Abstract
Mutations in transmembrane protease, serine 6 (TMPRSS6), encoding matriptase-2, are responsible for the familial anemia disorder iron-refractory iron deficiency anemia (IRIDA). Patients with IRIDA have inappropriately elevated levels of the iron regulatory hormone hepcidin, suggesting that TMPRSS6 is involved in negatively regulating hepcidin expression. Hepcidin is positively regulated by iron via the bone morphogenetic protein (BMP)-SMAD signaling pathway. In this study, we investigated whether BMP6 and iron also regulate TMPRSS6 expression. Here we demonstrate that, in vitro, treatment with BMP6 stimulates TMPRSS6 expression at the mRNA and protein levels and leads to an increase in matriptase-2 activity. Moreover, we identify that inhibitor of DNA binding 1 is the key element of the BMP-SMAD pathway to regulate TMPRSS6 expression in response to BMP6 treatment. Finally, we show that, in mice, Tmprss6 mRNA expression is stimulated by chronic iron treatment or BMP6 injection and is blocked by injection of neutralizing antibody against BMP6. Our results indicate that BMP6 and iron not only induce hepcidin expression but also induce TMPRSS6, a negative regulator of hepcidin expression. Modulation of TMPRSS6 expression could serve as a negative feedback inhibitor to avoid excessive hepcidin increases by iron to help maintain tight homeostatic balance of systemic iron levels.
Introduction
Transmembrane protease, serine 6 (TMPRSS6), encoding matriptase-2 (MTP-2), a transmembrane serine protease produced by the liver,1,2 was recently identified as a critical gene for iron homeostasis.3,4 In both humans and mice, mutations in the TMPRSS6 gene lead to a strong increase in hepcidin expression, resulting in a dramatic decrease in ferroportin expression, and severe iron deficiency anemia that is unresponsive to oral iron treatment but partially responsive to parenteral iron therapy (IRIDA).3-6 Moreover, genome-wide association studies identified common TMPRSS6 variants associated with hematologic parameters7-9 and serum iron concentration,8,10 highlighting that TMPRSS6 is important in the control of iron homeostasis and normal erythropoiesis. It was recently proposed that the mechanism by which TMPRSS6 inhibits hepcidin expression is by down-regulation of the bone morphogenetic protein (BMP)–SMAD signaling pathway via proteolytic cleavage of the BMP coreceptor hemojuvelin.11,12
Hepcidin is an iron-regulated hepatic peptide hormone that controls iron absorption at the intestinal level and iron release from macrophages and hepatocytes. Hepcidin binds to the plasma membrane iron exporter ferroportin and induces its endocytosis and proteolysis, preventing release of iron into plasma.13 Iron14 and inflammatory cytokines (eg, interleukin-6)15 stimulate hepcidin expression, leading to reduced plasma iron levels. In contrast, hypoxia, high erythropoietic activity, and iron deficiency inhibit hepcidin expression.16
The role of the BMP-SMAD signaling pathway in regulating hepcidin expression is now well established.17,18 BMP6, whose mRNA expression is regulated by iron in vivo,19 is critical in mice to activate this signaling cascade.20,21 BMP6 binds type I and type II BMP receptors (BMPR-I and -II) in the presence of the BMP coreceptor hemojuvelin, inducing the phosphorylation of BMPR-I by BMPR-II. The activated receptor complex, in turn, phosphorylates a subset of SMAD proteins (SMAD-1/5/8). These receptor-activated SMADs then form heteromeric complexes with the common mediator SMAD4, and these translocate to the nucleus where they regulate transcription of specific targets, such as hepcidin.22 The importance of the BMP-SMAD signaling pathway in regulating hepcidin expression and iron homeostasis in vivo is supported by the severe iron overload phenotype seen in humans and mice with mutations of genes in this pathway. For example, mutation or loss of the hemojuvelin23-25 gene (HFE2, also known as HJV) causes juvenile hemochromatosis with an indistinguishable phenotype from juvenile hemochromatosis because of mutations or loss of HAMP (encoding hepcidin)26,27 itself in both human patients and in mice. Global knockout of Bmp620,21 or liver-specific conditional knockout of Smad428 also results in a similar iron overload phenotype in mice.
The mechanisms by which TMPRSS6 expression is regulated are not fully understood. Previously, it has been shown that TMPRSS6 expression can be induced by known inhibitory stimuli of hepcidin expression. Indeed, TMPRSS6 mRNA expression has been demonstrated to be induced by erythropoietin29 and hypoxia30 and by acute iron deprivation at the protein level.31 In this study, we investigated whether TMPRSS6 expression is regulated in vitro and in vivo by one of the most important activators of hepcidin expression, BMP6. We also assessed whether the potent physiologic inducer of hepcidin, iron, can modulate TMPRSS6 mRNA levels in vivo.
Methods
Cell culture
The human hepatocarcinoma cell line Hep3B (HB-8064, ATCC) was cultured in ATCC-formulated Eagle Minimum Essential Medium (ATCC) supplemented with 10% fetal bovine serum (FBS; ATCC). Cell cultures were maintained at 37°C under 5% CO2.
Treatment of Hep3B cells with BMP6, LDN-193189, and cycloheximide
Hep3B cells (1.2 × 105 per well) were seeded onto 24-well plates. Twenty-four hours later, the culture medium was exchanged to 1% FBS medium. After 4 hours, cells were treated with recombinant human BMP6 (5, 25, and 50 ng/mL, 16 hours; R&D Systems) and then harvested for total RNA extraction.
For LDN-193189 experiments, the small molecule BMP inhibitor LDN-193189 (60nM) was custom synthesized as previously described32-34 and was added 30 minutes after serum starvation. Cells were then treated with BMP6 as previously described.
For cycloheximide experiments, cells were treated with 10 μg/mL of cycloheximide (Calbiochem) 2 hours before BMP6 treatment.
Time course of BMP6 treatment on hepcidin and TMPRSS6 expression in vitro
Hep3B cells (5 × 104 per well) were seeded onto 24-well plates. Seven hours later, the culture medium was exchanged to 1% FBS medium. After 16 hours, cells were treated with recombinant human BMP6 (25 ng/mL; R&D Systems) and then harvested for total RNA extraction 1, 4, 9, 16, 24, 32, and 48 hours after treatment.
Animals
The Institutional Animal Care and Use Committee at the Massachusetts General Hospital (MGH) approved all of the following animal protocols.
For BMP6 injection and BMP6 antibody injection experiments, C57BL/6 mice were purchased from Taconic at 7 weeks of age, then housed in the MGH facility and maintained on a standard rodent diet (Prolab RMH 3000 diet, 380 ppm iron) for 1 week before the start of treatment.
For BMP6 experiments, 8-week-old males received an intraperitoneal injection of BMP6 diluted in vehicle (20mM sodium acetate, 5% mannitol solution, pH 4.0) at 750 μg/kg or an equal volume of vehicle alone (n = 3 per group). Mice were killed and tissues harvested for analysis at 6 and 12 hours after injection. BMP6 was produced as previously described.35
For BMP6 antibody experiments, 8-week-old males received an intraperitoneal injection of monoclonal anti-BMP6 antibody (R&D Systems) 15 mg/kg daily for 1 week. Control mice received an equivalent volume of phosphate-buffered saline (n = 5 per group). Mice were killed and tissues harvested for analysis 6 hours after the last injection.
For chronic iron treatment experiments, 7-week-old C57BL/6 male mice received an iron-deficient diet (2-6 ppm iron, TD.80396, Harlan Teklad) for 12 to 14 days before iron administration. Then, mice were fed with a 2% carbonyl iron diet (TD.08496, Harlan Teklad). Mice were killed and tissue harvested at time zero (baseline), 24 hours, 48 hours, 72 hours, 1 week, or 2 weeks of iron enriched diet (n = 6 per group), as previously described.36
RNA extraction and quantitative real-time PCR
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN), with DNase I digestion by the RNase-Free DNase Set (QIAGEN) according to the manufacturer's instructions. Real-time quantification of mRNA transcripts was performed using 2-step reverse-transcribed polymerase chain reaction (RT-PCR) using the ABI Prism 7900HT Sequence Detection System. First-strand cDNA synthesis was performed using iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's instruction. Next, TMPRSS6, hepcidin, SMAD7, and inhibitor of DNA binding 1 (ID1) transcripts were amplified with specific primers (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and detected using the Power SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer's instructions. In parallel, RPL19 transcripts were amplified with specific primers (supplemental Table 1) and detected in a similar manner to serve as an internal control. Samples were analyzed in duplicate for each experiment, and results are reported as the ratio of mean values for TMPRSS6, hepcidin, SMAD7, and ID1 to RPL19.
Western blot analysis
Hep3B cells (3 × 106 per plate) were seeded in 10-cm dishes. During seeding, cells were reverse transfected with 20 μL of the liposomal transfection reagent lipofectamine 2000 (Invitrogen) and 5nM of control (ON-TARGET plus Nontargeting pool D-001810–10-05, Dharmacon) or siRNA TMPRSS6 (siGENOME SMART pool siRNA D-006052, Dharmacon) or 8 μg of human TMPRSS6-FLAG cDNA.37 Five hours later, culture medium was exchanged to stop the transfection. Twenty-four hours after seeding, cells were treated with BMP6 (25 ng/mL; R&D Systems) and then harvested for protein extraction 48 hours after treatment. Membrane proteins were extracted using proteoJET Membrane Protein Extraction Kit (Fermentas) according to the manufacturer's instructions. Membrane proteins were concentrated using a 3-kDa molecular weight cutoff ultrafiltration system (Amicon Ultra, Millipore). Proteins were quantified by using the BCA Protein Assay (Pierce); equal amounts of membrane protein (3.6 μg) were subjected to NuPAGE Novex 6% Tris-Glycine Gel (Invitrogen) and then transferred to Immun-Blot PVDF Membrane (Bio-Rad). Blots were blocked with stringent milk (10% milk, 1% bovine serum albumin, 1% Igepal, 0.25% Tween-20, phosphate-buffered saline 1 time) 1 hour at room temperature, incubated overnight 4°C with rabbit anti-TMPRSS6 (1:1000; Abcam). After washing with phosphate-buffered saline–Tween-20 0.1%, blots were incubated for 1 hour at room temperature with horseradish peroxidase–conjugated secondary anti–rabbit (Sigma-Aldrich) diluted 1:5000 in stringent milk and developed using a chemiluminescence detection kit (PerkinElmer). Membranes were then stripped with glycine buffer (0.5M, pH 2.5) and reprobed with rabbit anti–pan-cadherin (Abcam; 1:1000) in stringent milk for normalization.
Measurement of MTP-2 activity
Hep3B cells were treated as described in the previous paragraph for the Western blot experiments. Twenty-four hours after treatment with BMP6 (25 ng/mL) media was replaced with Optimem media (Invitrogen) again containing BMP6. After 24 hours, conditioned media were prepared by centrifugation (30 minutes 4000g) using a 3-kDa molecular weight cutoff ultrafiltration system (Amicon Ultra). Protein concentration in conditioned media was assessed by BCA test and 15 μg of protein was used for enzymatic assay.
As previously described,38 in brief, activity was assayed in Tris/saline buffer (50mM Tris, pH 8.0, and 150mM NaCl) at 37°C by monitoring the release of p-nitroaniline from the chromogenic substrate N-(tert-butoxycarbonyl)-Gln-Ala-Arg-p-nitroanilide (666μM; Bachem) at a wavelength of 405 nm for up to 20 minutes.
siRNA experiments
Hep3B cells (1.2 × 105 per well) were seeded onto 24-well plates. During seeding, cells were reverse-transfected with 1 μL of lipofectamine 2000 (Invitrogen) and 10nM of control (siGENOME SMART pool siRNA D-006052, Dharmacon) or human SMAD7 (siGENOME SMART pool siRNA D-020068, Dharmacon) or human ID1 siRNA (siGENOME SMART pool siRNA D-005051, Dharmacon). Five hours later, culture medium was exchanged to stop the transfection. Twenty-four hours after seeding, cells were serum starved with FBS 1% medium and 4 hours later treated with BMP6 (25 ng/mL, R&D Systems) and then harvested for RNA extraction 24 hours after treatment.
Results
TMPRSS6 expression is increased by BMP6
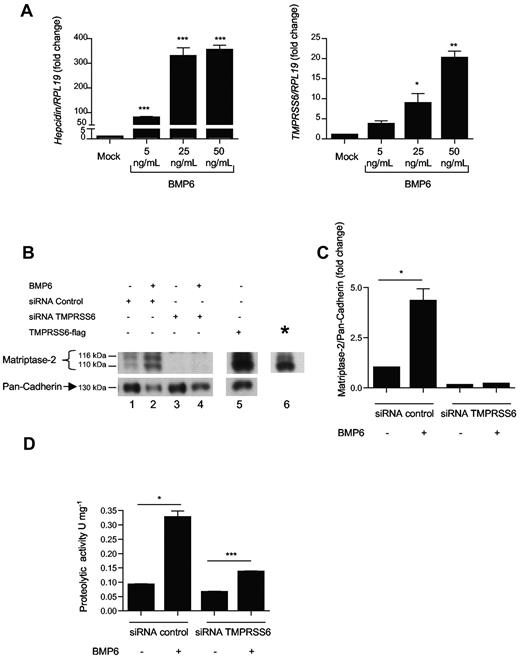
To determine whether BMP6, a known activator of hepcidin,18 can modulate TMPRSS6 expression levels, Hep3B cells were treated for 16 hours with 5, 25, or 50 ng/mL of BMP6, and hepcidin and TMPRSS6 mRNA expression were measured by quantitative real-time PCR. These data reveal that treatment with BMP6 induces a dose-dependent increase in hepcidin expression (Figure 1A). Interestingly, TMPRSS6 mRNA expression was also stimulated by BMP6 in a dose-dependent manner, by 4-fold with 5 ng/mL of BMP6, by 9-fold with 25 ng/mL, and by 20-fold with 50 ng/mL (Figure 1A). Similar increases were observed after treatment with other BMP ligands (BMP2, 4, 7, 9, and 11, supplemental Figure 1).
TMPRSS6 expression is induced by BMP6. (A) Hep3B cells were treated with 5, 25, and 50 ng/mL of human BMP6 for 16 hours and were analyzed for hepcidin and TMPRSS6 relative to RPL19 mRNA expression by quantitative real-time RT-PCR. The mean of 3 to 8 (depending of the dose) independent experiments is presented. Results are reported as the mean ± SEM for the fold change from mock, and significant changes represent the comparisons with mock. (B-C) Hep3B cells were transfected with siRNA control (5nM), siRNA TMPRSS6 (5nM), or TMPRSS6-FLAG (8 μg), and treated with 25 ng/mL of BMP6 for 48 hours. Cells were analyzed for matriptase-2 level relative to pan-cadherin protein by Western blot (B) followed by chemiluminescence quantification (C). (B) *A shorter exposure of lane 5 to better distinguish the 2 bands. (C) The mean of 3 experiments is presented, and results are reported as the mean ± SEM. (D) A total of 15 μg of protein from conditioned media of Hep3B cells transfected with siRNA control (5nM) and siRNA TMPRSS6 (5nM) and treated with BMP6 (25 ng/mL) for 48 hours were incubated with 666μM of N-(tert-butoxycarbonyl)-Gln-Ala-Arg-p-nitroanilide. Activity of matriptase-2 was assessed by measurement of the release of the dye p-nitroaniline during up to 20 minutes at a wavelength of 405 nm at 37°C using a spectrophotometer. The resulting activities (1 U corresponds to a release rate of 1 mmol of p-nitroaniline per minute) were measured in duplicate in 3 independent experiments. Results are reported as the mean ± SEM. (A,C-D) Significant changes are as follows: *P < .05; **P < .01; and ***P < .005.
TMPRSS6 expression is induced by BMP6. (A) Hep3B cells were treated with 5, 25, and 50 ng/mL of human BMP6 for 16 hours and were analyzed for hepcidin and TMPRSS6 relative to RPL19 mRNA expression by quantitative real-time RT-PCR. The mean of 3 to 8 (depending of the dose) independent experiments is presented. Results are reported as the mean ± SEM for the fold change from mock, and significant changes represent the comparisons with mock. (B-C) Hep3B cells were transfected with siRNA control (5nM), siRNA TMPRSS6 (5nM), or TMPRSS6-FLAG (8 μg), and treated with 25 ng/mL of BMP6 for 48 hours. Cells were analyzed for matriptase-2 level relative to pan-cadherin protein by Western blot (B) followed by chemiluminescence quantification (C). (B) *A shorter exposure of lane 5 to better distinguish the 2 bands. (C) The mean of 3 experiments is presented, and results are reported as the mean ± SEM. (D) A total of 15 μg of protein from conditioned media of Hep3B cells transfected with siRNA control (5nM) and siRNA TMPRSS6 (5nM) and treated with BMP6 (25 ng/mL) for 48 hours were incubated with 666μM of N-(tert-butoxycarbonyl)-Gln-Ala-Arg-p-nitroanilide. Activity of matriptase-2 was assessed by measurement of the release of the dye p-nitroaniline during up to 20 minutes at a wavelength of 405 nm at 37°C using a spectrophotometer. The resulting activities (1 U corresponds to a release rate of 1 mmol of p-nitroaniline per minute) were measured in duplicate in 3 independent experiments. Results are reported as the mean ± SEM. (A,C-D) Significant changes are as follows: *P < .05; **P < .01; and ***P < .005.
Using Western blot analysis, we assessed whether BMP6 treatment also led to an increase in MTP-2 protein. To assess the specificity of the anti–matriptase-2 antibody that we used for our experiments, we transfected Hep3B cells with previously published human TMPRSS6-FLAG cDNA37 as a positive control, and a pool of 4 siRNA TMPRSS6 was used as a negative control. Transfection of Hep3B cells with 5nM of siRNA TMPRSS6 reduces TMPRSS6 mRNA expression by 93% in nontreated cells and in cells treated with 25 ng/mL of BMP6 compared with Hep3B cells transfected with siRNA control (supplemental Figure 2A-B). We detected an increase by 4.3-fold in 2 specific bands for MTP-2 protein in the membrane protein fraction of cells treated with BMP6 (Figure 1B lane 2; Figure 1C), but not in the soluble cytosolic protein fraction. This pattern of MTP-2 expression is in agreement with results previously described2,11 and indicates that MTP-2 protein produced during BMP6 treatment is correctly targeted to the cell membrane and is likely differentially N-link glycosylated. The 2 bands that were increased with BMP6 treatment were abolished by transfection of TMPRSS6 siRNA (Figure 1B, lane 4; Figure 1C), and were induced by transfection with TMPRSS6-FLAG (Figure 1B, lane 5), confirming their specificity for MTP-2.
We next addressed whether the increase in MTP-2 protein quantity induced by BMP6 treatment resulted in an increase in MTP-2 activity. Under our experimental conditions, detection of cleaved soluble HJV protein by Western blot analysis in the conditioned media of Hep3B cells treated with BMP6 ligand was not a sensitive assay for assessing changes in MTP-2 activity (supplemental Figure 3). To measure MTP-2 protein activity, we determined the hydrolysis rate of N-(tert-butoxycarbonyl)-Gln-Ala-Arg-p-nitroanilide, a specific chromogenic substrate for trypsin-like proteases,38,39 in conditioned media derived from Hep3B cells treated with BMP6 and transfected with either control siRNA or TMPRSS6 siRNA. Treatment with BMP6 induced an increase by 3.6-fold of protease activity in the conditioned media (Figure 1D). Moreover, this increase in activity was inhibited by TMPRSS6 siRNA transfection. Thus, the increase in MTP-2 protein expression induced by BMP6 also leads to a concomitant increase in MTP-2 protease activity.
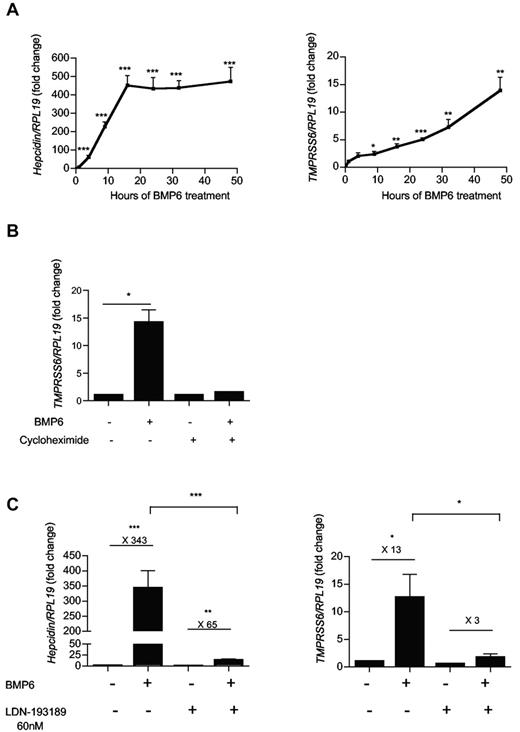
We then investigated whether the induction of TMPRSS6 and hepcidin mRNA by BMP6 were contemporaneous. Hep3B cells were treated with 25 ng/mL of BMP6 and total RNA was extracted after several time points between 1 and 48 hours of treatment. After 1 hour, hepcidin mRNA expression was already significantly increased by 5-fold to reach a plateau after 16 hours of treatment (∼ 450-fold; Figure 2A). However, TMPRSS6 mRNA expression became significantly increased only after 9 hours of treatment with BMP6, after which it progressively increased until 48 hours of treatment (Figure 2A). Because of this delay in time necessary to stimulate TMPRSS6 mRNA expression in response to BMP6 treatment, we next analyzed whether this regulation was direct or dependent on new protein synthesis. The presence of the protein synthesis inhibitor cycloheximide during treatment with BMP6 significantly reduced TMPRSS6 mRNA expression by 90% (Figure 2B) compared with treatment with BMP6 alone, suggesting the necessity of de novo protein synthesis to regulate TMPRSS6 mRNA expression in response to BMP6.
Regulation of TMPRSS6 mRNA expression in response to BMP6. Hep3B cells were treated with 25 ng/mL of BMP6 for several time points between 1 and 48 hours (A), and were analyzed for hepcidin and TMPRSS6 relative to RPL19 mRNA expression by quantitative real-time RT-PCR. (B-C) Hep3B cells received 10 μg/mL of cycloheximide (B) or 60nM of LDN-193189 (C) before BMP6 (25 ng/mL) treatment and were analyzed for gene expression relative to RPL19 mRNA expression by quantitative real-time RT-PCR. The mean of 6 (A-B) and 3 (C) independent experiments is presented. (B-C) Results are reported as the mean ± SEM for the fold change from mock (just before adding BMP6) and significant changes represent the comparisons with mock. Significant changes are as follows: *P < .05; **P < .01; and ***P < .005.
Regulation of TMPRSS6 mRNA expression in response to BMP6. Hep3B cells were treated with 25 ng/mL of BMP6 for several time points between 1 and 48 hours (A), and were analyzed for hepcidin and TMPRSS6 relative to RPL19 mRNA expression by quantitative real-time RT-PCR. (B-C) Hep3B cells received 10 μg/mL of cycloheximide (B) or 60nM of LDN-193189 (C) before BMP6 (25 ng/mL) treatment and were analyzed for gene expression relative to RPL19 mRNA expression by quantitative real-time RT-PCR. The mean of 6 (A-B) and 3 (C) independent experiments is presented. (B-C) Results are reported as the mean ± SEM for the fold change from mock (just before adding BMP6) and significant changes represent the comparisons with mock. Significant changes are as follows: *P < .05; **P < .01; and ***P < .005.
BMP6 induction of TMPRSS6 mRNA expression is dependent on BMP type I receptor kinase activity and ID1
Because BMP6 is a strong inducer of the BMP-SMAD pathway, we addressed whether an intact and functional BMP-SMAD pathway was necessary to permit an increase of TMPRSS6 mRNA by BMP6 treatment. Before treatment with 25 ng/mL of BMP6, Hep3B cells were first treated with LDN-193189, a small molecule inhibitor of the BMP-SMAD pathway that inhibits BMP type I receptor kinase activity.40 As already demonstrated with an analog of LDN-193189, Dorsomorphin,41 treatment with LDN-193189 alone induced a significant decrease of hepcidin mRNA compared with mock-treated cells by 4-fold. Moreover, the presence of LDN-193189 strongly diminished the increase of hepcidin mRNA caused by BMP6 treatment (Figure 2C). Treatment with LDN-193189 alone induced a trend toward a decrease in TMPRSS6 mRNA expression by 31%. Interestingly, LDN-193189 significantly reduced the increase of TMPRSS6 mRNA expression induced by BMP6 treatment by 83% (Figure 2C), highlighting the necessity of a functional BMP-SMAD pathway to regulate TMPRSS6 mRNA expression.
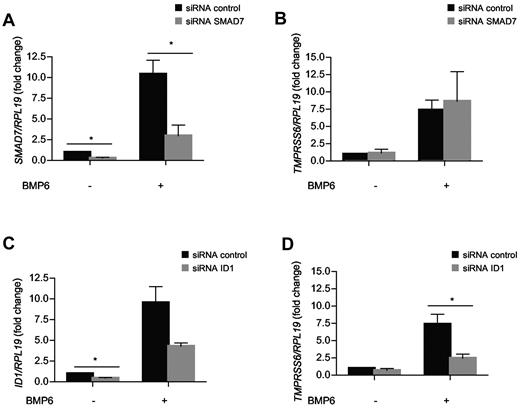
To explore the mechanism of regulation of the TMPRSS6 mRNA expression by BMP6, we analyzed the effect of silencing SMAD7 and ID1. We hypothesized that SMAD7 could participate in TMPRSS6 regulation because it was demonstrated that SMAD7 is an inhibitory SMAD protein that is induced by BMP6-SMAD signaling in hepatocytes to antagonize TGF-β/BMP signaling42-44 and to suppress steady-state hepcidin expression as well as hepcidin induction by BMP stimulation.45 To test whether SMAD7 is involved in TMPRSS6 mRNA regulation, we transfected Hep3B cells with a pool of siRNA directed against SMAD7 and a control siRNA pool. Treatment of Hep3B cells with BMP6 induced an increase in SMAD7 mRNA expression by 10-fold (Figure 3A black bars), whereas silencing of SMAD7 with siRNA transfection in nontreated cells or cells treated with 25 ng/mL of BMP6 leads to a reduction of SMAD7 mRNA level by 70% (Figure 3A gray bars). However, silencing of SMAD7 expression had no effect on TMPRSS6 mRNA expression in nontreated condition as well in response to BMP6 treatment (Figure 3B). This finding indicates that SMAD7 is not a participant in TMPRSS6 regulation by BMP6.
TMPRSS6 expression is controlled by ID1 in response to BMP6. Hep3B cells transfected with 10nM of siRNA control, siRNA SMAD7 (A-B), or siRNA ID1 (C-D), and treated in the absence or presence of 25 ng/mL of BMP6 for 24 hours were analyzed for TMPRSS6 (B,D) SMAD7 (A), and ID1 (C) mRNA expression relative to RPL19 mRNA by quantitative real-time RT-PCR. Results are reported as the mean ± SEM for the fold change from mock, and significant changes represent the comparisons with mock (siRNA control alone). Significant changes are as follows: *P < .05.
TMPRSS6 expression is controlled by ID1 in response to BMP6. Hep3B cells transfected with 10nM of siRNA control, siRNA SMAD7 (A-B), or siRNA ID1 (C-D), and treated in the absence or presence of 25 ng/mL of BMP6 for 24 hours were analyzed for TMPRSS6 (B,D) SMAD7 (A), and ID1 (C) mRNA expression relative to RPL19 mRNA by quantitative real-time RT-PCR. Results are reported as the mean ± SEM for the fold change from mock, and significant changes represent the comparisons with mock (siRNA control alone). Significant changes are as follows: *P < .05.
ID1 is a target transcript that is up-regulated by the BMP-SMAD pathway. It has been shown that, in vivo, Id1 mRNA is up-regulated in the liver in parallel with Bmp6 mRNA, phosphorylated SMAD1/5/8 protein, and hepcidin mRNA by iron.19,46 ID1 proteins are negative regulators of basic-helix-loop-helix transcription factors that can either activate47 or repress transcription.48 As shown in Figure 3C, treatment of Hep3B cells with 25 ng/mL of BMP6 increased ID1 mRNA expression by 10-fold. Transfection with an siRNA pool directed against ID1 leads to a decrease in ID1 expression by 50% either in nontreated cells or in cells treated with BMP6. Surprisingly, silencing ID1 also had a consequence on TMPRSS6 mRNA expression (Figure 3D). In Hep3B cells treated with BMP6, the increase of TMPRSS6 mRNA expression was significantly reduced by 67% by silencing ID1 (Figure 3D), and in nontreated cells, TMPRSS6 mRNA expression was reduced by 30% (nonsignificant). These results suggest that ID1 is a link between the BMP/SMAD signaling pathway and TMPRSS6 regulation in response to BMP6.
Modulation of TMPRSS6 mRNA expression in vivo by BMP6 and iron
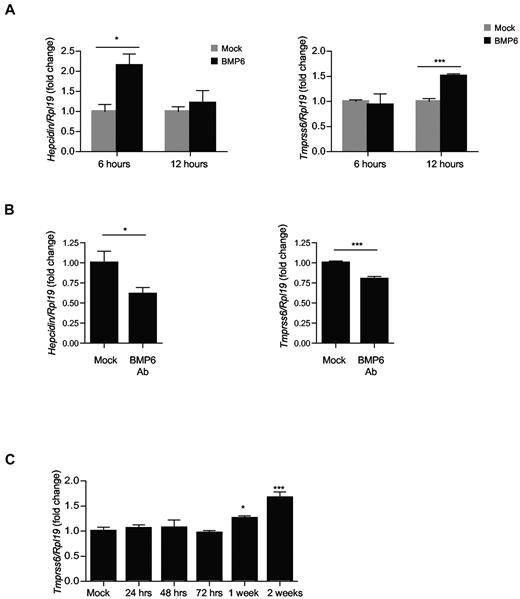
To investigate whether TMPRSS6 mRNA expression could be modulated by BMP6 in vivo, we injected 8-week-old C57BL/6 male mice with 750 μg/kg of BMP6 or vehicle alone. Six and twelve hours after injection, mice were killed and hepcidin and Tmprss6 mRNA expression was measured by quantitative real-time PCR in the liver. As previously shown,20 BMP6 injection led to a significant increase in hepatic hepcidin mRNA by 2-fold at 6 hours after injection (Figure 4A left). However, this increase was transient, and hepcidin mRNA expression returned to basal levels at 12 hours after injection (Figure 4A left). BMP6 injection induced a significant increase in Tmprss6 mRNA expression by 1.5-fold at 12 hours after injection, but there was no change at 6 hours after injection (Figure 4A right). This indicates that BMP6 regulates TMPRSS6 mRNA expression in vivo, and there is a delay between the up-regulation of hepcidin and up-regulation of Tmprss6 mRNA expression, similar to the delay seen in in vitro experiments (Figure 2A).
TMPRSS6 expression is up-regulated in vivo by BMP6 and chronic iron treatment. (A) Eight-week-old male C57BL/6 mice received an intraperitoneal injection of BMP6 750 μg/kg animal weight (BMP6, black bars) or vehicle alone (mock, gray bars; n = 3 per group) for 6 and 12 hours. (B) Eight-week-old male C57BL/6 mice received an intraperitoneal injection of neutralizing BMP6 antibody 15 mg/kg once a day for 1 week (n = 5 per group). (C) Seven-week-old male C57BL/6 mice were killed at time zero (baseline) or after initiation of a 2% carbonyl iron diet for 24 hours, 48 hours, 72 hours, 1 week, or 2 weeks (n = 6 per group). Tissues were analyzed for hepatic hepcidin and Tmprss6 relative to Rpl19 mRNA by quantitative real-time RT-PCR. Results are reported as the mean ± SEM for the fold change from mock and significant changes represent the comparisons with mock. Significant changes are as follows: *P < .05; and ***P < .005.
