hematopoietic pattern of AI467606 expression both in embryonic, fetal and adult hematopoiesis. AI467606 expression appears at the onset of hematopoiesis at E8.5 within a CD41+ population enriched in definitive hematopoietic progenitors and is maintained in fetal liver and adult hematopoietic cells, in contrast to CD41, which becomes more restricted to megakaryocytes at these stages. The next steps will be to investigate the function of AI467606 in the hematopoietic compartment. We have so far attempted knockdown with several shRNA but did not observe any striking hematopoietic phenotype. These negative results are however not fully conclusive as none of our shRNA results in a complete shutdown of AI467606 expression, and we cannot therefore exclude that the remaining expression is enough to support hematopoietic differentiation. A definitive evaluation of the functionality of AI467606 expression in the development of the hematopoietic system will require the generation of a knockout mouse model.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank laboratory members for critical reading of the manuscript.
This work was supported by Cancer Research UK and BBSRC.
Authorship
Contribution: C.F., C.L., and M.L. designed and performed the research, analyzed the data, and wrote the manuscript; and V.K. and G.L. designed and supervised the research project, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Georges Lacaud, Paterson Institute for Cancer Research, University of Manchester, Wilmslow Rd, M20 4BX Manchester, United Kingdom; e-mail: glacaud@picr.man.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal