Abstract
The nuclear factor of activated T cells (NFAT) family of transcription factors plays important roles in many biologic processes, including the development and function of the immune and vascular systems. Cells usually express more than one NFAT member, raising the question of whether NFATs play overlapping roles or if each member has selective functions. Using mRNA knock-down, we show that NFATc3 is specifically required for IL2 and cyclooxygenase-2 (COX2) gene expression in transformed and primary T cells and for T-cell proliferation. We also show that NFATc3 regulates COX2 in endothelial cells, where it is required for COX2, dependent migration and angiogenesis in vivo. These results indicate that individual NFAT members mediate specific functions through the differential regulation of the transcription of target genes. These effects, observed on short-term suppression by mRNA knock-down, are likely to have been masked by compensatory effects in gene-knockout studies.
Introduction
The nuclear factor of activated T cells (NFAT) family of transcription factors consists of 4 members regulated by the Ca2+-calmodulin–dependent phosphatase calcineurin (CN). NFAT proteins were initially identified in T cells, and their functions in the immune system have been analyzed extensively.1 In resting T cells, NFATs are present in the cytoplasm in a hyperphosphorylated and inactive form. Increases in the levels of intracellular Ca2+ activate CN, which dephosphorylates NFAT. This promotes the translocation of NFATs to the nucleus, where they generally cooperate with other transcription factors to regulate an array of genes involved in the functions of the immune system,2-5 including IL26 and cyclooxygenase 2 (COX2).7 Expression of NFATc1, NFATc2, and NFATc3 (but not NFATc4) has been detected in normal and transformed T cells.8 The co-occurrence of different NFAT members in the same cells raises the question of whether there is a degree of functional redundancy or if the immunoregulatory functions of each member are unique. The NFAT DNA-binding domain is highly conserved among members,2,3 and the immune phenotypes of mice deficient in a single NFAT member are relatively mild,1 which supports the existence of functional redundancy in T-cell activation. The use of Nfatc1 knockout (KO) cells in complementation experiments in Rag 2−/− mice indicated that NFATc1 plays a role in the expansion of mature peripheral B and T lymphocytes and in Th2 cytokine production, but is not required for earlier differentiation and activation events.9,10 Similarly, Nfatc2−/− mice have mild defects in the immune response11 and impaired Th2 differentiation due to overproduction of IL4.12,13 Nfatc3-deficient mice show impaired thymic development, characterized by a loss of CD4+/CD8+ double-positive cells through programmed cell death.14 More recently, the use of conditional NFATc3 KO mice has revealed a role in both positive selection and double-negative development, but these mice showed no detectable alteration in T-cell activation.15 Nfatc4 gene deletion causes no major detectable anomaly in the immune response.16
More severe phenotypes are found in mice lacking more than one NFAT. T-cell effector functions are markedly impaired in Nfatc1-c2 double KO mice, and these mice fail to produce IL2, IL4, INFγ, and IL5.17 Double deletion of Nfatc2 and Nfatc3 results in spontaneous T-cell differentiation into Th2 cells, even in the absence of IL4.9,18 Therefore, although the data obtained with mice lacking individual NFAT members indicate a certain degree of redundancy, when taken together with findings from double KO mice they suggest that NFATc1 and NFATc2 are important for T-cell commitment and activation, whereas NFATc3 is important for T-cell development.
NFATs are also expressed in endothelial cells, where they regulate genes important for cell activation and angiogenesis.19,20 Activation of endothelial cells by the proangiogenic factor VEGF results in the NFAT-dependent expression of COX2.21 COX2 is involved in angiogenesis, and its transcriptional inhibition with the CN inhibitor cyclosporin A results in the inhibition of VEGF-dependent angiogenesis in vivo and of prostacyclin production and capillary morphogenesis in cultured endothelial cells.21 No significant defects in endothelial cell activation or angiogenesis have been reported in mice lacking individual NFAT members. As occurs with T cells, overt phenotypic defects are observed only in double KO mice; for example, mouse embryos lacking Nfats c3 and c4 are defective for vessel assembly and vascular patterning.16
One of the limitations of studies with KO mice is compensation of the role of the deficient gene by related genes, masking the true impact of a gene deletion.22 To avoid this limitation, we knocked down NFATc3 and NFATc1 individually and compared the effects on the transcriptional regulation of known NFAT target genes, obtaining novel findings not previously observed in KO studies. NFATc3 knock-down blocks IL2 and COX2 gene expression in human T cells and prevents their activation, whereas NFATc1 knock-down up-regulates the expression of IL2 and several other inflammatory modulators. Furthermore, NFAT member–dependent regulation is not restricted to this cell type, because in primary endothelial cells, VEGF-induced expression of COX2, proliferation, migration, and in vivo angiogenesis are also NFATc3 dependent.
Methods
Cell culture and transient transfection
HEK-293 cells were cultured in DMEM containing 10% FBS (Sigma-Aldrich) and l-glutamine plus antibiotics (100 units/mL of penicillin and 100 μg/mL of streptomycin). HEK cells were transfected by the calcium phosphate method.23 Jurkat cells were cultured in RPMI-1640 medium (GIBCO-Invitrogen) containing 10% FBS supplemented with l-glutamine plus antibiotics. Jurkat cells were transfected for 4 hours using Lipofectamine-PLUS (Invitrogen). Where indicated, cells were stimulated with 20 ng/mL of phorbol myristate acetate (PMA; Sigma-Aldrich) plus 1μM calcium ionophore A23187 (Calbiochem) and 0.3mM CaCl2 (PMA + Io treatment).
Peripheral blood lymphocytes (PBLs) were isolated from human blood as described previously,24 and then stimulated with PHA (5μg/mL) and cultured in RPMI medium supplemented with 10% FCS, 20 mM HEPES, l-glutamine, antibiotics, and 50 U/mL of IL2. After 12 days, cells were transferred to medium without IL2, and 2 days later blast cells were stimulated with plate-bound anti-CD3 antibody (OKT3; 1 μg/mL). CD4+ and CD8+ T cells were obtained from PBLs by negative selection using human CD4 or CD8 Cell Recovery Column Kits (Cedarlane). The purity of human CD4+ and CD8+ T cells was ≥ 90% and ≥ 70%, respectively, as shown by flow cytometric analysis (FACSCanto; BD Biosciences). After 2 days, T-cell subsets were used for proliferation assays. Where indicated, T-cell subsets were stimulated with 2 μg/mL of plate-bound anti-CD3 antibody and 2 μg/mL of CD28 antibody (AB85986; Abcam) with or without pretreatment for 30 minutes with 10μM etoricoxib (Amirall Prodesfarma).
HUVECs were cultured as described previously.21 Where indicated, HUVECs were stimulated with VEGF (Peprotech; 50 ng/mL) with or without pretreatment for 30 minutes with etoricoxib.
Plasmids
pcDNA3.1-NFATc1α-Myc was generated by in-frame cloning PCR-amplified full-length NFATc1 cDNA from the pSH107c-NFATc1 plasmid (provided by Dr G. Crabtree, Stanford University, Stanford, CA) into the pcDNA3.1myc-His vector (Invitrogen). PCR primer sequences were as follows: forward (5′-3′): GGGGGATCCAGGATGCCAAGCACCAGCTTTCCA; reverse (5′-3′): GGGTCTCTAGACTGAAAAAGCACCCCACGCGCTC.
cDNA encoding human NFATc3 (transcript variant 1) was PCR amplified from pECE-NFATc3 (provided by Dr L. de Windt, Maastricht University, Maastricht, The Netherlands). The PCR product was in-frame cloned into pcDNA3.1 myc/His and sequence verified. To generate knock-down–resistant NFATc3 (c3R), nucleotide changes were made by oligonucleotide-directed mutagenesis using the QuickChange Site Directed Mutagenesis kit (Stratagene). Oligonucleotide sequences used for site-directed mutagenesis were as follows: forward (5′-3′): CATCGAATCA CTGGGAAGAC AGTgGCaACa GCtAGCCAAG AGATAATAAT TG; reverse (5′-3′): CAATTATTAT CTCTTGGCTa GCtGTtGCcA CTGTCTTCCC AGTGATTCGA TG.
Human COX2 cDNA was amplified by PCR and cloned into an EcoRI-NotI lentiviral (LV) vector (pHRsin-hCOX2). The following primers were used: COX2-BamHI forward (5′-3′): CGCGGATCCGCGGTGAGAACCGTTTAC; COX2 reverse (5′-3′): GCGAGGAAGCGGAA-GAGTCTAGAGTCGACC.
Knockdown (KD) sequences were designed using Oligoengine Workstation 2 software (www.oligoengine.com). Oligonucleotides were annealed and then cloned into a BglII-NotI–digested pSuperRetro plasmid (Oligoengine). DNA molecules containing the RNA pol III–dependent H1 promoter and the KD sequences were further cloned into an EcoRI-NotI LV-Ubq-GFP LV vector. All plasmids were sequence verified. The KD sequences used were as follows: KDSc (5′-3′): GATCCGGCAACAAGATGAAGAGCACCTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTGTTTAAACGC; KD-c1 (5′-3′): GATCCCCGCCAGTACCAGCGTTTCACTTCAAGAGAGTGCGCTGGTACTGGCTTTTTGGAAC; KD-c3 (5′-3′): GATCCCCCATTGACTGCGCGGGAATCTTCAAGAGAGATTCCCGCGCAGT0CAAT-GTTTTTGGAAC.
The COX2-luc plasmid contains the human COX2 promoter sequence (−1796; +104) and the IL2-luc plasmid contains the minimal promoter region (−326; +45) of the human IL2 gene.7
LV production, titration, and infection
Lentiviruses expressing KD sequences were obtained by transient calcium phosphate transfection of HEK-293 cells with a 3 plasmid HIV-derived and VSV pseudotyped LV system kindly provided by M.K. Collins (University College London, United Kingdom). The supernatant containing the LV particles was collected 48 hours after removal of the calcium phosphate precipitate, and was then filtered through a 45-μM PVDF membrane (Steriflip; Millipore) and ultracentrifuged for 2 hours at 121 986g at 4°C (Ultraclear Tubes, SW28 rotor and Optima L-100 XP Ultracentrifuge; Beckman). Viruses were collected by adding cold sterile DMEM and titrated by quantitative PCR (qPCR), as described by Scherr et al.25
Jurkat cells were infected by adding virus (multiplicity of infectious units [MOI] = 5) and incubating for 5 hours at 37°C. HUVECs were infected by incubating in the presence of LV particles (MOI = 6) for 5 hours at 37°C in complete medium 199 containing 10% FBS and antibiotics.
Blast cells were infected 10 days after PHA stimulation by adding LV particles (MOI = 10) and incubating for 48 hours. Subsequently, virus-containing medium was replaced with fresh RPMI medium containing 10% FBS and l-glutamine plus antibiotics. Infection efficiency (green fluorescent protein [GFP] expression) and cell death (propidium iodide staining) were monitored by flow cytometry. At this time, blast cells were ≥ 98% positive for CD3 expression as shown by flow cytometric analysis. After infection, CD4+ T-cell blasts were purified by positive selection using magnetic microbeads (Flow Comp Human CD4 Dynabeads; Invitrogen). Purity of the CD4+ T-cell blasts was ≥ 98%. To purify CD8+ T-cell blasts, unbound cells were reincubated with anti-CD4–loaded magnetic beads. After 30 minutes at 4°C, unattached T-cell blasts were recovered and checked for CD8 expression by flow cytometry (CD8+ ≥ 90%).
Luciferase assay
For the transcriptional regulation experiments, cells were transfected with 0.5 μg of either COX-luc or the IL2-luc construct plus 0.5 μg of pECFP-C1 expression plasmid (Clontech) to monitor the transfection efficiency by flow cytometry. To determine the impact of NFAT overexpression on COX2 promoter activity, Jurkat cells (2 × 105) were transfected with each pcDNA3.1 construct (250 ng) together with the COX-luc reporter plasmid (250 ng). All cells were transfected using Lipofectamine-PLUS (Invitrogen). After transfection, cells were stimulated with PMA + Io for 5 hours and lysed with lysis buffer (Promega). Luciferase activity was determined with the Luciferase Assay System kit (Promega) and a Syrius luminometer (Berthold Technologies).
Cytokine production
Jurkat cells (106/mL) were stimulated with PMA + Io for 8 hours, and IL2 accumulated in the supernatant was measured with an ELISA kit (Diaclone).
CD4+ and CD8+ T cells were stimulated with plate-bound anti-CD3 (2 μg/mL) and CD28 (2 μg/mL) for different times and pretreated with etoricoxib (10μM) for 30 minutes where indicated. Cytokines released in the supernatant were measured with the Human Th1/Th2 11plex Kit FlowCytomix (eBioscience).
Proliferation assays
Proliferation of infected T-cell blasts and CD4+ and CD8+ T cells was analyzed with the CellTrace Violet Cell Proliferation Kit (Invitrogen). Infected blasts or noninfected primary CD4+ and CD8+ cells were labeled with CellTrace Violet and stimulated with plate-bound anti-CD3 (1-2 μg/mL) and CD28 (1-2 μg/mL) for 4 and 6 days. CellTrace Violet staining was determined by flow cytometry in viable cells at days 0, 1, 4, and 6 of stimulation. Data were analyzed with FlowJo 7.6 software (TreeStar).
Infected HUVECs were seeded at 20% confluence and serum-starved (0.5% FCS) overnight. Cells were then treated with 50 ng/mL of VEGF for 48 hours in the presence of 1 μCi/mL of 3H-thymidine. Cells were washed with cold PBS, and 3H-thymidine-labeled DNA was extracted through sequential incubations with 5% trichloroacetic acid and 0.5N NaOH. Incorporated 3H-thymidine was counted in a β-scintillation counter.
PGE2 assay
HUVECs (40 000 cells/well) cultured in gelatin-coated 24-well plates were serum starved for 16 hours. Where indicated, cells were pretreated for 30 minutes with etoricoxib (10μM) and stimulated for 8 hours with VEGF (50 ng/mL). Cell supernatants were collected and PGE2 was determined with an EIA kit (Cayman).
Migration assay
Infected HUVECs were plated at 90% confluence in p35 dishes. After 24 hours, monolayers were scratched with a p200 pipette tip. Cells were then washed and stimulated with VEGF (100 ng/mL) in medium containing 1% serum and no growth factors. Where indicated, infected cells were pretreated with etoricoxib for 30 minutes. Cells were placed in the incubator and wound healing was monitored by time-lapse microscopy with a Ti-Eclipse inverted microscope (Nikon) fitted with a 4× objective lens. Images were acquired with Nikon Nd2 Viewer and NIS Elements AR 3.0, and the cell-free area was calculated with Image J software.
In vivo Matrigel plug assay
Infected HUVECs were mixed with growth-factor reduced Matrigel (BD Bioscience) containing VEGF (500 ng/mL) and heparin (376 μg/mL). The HUVEC-loaded Matrigel (300 μL per mouse) was implanted subcutaneously in 4-week old female nude mice (Harlan). After 9 days mice were killed and matrigel plugs collected and split into 2 portions. One portion was weighed and disaggregated in water (300 μL). After centrifugation, hemoglobin content was determined by mixing the supernatant 1:1 with 3,3′, 5,5′-tetramethylbenzidine (TMB) liquid substrate system (Sigma-Aldrich) and measuring the absorbance at 605 nm. The other portion was paraffin-embedded, and sections stained with either hematoxylin-eosin or anti-GFP antibody (Invitrogen, A11122). The number of vessel-like structures per 20× field was quantified. Some plugs were fixed in 10% formalin and whole-mount stained with anti-GFP antibody. Three-dimensional images were acquired with a Bioptonics 3001 OPT scanner and analyzed with Skyscan 3001 software. All animal experiments were supervised and approved by the Bioethic Committee of Centro Nacional de Investigaciones Cardiovasculares.
Statistical analysis
All data are means ± SD of at least 3 independent experiments. Data were analyzed by 1- or 2-way ANOVA and Bonferroni post test except for hemoglobin detection experiments, where data were analyzed by t test. Differences were considered significant at P < .05.
Results
NFATc1 and NFATc3 differentially regulate expression of the IL2 and COX2 genes during T-cell activation
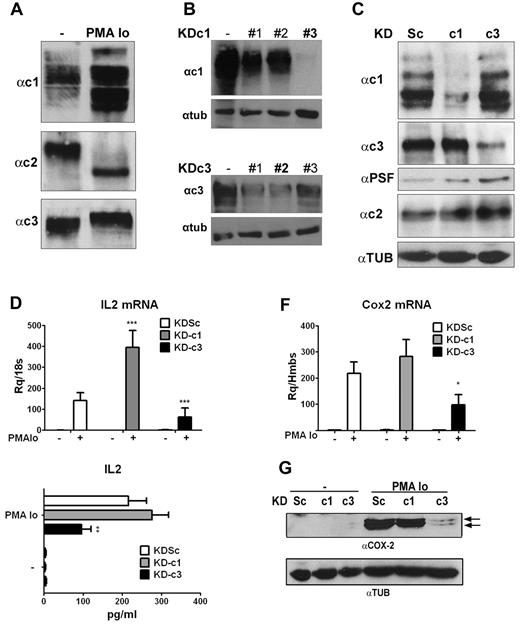
To determine whether individual NFAT members play nonoverlapping roles during T-cell activation, we designed shRNAs to individually knock down each NFAT member expressed in T lymphocytes. These were initially tested in the Jurkat T-cell line, and we first confirmed the expression of NFATc1, NFATc2, and NFATc3 in resting and activated cells8 (Figure 1A).
Knockdown of NFATc1 and NFATc3 has differential effects on induced expression of IL2 and COX2 in Jurkat cells. (A) Representative Western blot showing expression of NFATc1, NFATc2, and NFATc3 in total extracts of resting Jurkat cells or cells activated with PMA + Io. (B) HEK cells were transfected with pcDNA3.1 NFATc1 or c3 alone (−) or together with pSupeRetro shRNA sequences targeting NFATc1 (KDc1) and NFATc3 (KDc3). KDc1 shRNA #3 and KDc3 shRNA #2 were selected. (C) Jurkat cells were infected with the indicated knock-down LV vectors: Sc (control), c1 (NFATc1), and c3 (NFATc3). Expression of NFATc1, NFATc2, and NFATc3 protein was detected in lentivirus-infected Jurkat cells stimulated with PMA + Io for 5 hours. PTB-associated splicing Factor expression was monitored as a loading control for NFATc1 and NFATc3; tubulin expression was monitored as a loading control for NFATc2. (D) IL2 mRNA expression determined by qPCR after 2 hours of stimulation. (E) IL2 release measured by ELISA after 8 hours of stimulation. (F) COX2 mRNA levels determined by qPCR after 2 hours of stimulation. (G) Representative Western blot showing COX2 protein expression after 24 hours of stimulation. Tubulin (TUB) was used as a loading control. *P < .05, **P < .01, and ***P < .001 compared with control (KDSc) (n = 3).
Knockdown of NFATc1 and NFATc3 has differential effects on induced expression of IL2 and COX2 in Jurkat cells. (A) Representative Western blot showing expression of NFATc1, NFATc2, and NFATc3 in total extracts of resting Jurkat cells or cells activated with PMA + Io. (B) HEK cells were transfected with pcDNA3.1 NFATc1 or c3 alone (−) or together with pSupeRetro shRNA sequences targeting NFATc1 (KDc1) and NFATc3 (KDc3). KDc1 shRNA #3 and KDc3 shRNA #2 were selected. (C) Jurkat cells were infected with the indicated knock-down LV vectors: Sc (control), c1 (NFATc1), and c3 (NFATc3). Expression of NFATc1, NFATc2, and NFATc3 protein was detected in lentivirus-infected Jurkat cells stimulated with PMA + Io for 5 hours. PTB-associated splicing Factor expression was monitored as a loading control for NFATc1 and NFATc3; tubulin expression was monitored as a loading control for NFATc2. (D) IL2 mRNA expression determined by qPCR after 2 hours of stimulation. (E) IL2 release measured by ELISA after 8 hours of stimulation. (F) COX2 mRNA levels determined by qPCR after 2 hours of stimulation. (G) Representative Western blot showing COX2 protein expression after 24 hours of stimulation. Tubulin (TUB) was used as a loading control. *P < .05, **P < .01, and ***P < .001 compared with control (KDSc) (n = 3).
Because NFATc2 is essential for the inducible expression of NFATc1,26 its knock-down also interfered with the basal and inducible expression of NFATc1, making it impossible to assign observed effects to the lack of NFATc2. We therefore limited our analysis to NFATc1 and NFATc3. We designed a series of shRNA sequences directed at NFATc1 (KD-c1) and NFATc3 (KD-c3), as well as control shRNA sequences (scrambled; KDSc), and tested them for knock-down efficiency (Figure 1B). The selected shRNAs (shRNA #3 for NFATc1 and shRNA #2 for NFATc3) were cloned in LV vectors together with a GFP reporter transgene. Infection of Jurkat T cells was highly efficient (> 95% GFP+ cells, supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and confirmed the ability of the selected sequences to selectively and efficiently knock down NFATc1 or NFATc3 without affecting NFATc2 expression (Figure 1C and supplemental Figure 1B).
To determine the role of NFATc1 and NFATc3 in the regulation of genes involved in T-cell activation, infected Jurkat cells were activated with PMA + Io, a pharmacologic combination that mimics signaling induced by TCR and CD28 engagement, and mRNA expression was analyzed by qPCR on a low-density array. Knock-down of NFATc3 (KD-c3) specifically down-regulated mRNA expression of several genes involved in the immune response, including IL2 and COX2. Silencing of NFATc1 led to the transcriptional up-regulation of several genes, including IL2 and IL3 (Table 1). Individual qPCR analysis confirmed that knock-down of NFATc3 and NFATc1 had the opposite effects on the PMA + Io–induced expression of IL2 mRNA, which was paralleled by corresponding changes in IL2 content in cell supernatants (Figure 1D-E). As with IL2, analysis of COX2 expression in shRNA-transduced Jurkat cells confirmed the array data, showing that silencing of NFATc3 (but not NFATc1) inhibited PMA + Io–induced expression of COX2 mRNA (Figure 1F) and protein (Figure 1G). This result strongly suggests a selective regulation of COX2 gene transcription by NFATc3.
Gene expression after knockdown of NFATc1 or NFATc3
| Gene target . | Symbol . | KD-c1 . | KD-c3 . |
|---|---|---|---|
| Interleukin 2 | IL2 | 3.42 ± 0.15 | 0.45 ± 0.03 |
| Interleukin 3 | IL3 | 3.55 ± 0.33 | 0.98 ± 0.20 |
| Cyclooxygenase 2 | COX2 | 1.75 ± 0.34 | 0.29 ± 0.06 |
| Perforin 1 | PRF1 | 0.84 ± 0.08 | 0.74 ± 0.02 |
| TNF receptor member 6 | FAS | 1.22 ± 0.03 | 0.78 ± 0.18 |
| FAS ligand | FASLG | 1.89 ± 0.00 | 0.47 ± 0.09 |
| Transferrin receptor* | TFRC | 1.10 ± 0.00 | 1.05 ± 0.30 |
| Beta-glucuronidase* | GUSB | 1.07 ± 0.01 | 0.82 ± 0.09 |
| Gene target . | Symbol . | KD-c1 . | KD-c3 . |
|---|---|---|---|
| Interleukin 2 | IL2 | 3.42 ± 0.15 | 0.45 ± 0.03 |
| Interleukin 3 | IL3 | 3.55 ± 0.33 | 0.98 ± 0.20 |
| Cyclooxygenase 2 | COX2 | 1.75 ± 0.34 | 0.29 ± 0.06 |
| Perforin 1 | PRF1 | 0.84 ± 0.08 | 0.74 ± 0.02 |
| TNF receptor member 6 | FAS | 1.22 ± 0.03 | 0.78 ± 0.18 |
| FAS ligand | FASLG | 1.89 ± 0.00 | 0.47 ± 0.09 |
| Transferrin receptor* | TFRC | 1.10 ± 0.00 | 1.05 ± 0.30 |
| Beta-glucuronidase* | GUSB | 1.07 ± 0.01 | 0.82 ± 0.09 |
Jurkat cells transformed with lentiviral vectors encoding shRNAs for NFATc1 (KD-c1) or NFATc3 (KD-c3) were stimulated with PMA + Io for 4 hours. Total RNA was extracted and analyzed by gene expression array. Values correspond to the -fold induction (mean ± SD) relative to the expression in control cells transformed with scrambled shRNA (KDSc).
Endogenous control.
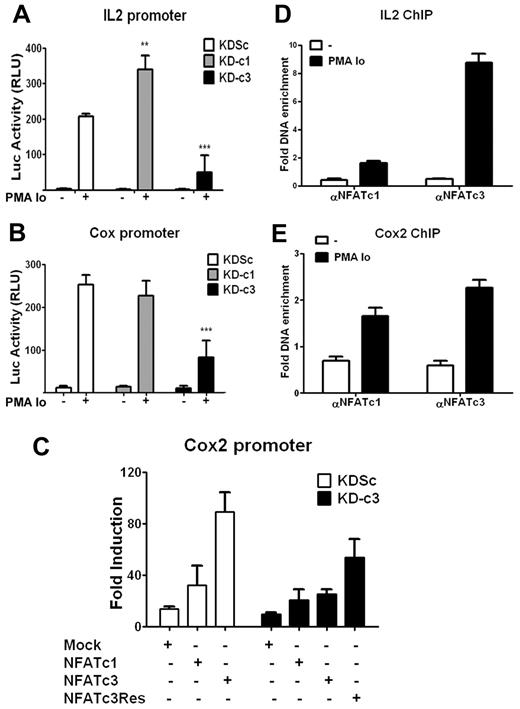
Because the IL2 and COX2 gene promoter regions both contain NFAT-binding sites, we investigated whether the specificity of NFATc1 and NFATc3 knock-down effects reflected differences in promoter activity. KDc1 or KDc3 Jurkat cells were transiently transfected with luciferase reporter constructs under the control of either the IL2 or COX2 promoter and stimulated with PMA + Io. Consistent with the effect on endogenous mRNA and protein expression, knock-down of NFATc3 strongly inhibited the activation of the IL2 and COX2 promoters, whereas knock-down of NFATc1 increased the transcriptional activity of the IL2 promoter but had no significant effect on inducible COX2 promoter activity (Figure 2A-B).
NFATc3 up-regulates the transcriptional activity of IL2 and COX2 promoters. (A-B) Jurkat cells were infected with the indicated knock-down LV vectors, and 48 hours later were transiently transfected with a luciferase reporter construct containing the IL2 (A) or the COX2 (B) promoter. After 16 hours, cells were stimulated for 5 hours with PMA + Io and luciferase activity was measured. Luciferase activity is shown in relative light units. Data are means ± SD (n ≥ 3); **P < .01 and ***P < .001 compared with control (KDSc). (C) Jurkat cells expressing KDSc or KD-c3 were transfected with the COX2-luc reporter construct plus an expression plasmid encoding wild-type NFATc1 (c1), wild-type NFATc3 (c3), or knock-down–resistant NFATc3 (c3Res). After 16 hours, cells were stimulated with PMA + Io for 5 hours and luciferase activity was measured. Luciferase activity is shown as the -fold induction relative to nonstimulated cells. One representative experiment of 4 is shown. (D-E) ChIP assays of NFATc1 and NFATc3 in Jurkat cells stimulated for 30 minutes with PMA + Io; histograms show the increased binding of both transcription factors to the endogenous promoters of IL2 (D) and COX2 (E). Data represent the amount of chromatin precipitated with αNFATc1 or αNFATc3 antibody compared with the input and are normalized to values obtained with IgG control antibodies. The results shown are representative of at least 3 experiments.
NFATc3 up-regulates the transcriptional activity of IL2 and COX2 promoters. (A-B) Jurkat cells were infected with the indicated knock-down LV vectors, and 48 hours later were transiently transfected with a luciferase reporter construct containing the IL2 (A) or the COX2 (B) promoter. After 16 hours, cells were stimulated for 5 hours with PMA + Io and luciferase activity was measured. Luciferase activity is shown in relative light units. Data are means ± SD (n ≥ 3); **P < .01 and ***P < .001 compared with control (KDSc). (C) Jurkat cells expressing KDSc or KD-c3 were transfected with the COX2-luc reporter construct plus an expression plasmid encoding wild-type NFATc1 (c1), wild-type NFATc3 (c3), or knock-down–resistant NFATc3 (c3Res). After 16 hours, cells were stimulated with PMA + Io for 5 hours and luciferase activity was measured. Luciferase activity is shown as the -fold induction relative to nonstimulated cells. One representative experiment of 4 is shown. (D-E) ChIP assays of NFATc1 and NFATc3 in Jurkat cells stimulated for 30 minutes with PMA + Io; histograms show the increased binding of both transcription factors to the endogenous promoters of IL2 (D) and COX2 (E). Data represent the amount of chromatin precipitated with αNFATc1 or αNFATc3 antibody compared with the input and are normalized to values obtained with IgG control antibodies. The results shown are representative of at least 3 experiments.
To corroborate NFATc3-dependent regulation of COX2, Jurkat cells infected with KD-c3 or KDSc were cotransfected with the COX2-Luc promoter reporter construct together with a plasmid expressing either wild-type NFATc1 (c1), NFATc3 (c3), or a knock-down–resistant form of NFATc3 (c3R) (supplemental Figure 1C). In agreement with the earlier results, overexpression of NFATc3 in cells expressing KDSc strongly increased PMA + Io–stimulated COX2 promoter activity, whereas overexpression of NFATc1 had a weaker effect. The NFATc3-specific increase was blocked in cells expressing KD-c3, and this blockade was partially reversed in cells overexpressing knock-down–resistant NFATc3 (c3R) (Figure 2C). In stimulated Jurkat cells, chromatin immunoprecipitation (ChIP) coprecipitated IL2 and COX2 promoter sequences with both NFATc1 and NFATc3, confirming physical binding of NFATc1 and NFATc3 to the endogenous COX2 and IL2 promoters (Figure 2D-E).
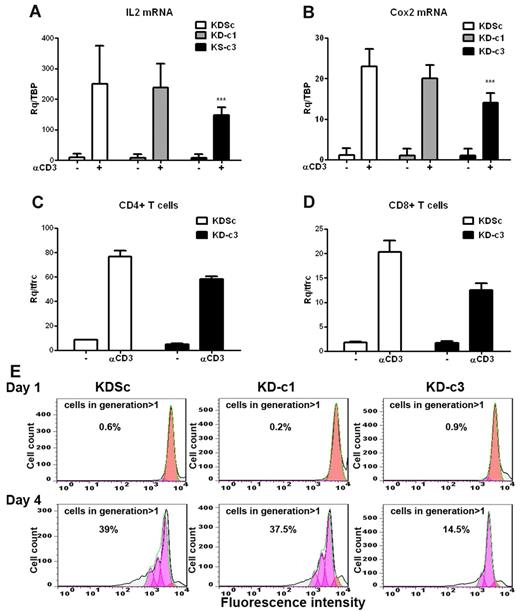
We next investigated whether the differential actions of NFATc1 and NFATc3 on IL2 and COX2 gene expression in Jurkat cells also operate in primary T-cell cultures. Given the low efficiency of LV infection in quiescent primary T lymphocytes, we first generated T-cell blasts by activating human PBLs with PHA and IL2. Infection of T-cell blasts during the mitogenic phase yielded an infection rate of 60% GFP+ cells, enabling us to obtain significant knock-down of NFATc1 or NFATc3 (supplemental Figure 2A). Knock-down of NFATc3 in these cells readily diminished the expression of anti-CD3–induced COX2 and IL2 mRNAs (Figure 3A-B). However, unlike Jurkat cells, NFATc1 silencing alone had no significant effect on IL2 mRNA expression. Unexpectedly, NFATc3 silencing in T-cell blasts impaired the activation-induced increase in NFATc1 expression (supplemental Figure 2A). To assess whether NFATc3 regulates COX2 transcription in mature T cells, we analyzed the response to CD3 engagement of NFATc3-silenced CD4+ and CD8+ populations. Induced COX2 mRNA expression was lower in both T-cell subsets (Figure 3C-D).
COX2 expression and cell proliferation are NFATc3 dependent in primary T blasts. Primary T cells were stimulated with PHA (5 μg/mL) and maintained in medium containing IL2 (50 U/mL) for 10 days. The cells were then infected with KDSc, KD-c1, or KD-c3 LV vectors as indicated. (A) shRNA-infected T cells were stimulated by plating onto anti-CD3–coated wells (1μg/mL) for 2 hours, and the expression of COX2 (A) (n = 7) and IL2 (B) (n = 8) mRNA was determined by qPCR. ***P < .001. CD4+ (C) and CD8+ (D) blasts were isolated from the infected T-cell blast population and stimulated with plate-bound anti-CD3 for 2 hours. COX2 mRNA expression was analyzed by qPCR. Data are from 1 representative experiment of 3 performed. (E) Infected T-cell blasts were labeled with CellTrace Violet, plated onto wells coated with anti-CD3 plus anti-CD28 (1 μg/mL of each), and proliferation was analyzed by flow cytometry 1 and 4 days after treatment. The proliferating cells lose fluorescent compound in each division. Numbers indicate the per-centage of proliferating cells (generation > 1) after 1 or 4 days. A representative experiment is shown.
COX2 expression and cell proliferation are NFATc3 dependent in primary T blasts. Primary T cells were stimulated with PHA (5 μg/mL) and maintained in medium containing IL2 (50 U/mL) for 10 days. The cells were then infected with KDSc, KD-c1, or KD-c3 LV vectors as indicated. (A) shRNA-infected T cells were stimulated by plating onto anti-CD3–coated wells (1μg/mL) for 2 hours, and the expression of COX2 (A) (n = 7) and IL2 (B) (n = 8) mRNA was determined by qPCR. ***P < .001. CD4+ (C) and CD8+ (D) blasts were isolated from the infected T-cell blast population and stimulated with plate-bound anti-CD3 for 2 hours. COX2 mRNA expression was analyzed by qPCR. Data are from 1 representative experiment of 3 performed. (E) Infected T-cell blasts were labeled with CellTrace Violet, plated onto wells coated with anti-CD3 plus anti-CD28 (1 μg/mL of each), and proliferation was analyzed by flow cytometry 1 and 4 days after treatment. The proliferating cells lose fluorescent compound in each division. Numbers indicate the per-centage of proliferating cells (generation > 1) after 1 or 4 days. A representative experiment is shown.
The Cox2-specific inhibitor NS-398 negatively regulates T-cell proliferation and production of IL2, TNFα, and IFNγ.24 We therefore studied the effect of NFATc3 knock-down on T-cell proliferation induced by CD3 + CD28 stimulation. We found that the absence of NFATc3 severely affected primary T-cell proliferation (Figure 3E). Consistently, etoricoxib, a Cox2-specific inhibitor, significantly decreased the proliferation rate of CD3 + CD28–stimulated primary CD4+ and CD8+ subpopulations (supplemental Figure 2B). Furthermore, etoricoxib severely reduced the production of cytokines secreted after CD3 + CD28 treatment in both lymphocyte subsets, as IL2, IFN-γ, IL5, and TNFα (supplemental Figure 2C).
These results show that NFATc1 and NFATc3 have opposing actions on the regulation of IL2 and COX2 gene expression and, most importantly, indicate that COX2 is specifically regulated by NFATc3 during T-cell activation. Moreover, in Jurkat cells, these selective NFAT effects appear to be regulated at the promoter level.
NFATc3 regulates Cox2 gene expression in endothelial cells
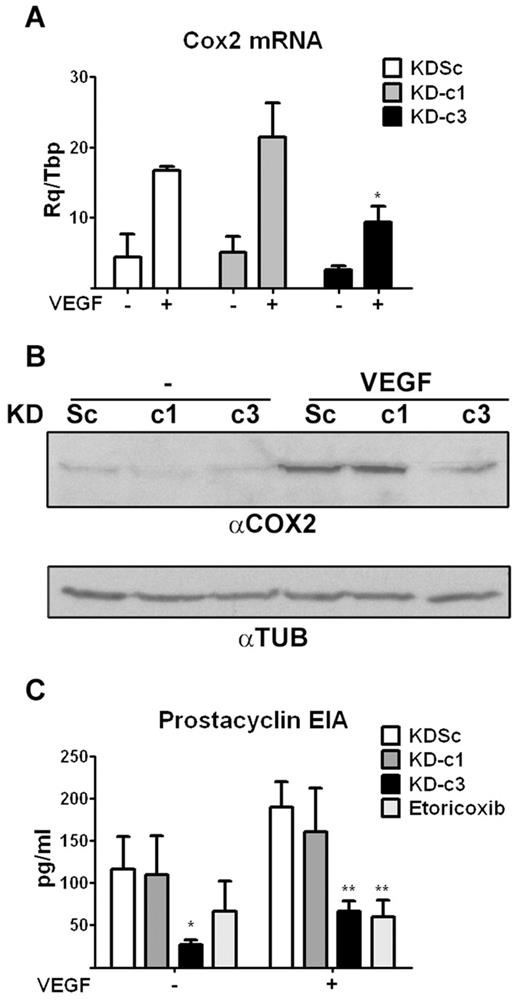
We next analyzed whether NFATc3-mediated regulation of COX2 expression occurs in cell types other than T cells. Because COX2 also plays an important role in the VEGF-mediated activation of NFAT in endothelial cells,21 we addressed this issue in primary HUVECs. Expression of NFATc1 and NFATc3 in these cells was efficiently and specifically knocked down by infection with KD-c1 and KD-c3 LV shRNA vectors (supplemental Figure 3). However, only NFATc3 knock-down inhibited the VEGF-induced up-regulation of COX2 RNA and protein expression (Figure 4A-B). Consistently, NFATc3 knock-down also inhibited COX2-mediated synthesis of prostacyclin in VEGF-stimulated cells, to a similar extent as the COX2-specific inhibitor etoricoxib (Figure 4C). NFATc3 knock-down also decreased prostacyclin content in unstimulated cells, probably reflecting inhibition of basal COX2 expression in cultured endothelial cells.27 Consistent with the results in lymphocytes, these findings indicate that NFATc3 is essential for the up-regulation of COX2 expression by VEGF in endothelial cells.
NFATc3-dependent COX2 expression and activity in endothelial cells. (A) shRNA-transformed HUVECs were treated with VEGF (50 ng/mL) for 5 hours and the expression of COX2 mRNA analyzed by qPCR. *P < .05 compared with control (KDSc) (n = 3). (B) Representative Western blot of COX2 protein expression in VEGF-stimulated shRNA-transformed HUVECs. Tubulin (TUB) expression was analyzed as a loading control. (C) EIA detection of prostacyclin release, an index of COX2 activity, in VEGF-stimulated shRNA-transformed HUVECs. Treatment of KDSc-expressing cells with the COX2-specific inhibitor etoricoxib (10μM) was used as a positive control of inhibition. *P < .05 and **P < .01 compared with control (KDSc) (n = 3).
NFATc3-dependent COX2 expression and activity in endothelial cells. (A) shRNA-transformed HUVECs were treated with VEGF (50 ng/mL) for 5 hours and the expression of COX2 mRNA analyzed by qPCR. *P < .05 compared with control (KDSc) (n = 3). (B) Representative Western blot of COX2 protein expression in VEGF-stimulated shRNA-transformed HUVECs. Tubulin (TUB) expression was analyzed as a loading control. (C) EIA detection of prostacyclin release, an index of COX2 activity, in VEGF-stimulated shRNA-transformed HUVECs. Treatment of KDSc-expressing cells with the COX2-specific inhibitor etoricoxib (10μM) was used as a positive control of inhibition. *P < .05 and **P < .01 compared with control (KDSc) (n = 3).
NFATc3 is required for endothelial cell migration, proliferation, and angiogenesis in vivo
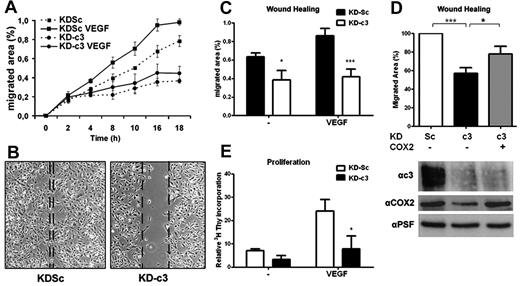
Because prostanoids regulate endothelial cell migration and angiogenesis,28,29 we investigated whether the inhibition of COX2-dependent prostanoid synthesis in NFATc3-silenced HUVECs affects cell migration in wound-healing experiments. NFATc3 knock-down severely impaired endothelial cell migration both in the absence and the presence of VEGF (Figure 5A-C). The inhibitory effect of NFATc3 silencing was similar to that of etoricoxib (supplemental Figure 4A), suggesting a role for COX2 expression and activity in the action of NFATc3 during endothelial cell migration. LV-mediated overexpression of human COX2 in KDc3-HUVECs partially restored the capacity of these cells to migrate in response to VEGF (Figure 5D).
NFATc3 knockdown blocks COX2-dependent endothelial cell migration and proliferation. (A) Time course of HUVEC migration in wound-healing experiments. HUVEC monolayers in medium containing 1% serum were scratched, and VEGF (100 ng/mL) was added to stimulate endothelial cell migration. The migrated area was measured from captured images at the indicated times. The graphic shows 1 representative experiment out of 3 performed. (B) HUVECs expressing KDSc or KD-c3 were analyzed with an in vitro wound-healing assay. Shown is a photomicrograph of representative fields after 14 hours of stimulation. Dotted lines mark the limits of the unpopulated area. (C) Quantification of migrated area (the proportion of the denuded area repopulated by migrating cells) at 14 hours (mean ± SD; n = 3). (D) Wound-healing assay with VEGF-stimulated HUVECs expressing KDSc, KD-c3, or KD-c3 + COX2. Knockdown of NFATc3 and overexpression of human Cox2 were detected by Western blot (bottom). (E) Endothelial proliferation assay. HUVECs were incubated with VEGF (50 ng/mL) and 3H-thymidine (1μCi/mL) for 48 hours. Values of incorporated 3H-thymidine for each condition were normalized to the within-experiment mean. Data are the means ± SD of 3 independent experiments performed at least in triplicate. *P < .05, **P < .01, and ***P < .001 compared with control (KDSc).
NFATc3 knockdown blocks COX2-dependent endothelial cell migration and proliferation. (A) Time course of HUVEC migration in wound-healing experiments. HUVEC monolayers in medium containing 1% serum were scratched, and VEGF (100 ng/mL) was added to stimulate endothelial cell migration. The migrated area was measured from captured images at the indicated times. The graphic shows 1 representative experiment out of 3 performed. (B) HUVECs expressing KDSc or KD-c3 were analyzed with an in vitro wound-healing assay. Shown is a photomicrograph of representative fields after 14 hours of stimulation. Dotted lines mark the limits of the unpopulated area. (C) Quantification of migrated area (the proportion of the denuded area repopulated by migrating cells) at 14 hours (mean ± SD; n = 3). (D) Wound-healing assay with VEGF-stimulated HUVECs expressing KDSc, KD-c3, or KD-c3 + COX2. Knockdown of NFATc3 and overexpression of human Cox2 were detected by Western blot (bottom). (E) Endothelial proliferation assay. HUVECs were incubated with VEGF (50 ng/mL) and 3H-thymidine (1μCi/mL) for 48 hours. Values of incorporated 3H-thymidine for each condition were normalized to the within-experiment mean. Data are the means ± SD of 3 independent experiments performed at least in triplicate. *P < .05, **P < .01, and ***P < .001 compared with control (KDSc).