TMPRSS6 expression is up-regulated in vivo by BMP6 and chronic iron treatment. (A) Eight-week-old male C57BL/6 mice received an intraperitoneal injection of BMP6 750 μg/kg animal weight (BMP6, black bars) or vehicle alone (mock, gray bars; n = 3 per group) for 6 and 12 hours. (B) Eight-week-old male C57BL/6 mice received an intraperitoneal injection of neutralizing BMP6 antibody 15 mg/kg once a day for 1 week (n = 5 per group). (C) Seven-week-old male C57BL/6 mice were killed at time zero (baseline) or after initiation of a 2% carbonyl iron diet for 24 hours, 48 hours, 72 hours, 1 week, or 2 weeks (n = 6 per group). Tissues were analyzed for hepatic hepcidin and Tmprss6 relative to Rpl19 mRNA by quantitative real-time RT-PCR. Results are reported as the mean ± SEM for the fold change from mock and significant changes represent the comparisons with mock. Significant changes are as follows: *P < .05; and ***P < .005.
To further validate the importance of the endogenous BMP6-SMAD pathway in the regulation of TMPRSS6 expression in vivo, 8-week-old C57BL/6 mice were injected with 15 mg/kg of neutralizing BMP6 antibody once a day for one week to inhibit the BMP6-SMAD pathway. In accordance with results already published,20 anti-BMP6 treatment reduced significantly hepcidin mRNA levels by 39% (Figure 4B left). Anti-BMP6 treatment also caused a significant reduction in hepatic expression of Tmprss6 mRNA by 20% (Figure 4C right), indicating that Tmprss6 mRNA expression in vivo is regulating by the BMP-SMAD pathway.
Because chronic dietary iron loading significantly increases hepatic Bmp6 mRNA expression in mice,19,46,49 we investigated whether chronic dietary iron loading in mice also induces Tmprss6 mRNA expression. As previously described,36 we fed 8-week-old C57BL/6 male mice with 2% carbonyl iron diet for 2 weeks. Under these conditions, serum iron, transferrin saturation, and liver iron content were all significantly increased by 24 hours and for the entire 2-week time course.36 Hepcidin mRNA expression was significantly increased by 2.7-fold after 24 hours and up to 3.5-fold by 48 hours above baseline, after which hepcidin mRNA expression plateaued at 3- to 3.5-fold above baseline for the remaining 2 weeks on the 2% carbonyl iron diet.36 ID1 was weakly induced by 2- to 3-fold above baseline after 24 to 72 hours, and more strongly induced to 6.7-fold by 1 week and 10-fold by 2 weeks on the high-iron diet.36 Tmprss6 mRNA expression remained unchanged at 24, 48, and 72 hours on the 2% carbonyl iron diet and but became significantly increased after 1 week of iron-enriched diet, reaching a peak after 2 weeks (1.7-fold; Figure 4C).
Discussion
Iron is an essential nutrient, but free iron can form reactive oxygen species leading to cell damage. Thus, iron homeostasis needs to be tightly regulated. Because there is no physiologic means to excrete iron from the body, this regulation occurs at the level of intestinal iron absorption, and macrophage and hepatocyte iron release by modulation of the iron exporter ferroportin and its ligand hepcidin. There is a complex interplay between regulatory networks that activate hepcidin expression in response to iron14,19 and inflammation,15 and those that inhibit hepcidin expression in response to hypoxia, anemia, and iron deficiency.16 Recently, TMPRSS6, a gene that encodes the membrane bound serine protease protein MTP-2 and is mainly expressed in the liver, was identified as an important inhibitor of hepcidin expression to control iron homeostasis and normal erythropoiesis.7-9 Known inhibitors of hepcidin expression, hypoxia and erythropoietin, have been shown to induce an up-regulation of TMPRSS6 expression.29,30 Here, we investigated whether known activators of hepcidin expression, including BMP6 and iron, could also regulate TMPRSS6 expression.
We show that TMPRSS6 mRNA expression in Hep3B cells, a widely studied hepatocyte derived hepatoma cell line, is stimulated by BMP6. TMPRSS6 mRNA expression could also be induced by several other BMPs which have previously been shown to strongly stimulate hepcidin mRNA expression,18 confirming the link between the BMP-SMAD pathway and the up-regulation of TMPRSS6 mRNA expression. Importantly, we further show that the up-regulation of TMPRSS6 mRNA levels induced by BMP6 was almost completely abolished by treatment with LDN-193189, a specific inhibitor of the BMP-SMAD signaling pathway, in Hep3B cells. These experiments indicate that the BMP signaling pathway is a potent activator of TMPRSS6 mRNA expression in vitro.
We also demonstrate a corresponding increase in MTP-2 protein levels in Hep3B cells in response to BMP6 treatment. We used a polyclonal anti–MTP-2 antibody that was able to recognize human MTP-2 protein bands of approximately 110 and 116 kDa in size. Of note, 2 protein bands of similar size, which probably represent differentially glycosylated MTP-2 protein, have been previously reported by several groups.2,39 The specificity of the anti–MTP-2 antibody for these protein bands was confirmed in studies using TMPRSS6-FLAG cDNA as a positive control and by the silencing of TMPRSS6 using specific siRNA treatment as a negative control. The anti–MTP-2 antibody could recognize the 2 protein bands when TMPRSS6-FLAG was overexpressed in Hep3B cells, confirming that the anti–MTP-2 antibody could recognize full-length human MTP-2 protein. In Hep3B cells, silencing of TMPRSS6 with specific siRNA leads to loss of the 2 protein bands (Figure 1B, lanes 3-4), indicating that these 2 bands probably represented MTP-2 protein.
Importantly, the increase in MTP-2 protein level induced by BMP6 results in an increase in MTP-2 activity measured in an assay using a specific peptidic substrate.39 We were unable to detect cleavage of soluble HJV protein by MTP-2 activity induced by BMP6 under the conditions tested. Instead, we used a highly sensitive MTP-2 activity assay using N-(tert-butoxycarbonyl)-Gln-Ala-Arg-p-nitroanilide as a chromogenic substrate for trypsin-like proteases that is also suitable for MTP-2.2,39,50 The liberation of the dye p-nitroaniline by proteolytic cleavage leads to an increase of absorption at 405 nm, which corresponds to the protease activity. In transfected HEK cells, MTP-2 is autocatalytically activated, and the active fragment is released into the conditioned media.39 The activity of MTP-2 can then be measured in the conditioned media using the chromogenic substrate assay. We show that treatment with BMP6 leads to an increase in the absorbance at 405 nm in the conditioned media of Hep3B cells. This signal could be the result of MTP-2 or other trypsin-like protease activity. However, we show that silencing of TMPRSS6 with siRNA leads to near-complete abolition of this increase in the 405-nm absorbance signal. This result indicates that increase induced by BMP6 was indeed mostly the result of MTP-2 activity, although there could be a participation of another trypsin-like protease that is induced. It is more probable that the inability to completely block the increase in the 405-nm absorbance signal could be the result of incomplete silencing by the TMPRSS6 siRNA. Nevertheless, BMP6 treatment clearly leads to an increased cleavage of the MTP-2 peptidic substrate.
Interestingly, the induction of TMPRSS6 mRNA is dependent on de novo protein synthesis because the protein translation inhibitor cycloheximide could block the increase induced by BMP6. Indeed, the induction of TMPRSS6 mRNA expression requires at least 9 hours of BMP6 treatment before a significant effect is seen. This time delay is unlike the time course of other BMP-SMAD targets, such as hepcidin and ID1, that can rapidly increase their expression within 1 hour of BMP-SMAD pathway activation. With promoter analysis software, we did not identify in the TMPRSS6 promoter any BMP responsive elements (BMP-RE) with sequences identical to the ones previously identified in the hepcidin,51 ID1,52 and SMAD745 promoters. Thus, it is unlikely that the BMP-SMAD pathway directly regulates TMPRSS6 mRNA through a BMP-RE in the TMPRSS6 promoter.
Because BMP6 treatment leads to an early induction of downstream signaling molecules and transcription modulators, such as SMAD7 and ID1, we tested whether SMAD7 or ID1 was involved in the later up-regulation of TMPRSS6 mRNA. We found that, whereas the silencing of SMAD7 had no effect on TMPRSS6 induction by BMP6, silencing of ID1 significantly and strongly blocked the increase of TMPRSS6 mRNA expression induced by BMP6 treatment. Thus, ID1 expression appears to be important for the up-regulation of TMPRSS6 by BMP6. This is the first description that ID1 has a role in up-regulating TMPRSS6, and this could subsequently lead to an indirect inhibition of hepcidin expression (Figure 5). ID1 does not bind to DNA directly but instead acts to prevent the formation of transcriptional activator complexes.47,48 The specific transcriptional activator that is important for the regulation of TMPRSS6 by ID1 is an important question for future study.
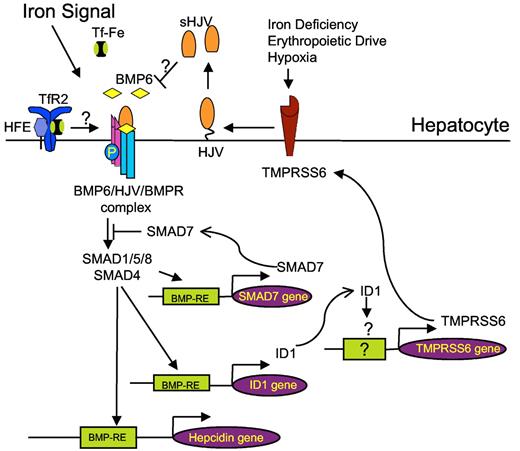
Schematic representation showing proposed role of TMPRSS6 regulation by the BMP6-SMAD signaling pathway and iron via ID1. We propose that, in addition to being stimulated by several signals that inhibit hepcidin, such as iron deficiency, erythropoeitic drive, and hypoxia, TMPRSS6 expression is also stimulated indirectly by the hepcidin activators BMP6 and iron. Stimulation by BMP6 and/or iron induces an increase of the BMP6-HJV-SMAD pathway activity, possibly through a mechanism involving HFE and transferrin receptor 2 (TFR2), leading to binding of SMAD complexes to BMP-responsive elements (BMP-REs) on the hepcidin promoter and up-regulation of hepcidin transcription. In parallel, BMP6-SMAD pathway directly up-regulates SMAD7 and ID1 transcription. ID1 induction leads to the up-regulation of TMPRSS6 expression. TMPRSS6 then serves as a negative feedback inhibitor of BMP-SMAD pathway activity and hepcidin expression by cleaving the BMP coreceptor HJV. Inhibitory SMAD7 can also act as a negative feedback inhibitor by blocking SMAD activation. By acting as negative feedback inhibitors, TMPRSS6 and SMAD7 are important to prevent excessive hepcidin increases in response to BMP6 and iron, thereby maintaining tight control of iron homeostasis. Abbreviations: BMPR indicates BMP receptor; TF-Fe, holotransferrin; sHJV, soluble hemojuvelin; and BMP-RE, BMP responsive element.
Schematic representation showing proposed role of TMPRSS6 regulation by the BMP6-SMAD signaling pathway and iron via ID1. We propose that, in addition to being stimulated by several signals that inhibit hepcidin, such as iron deficiency, erythropoeitic drive, and hypoxia, TMPRSS6 expression is also stimulated indirectly by the hepcidin activators BMP6 and iron. Stimulation by BMP6 and/or iron induces an increase of the BMP6-HJV-SMAD pathway activity, possibly through a mechanism involving HFE and transferrin receptor 2 (TFR2), leading to binding of SMAD complexes to BMP-responsive elements (BMP-REs) on the hepcidin promoter and up-regulation of hepcidin transcription. In parallel, BMP6-SMAD pathway directly up-regulates SMAD7 and ID1 transcription. ID1 induction leads to the up-regulation of TMPRSS6 expression. TMPRSS6 then serves as a negative feedback inhibitor of BMP-SMAD pathway activity and hepcidin expression by cleaving the BMP coreceptor HJV. Inhibitory SMAD7 can also act as a negative feedback inhibitor by blocking SMAD activation. By acting as negative feedback inhibitors, TMPRSS6 and SMAD7 are important to prevent excessive hepcidin increases in response to BMP6 and iron, thereby maintaining tight control of iron homeostasis. Abbreviations: BMPR indicates BMP receptor; TF-Fe, holotransferrin; sHJV, soluble hemojuvelin; and BMP-RE, BMP responsive element.