Because endothelial proliferation is an essential step in angiogenesis, and in view of the results obtained in primary T cells, we analyzed the impact of NFATc3 knockdown on VEGF-induced endothelial proliferation. HUVEC proliferation was markedly reduced in the absence of NFATc3 (Figure 5E). Unlike migration, proliferation was not rescued by overexpression of COX2 in KDc3-HUVECs (data not shown). These results suggest that NFATc3 regulates COX2-dependent and -independent pathways that contribute to VEGF-mediated endothelial migration and proliferation. Accordingly, analysis of a set of angiogenesis-associated genes showed that NFATc3 knock-down also inhibited expression of other NFAT-dependent genes such as RCAN1.420 and IL830 (supplemental Figure 4B).
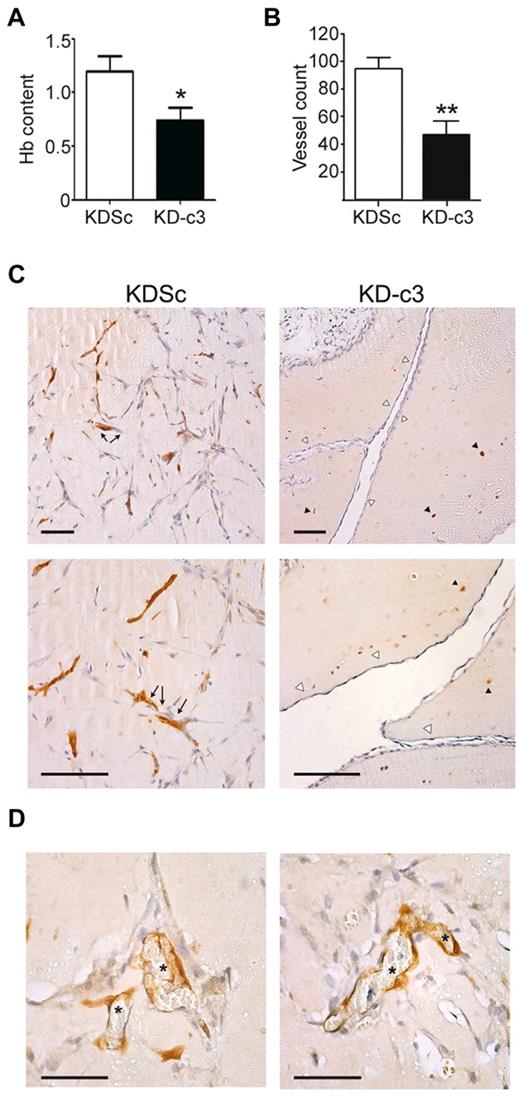
Given the importance of endothelial cell migration and proliferation in VEGF-mediated angiogenesis,31 and because of the lack of a significant effect of KD-c1 on HUVEC migration (supplemental Figure 4C), we focused on NFATc3 and explored its role in the VEGF-induced formation of new vessels. NFATc3-silenced HUVECs were embedded in Matrigel plugs and implanted subcutaneously in nude mice. These plugs produced fewer vessels and had lower hemoglobin content than control implants containing KDSc-expressing HUVECs, indicating an impaired angiogenic capability (Figure 6A-B). To determine the origin of the endothelial cells present in these vessels, we stained the plugs with anti-GFP antibody. HUVECs expressing KDSc formed vessel-like structures with an obvious lumen that contained blood cells. In contrast, KD-c3–expressing cells were unable to assemble into a vascular network, and vessels present in KD-c3 plugs originated mainly from the host (Figure 6C-D and supplemental Video 1).
NFATc3 knock-down blocks COX-2-dependent in vivo angiogenesis. Matrigel plugs embedded with VEGF (500 ng/mL) and KDSc and KD-c3 human endothelial cells (2.5 × 105/mouse) were implanted in nude mice and 10 days later were collected and analyzed. (A) Hemoglobin was measured spectrophotometrically at 605 nm. Graph shows means ± SD from 3 independent experiments. (B) Matrigel plugs containing KDSc or KD-c3 HUVECs were paraffin embedded and sectioned. Vessel count is the number of vessels counted in H&E-stained sections (means ± SD; n = 3). (C) Paraffin sections of Matrigel plugs containing either KDSc or KD-c3 HUVECs were stained with anti-GFP antibody. Left panels are micrographs showing vessel-like structures formed by KDSc-expressing HUVECs (arrows). Right panels are KD-c3-expressing HUVECs unable to form vascular structures (black arrowheads); instead, vessels present in these plugs were mainly composed of GFP− host cells (white arrowheads). Bars indicate 100μm. (D) Micrographs showing vessels formed by GFP+ endothelium (left) with an evident lumen containing blood cells (asterisks). Bars indicate 50μm. *P < .05 and **P < .01 compared with control (KDSc).
NFATc3 knock-down blocks COX-2-dependent in vivo angiogenesis. Matrigel plugs embedded with VEGF (500 ng/mL) and KDSc and KD-c3 human endothelial cells (2.5 × 105/mouse) were implanted in nude mice and 10 days later were collected and analyzed. (A) Hemoglobin was measured spectrophotometrically at 605 nm. Graph shows means ± SD from 3 independent experiments. (B) Matrigel plugs containing KDSc or KD-c3 HUVECs were paraffin embedded and sectioned. Vessel count is the number of vessels counted in H&E-stained sections (means ± SD; n = 3). (C) Paraffin sections of Matrigel plugs containing either KDSc or KD-c3 HUVECs were stained with anti-GFP antibody. Left panels are micrographs showing vessel-like structures formed by KDSc-expressing HUVECs (arrows). Right panels are KD-c3-expressing HUVECs unable to form vascular structures (black arrowheads); instead, vessels present in these plugs were mainly composed of GFP− host cells (white arrowheads). Bars indicate 100μm. (D) Micrographs showing vessels formed by GFP+ endothelium (left) with an evident lumen containing blood cells (asterisks). Bars indicate 50μm. *P < .05 and **P < .01 compared with control (KDSc).
Discussion
In the present study, we show that NFATc3 specifically regulates IL2 and COX2 gene expression in transformed and primary T cells. Similarly, NFATc3 controls COX2 expression in endothelial cells and is required for endothelial cell migration, proliferation, and angiogenesis in vivo.