BMP6 is also a positive regulator of hepatic Tmprss6 mRNA expression in vivo in mice. Injection of BMP6 into mice leads to an increase in hepatic hepcidin mRNA expression and also leads to an increase in Tmprss6 mRNA levels in the liver. Interestingly, the induction in Tmprss6 mRNA levels was delayed compared with the timing of hepcidin mRNA induction, a result that mimics the time delay seen in vitro in Hep3B cells. Importantly, inhibition of endogenous BMP6 in mice by administration of an anti-BMP6 monoclonal antibody significantly decreased baseline hepatic Tmprss6 mRNA expression. These data suggest that BMP6 is a physiologic regulator of Tmprss6 mRNA levels in vivo.
Chronic dietary iron loading is associated with increased hepatic Bmp6 mRNA expression and Smad1/5/8 protein phosphorylation after as early as 24 hours, and hepcidin induction by chronic dietary iron loading is inhibited by coadministration with an anti-BMP6 antibody,36 suggesting that stimulation of the BMP6-SMAD pathway activity is a main mechanism by which iron stimulates hepcidin expression. Here, we show that, after one week of an iron-enriched diet, hepatic Tmprss6 mRNA expression was significantly up-regulated, indicating that this gene is also physiologically regulated in response to iron overload. Given our data showing that BMP6 up-regulates TMPRSS6 expression both in vitro and in vivo, we hypothesize that the mechanism by which dietary iron loading up-regulates hepatic Tmprss6 mRNA expression is by up-regulating hepatic Bmp6 mRNA expression. The delay between the increase in Bmp6 mRNA expression and Tmprss6 may be the result of the indirect nature of TMRPSS6 induction by BMP6 or the necessity of a high level of BMP6 expression or an accumulation of BMP6 protein to stimulate Tmprss6 mRNA expression.
With chronic dietary iron loading, Id1 mRNA expression also begins to be significantly up-regulated after 24 hours but is very strongly increased after 1 week to reach the maximal expression at 2 weeks.36 This time delay parallels the time delay seen in the induction of Tmprss6 mRNA, reinforcing the notion that ID1 is the main regulator of TMPRSS6 expression in response to BMP6. Interestingly, Lakhal et al30 demonstrated that hypoxia induces TMPRSS6 mRNA expression, and Peng et al29 showed that EPO treatment in mice also increases TMPRSS6 mRNA expression. These studies did not examine the role of the BMP-SMAD pathway or present information regarding a time delay in TMPRSS6 mRNA regulation. It would be of interest to study the involvement of BMP6 and ID1 in TMPRSS6 regulation under conditions of hypoxia or erythropoietic drive to determine whether ID1 regulation of TMPRSS6 is a broad mechanism or is only necessary in response to BMP6 signaling and iron loading.
The induction of TMPRSS6 expression could be an important part of a counter-regulatory feedback mechanism necessary to prevent overshooting of hepcidin expression in response to positive stimuli, such as iron. Indeed, it has been shown that overexpression of TMPRSS6 cDNA in cells could prevent BMP-mediated induction of hepcidin mRNA expression.3 In our current experiments, this notion is supported by the observation that chronic dietary iron loading leads to hepcidin mRNA expression that reaches a plateau after 48 hours, and this plateau lasts for up to 2 weeks.36 During this time period, any increase in TMPRSS6 expression could be involved in preventing further rises in hepcidin levels, thus maintaining the plateau in hepcidin levels. Therefore, an increase in TMPRSS6 expression may have a similar inhibitory function on hepcidin levels as has been postulated for SMAD7, which is also stimulated by chronic dietary iron loading19 and BMP-SMAD signaling pathway activity in the liver,45 and functions as a feedback inhibitor of BMP-SMAD pathway activity to down-regulate hepcidin expression.45 Although TMPRSS6 and SMAD7 act on hepcidin expression as feedback inhibitors, it possible that their roles are different. SMAD7 expression, similar to hepcidin expression, is up regulated after 24 hours under chronic iron treatment, whereas TMPRSS6 mRNA expression is significantly increased after only 1 week of iron-enriched treatment. These results suggest that, in response to iron, SMAD7 may participate in the establishment of the plateau phase of hepcidin expression after 48 hours, whereas TMPRSS6 could be involved in the maintenance of this plateau as a secondary mechanism.
In conclusion, we demonstrate here that TMPRSS6 is regulated by known hepcidin stimulators, such as BMP6 and iron, possibly via ID1. This regulation of TMPRSS6 is in addition to its known regulation in response to iron deficiency, anemia, and hypoxia. These data indicate that TMPRSS6 may be involved in fine-tuning hepcidin expression by limiting hepcidin induction over the long-term (ie, over weeks) by BMP6 and iron. Understanding the role and regulation of TMPRSS6 in hepcidin regulation may ultimately lead to new therapeutic strategies to treat diseases where hepcidin levels are dysregulated, such as anemia of chronic disease or hemochromatosis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
D.M. was supported by the MGH Fund for Medical Discovery (Postdoctoral Fellowship Award). E.C. was supported in part by the Massachusetts Biomedical Research Corporation at Massachusetts General Hospital (Tosteson Postdoctoral Fellowship Awards). J.L.B. was supported in part by the National Institutes of Health (grants K08 DK075846 and RO1 DK087727) and the Massachusetts General Hospital (Claflin Distinguished Scholar Award). H.Y.L. was supported in part by the National Institutes of Health (grants RO1 DK-069533 and RO1 DK-071837).
National Institutes of Health
Authorship
Contribution: D.M., J.L.B., and H.Y.L. designed the experiments and analyzed and interpreted the data; D.M., V.V., C.C.S., E.C., and S.C. performed the experiments; C.L.-O., L.G., C.C.H., and S.V. supplied vital reagents; M.S. and M.G. provided scientific expertise about matriptase-2 activity; D.M. and H.Y.L. wrote the manuscript; V.V., C.C.S., E.C., C.L.-O., M.S., M.G., S.V., and J.L.B. edited the manuscript; and H.Y.L. supervised the project.
Conflict-of-interest disclosure: J.L.B. and H.Y.L. have ownership interest in a startup company Ferrumax Pharmaceuticals, which has licensed technology from the Massachusetts General Hospital based on their work. The remaining authors declare no competing financial interests.
Correspondence: Herbert Y. Lin, Program in Membrane Biology, Division of Nephrology, Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge St, CPZN-8216, Boston, MA 02114; e-mail: lin.herbert@mgh.harvard.edu.