The relatively mild impact of gene deletion of single NFAT members on the immune response1 has been interpreted as indicating functional redundancy among NFAT proteins. However, our knockdown analysis shows that NFATc1 and NFATc3 can have nonoverlapping functions and differentially regulate specific sets of genes involved in the activation of transformed and primary T cells. Furthermore, our results indicate that the silencing of NFATc1 isoforms may suppress the repressive functions previously described for some of them. Among NFATc1 isoforms,4 NFATc1/αA is strongly induced by TCR stimulation in a calcineurin-dependent manner and induces IL2 production when overexpressed in human T cells. In contrast, the 4 long NFATc1 isoforms suppress IL2 promoter activation by recruiting HDACs through their sumoylatable C-terminal region.32 The coexistence of differently regulated isoforms with opposite transcriptional effects complicates investigation of the role of NFATc1 in the regulation of a given gene, and our experimental approach did not allow us to dissect out the specific contribution of each NFATc1 isoform to IL2 and COX2 transcriptional regulation. We can therefore only conclude that silencing all NFATc1 isoforms has an overall activating outcome on IL2 transcription, and that they are not required for COX2 induction, even though some of them can interact with the endogenous COX2 promoter, as shown in ChIP experiments (Figure 2E). Given that NFATc1 knock-down had no effect on COX2 mRNA expression or promoter activity, we conclude that the binding of NFATc1 to this promoter in ChIP assays is an example of nonfunctional promoter binding, as has been reported for numerous transcription factors.33-36
We found that NFATc3 regulates COX2 and IL2 gene expression in both transformed Jurkat and primary human T cells. Furthermore, NFATc3 knock-down impairs the CD3-induced increase in NFATc1 expression in T-cell blasts. Although we cannot rule out a possible nonspecific effect, these data suggest that, as described previously for NFATc2,26 NFATc3 may regulate the inducible expression of NFATc1 in primary T cells and that NFATc3, either alone or in cooperation with NFATc1, is required for the regulation of IL2 and COX2.
The lack of an action of NFATc1 on COX2 mRNA expression highlights a selective role of NFATc3 in COX2 transcriptional regulation. NFATc3 is also essential for VEGF-induced migration, proliferation, and angiogenesis, processes known to be regulated at least in part by COX2-generated prostanoids.37 Moreover, the COX2 overexpression experiments (Figure 5D) indicate a role for COX2 in NFATc3-mediated endothelial cell migration. However, exogenous COX2 only partially rescued migratory capacity in NFATc3-silenced cells and had no effect on proliferation. These results suggest the existence of additional NFATc3 targets that could be involved in VEGF-induced endothelial migration and proliferation, and our analysis showed that NFATc3 knock-down impaired the up-regulation of IL8 and RCAN 1.4 in response to VEGF (supplemental Figure 4B). IL8 is a proangiogenic chemokine with a well-known chemotactic activity.38 The role of the calcineurin regulator RCAN1.4 in angiogenesis is controversial20,39 ; however, some reports provide evidence for a positive role of RCAN1.4 in VEGF-induced endothelial cell migration and in vitro tube formation.40,41 Therefore, in addition to COX2, IL8 and RCAN1.4 may contribute to NFATc3-mediated endothelial migration.
An involvement of NFATc3 in VEGF-induced angiogenesis is consistent with recent reports proposing a role for this transcription factor in angiogenesis promoted by other stimuli such as secreted frizzle-related protein 2 (SFRP2) and activated peroxisome proliferator–activated receptorγ (PPARγ).42,43 However, angiogenesis remains a highly complex, multistep program and therefore the participation of NFAT members other than NFATc3 in this process cannot be excluded.
Given the major role of VEGF in angiogenesis in mice and humans, it was surprising that Nfatc3 KO mice showed no alterations in physiologic or pathologic angiogenesis. Our results suggest that individual NFAT members play specific and selective roles in T-cell and endothelial cell activation. Selective and differential regulation of specific gene sets by different NFAT members is also supported by studies showing that ectopic expression of NFATc2, but not NFATc1, increases TNF-α synthesis in T cells,44 and by other studies showing that NFATc4 specifically controls MyHC-2B expression in skeletal muscle.45 However, this selectivity is not reflected in the phenotypes of KO mice deficient for individual NFAT members. We propose that under normal conditions, NFATs specifically and selectively regulate a given set of genes, and that the prolonged absence of one member triggers compensatory mechanisms involving other NFAT members coexpressed in the same cell. This issue might be clarified by future analysis with conditional KO mice, allowing inducible, tissue-specific, and short-duration deletion of individual NFAT members.
An important question arising from this study is what mechanisms might underlie selective NFAT member–dependent transcription and how they can be bypassed. NFAT members differ in their affinity for CN,46 which raises the possibility that differential phosphorylation of NFAT members in the cytosol might provide a means of controlling their relative concentrations in the nucleus, resulting in differences in the kinetics of target gene activation. NFATs also contain 2 transactivation domains located at the N-terminal and C-terminal regions. Sequence homology among these domains is very low,3,44 and it is possible that the transactivation domain of each member might confer interaction with specific transactivation partners. Likewise, the transcriptional activities of individual NFAT members might be affected by specific epigenetic modifications, such as those described recently for NFATc132 and NFATc2.47 NFAT selectivity might also be influenced by recently identified regulators of NFAT activation, such as NRON miRNA and the scaffold proteins HOMER2 and HOMER3, which regulate NFAT subcellular localization, and Caspase 3, which modulates the expression of specific NFAT members.48
Our results show that short-term suppression by mRNA knock-down reveals specific functions for individual NFAT members. NFATc3, which was previously thought not to play an important role in T-cell activation and inflammation, is specifically required for the transcriptional up-regulation of the important inflammatory factors IL2 and COX2. Moreover, NFATc3-dependent COX2 regulation plays an important role in endothelial cell migration and angiogenesis. These results strongly suggest that individual NFAT members have differential effects on the transcription of target genes that are likely masked by compensatory effects in KO studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Balbino Alarcon for critical reading of the manuscript; Dr S. Bartlett for English editing; B. Ornes and A. Guio for technical support; and the Centro Nacional de Investigaciones Cardiovasculares's Cellomics Unit for technical support with flow cytometry.
J.M.R. is supported by the Spanish Ministry of Science and Innovation (Ministerio de Ciencia e Innovación; SAF2009-10708), the Comunidad de Madrid (CAM-S2006/BIO-0194), Fundación La Marató TV3 (080731), Fundación Genoma España (GENOMA), the Spanish Ministry of Health (Ministerio de Sanidad y Consumo), and Red Temática de Investigación Cooperativa en Enfermedades Cardiovasculares (RECAVA) (RD06/0014/0005). A.R. is supported by the Spanish Ministry of Science and Innovation (SAF2009-10691) and the Comunidad de Madrid (S2006/BIO-0236). The Centro Nacional de Investigaciones Cardiovasculares is supported by the Spanish Ministry of Science and Innovation and the Pro-CNIC (Centro Nacional de Investigaciones Cardiovasculares) Foundation. K.U. holds a Fondo de Investigaciones Sanitarias fellowship.
Authorship
Contribution: K.U. performed and analyzed most experiments and generated the figures; S.M.-M. performed and interpreted the T-cell proliferation experiments with the help of A.E. (Figure 3C); A.E. carried out cytokine quantitation; A.A. performed and interpreted endothelial proliferation assays and in vivo Matrigel experiments (Figure 6); I.O. and A.R. designed siRNA sequences and made the in vitro selection (Figure 1B); and A.A., A.R., and J.M.R. supervised the experiments, interpreted and analyzed results, and with the assistance of K.U. and S.M.-M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for I.O. is Pitié-Salpêtrière Hospital, Paris, France.
Correspondence: Antonio Rodríguez, Department of Molecular Biology, Universidad Autónoma de Madrid, Campus universitario de Cantoblanco, E-28049 Madrid, Spain; e-mail: a.rodriguez@uam.es; or Juan Miguel Redondo, Department of Vascular Biology and Inflammation, Centro Nacional de Investigaciones Cardiovasculares, Melchor Fernández Almagro 3, E-28029 Madrid, Spain; e-mail: jmredondo@cnic.es.