Abstract
The biologic effects of IFNγ are mediated by the transcription factor STAT1. The activity of STAT1 is inhibited by small ubiquitin-like modifier (SUMO) conjugation. This occurs both directly through decreasing STAT1 tyrosine phosphorylation and indirectly by facilitating STAT1 dephosphorylation consequential to increased STAT1 solubility because of suppressed paracrystal assembly. However, the physiologic implications of SUMO conjugation have remained unclear. Here, we used fibroblasts and bone marrow–derived macrophages (BMMs) from knockin mice expressing SUMO-free STAT1 to explore the consequences of STAT1 sumoylation for IFNγ signaling. Our experiments demonstrated buffer property of paracrystals for activated STAT1, such that SUMO-mediated paracrystal dispersal profoundly reduced phosphorylation of STAT1, which affected both the activating tyrosine 701 and the transcription-enhancing serine 727. Accordingly, the curtailed STAT1 activity in the nucleus caused by SUMO conjugation resulted in diminished transcription of IFNγ-responsive genes; and increased the IFNγ concentration more than 100-fold required to trigger lipopolysaccharide-induced cytotoxicity in bone marrow–derived macrophages. These experiments identify SUMO conjugation of STAT1 as a mechanism to permanently attenuate the IFNγ sensitivity of cells, which prevents hyperresponsiveness to this cytokine and its potentially self-destructive consequences. This sets the mode of SUMO-mediated inhibition apart from the other negative STAT regulators known to date.
Introduction
IFNγ fulfills multiple roles in immunity by regulating gene expression.1 Its actions are largely dependent on STAT1 activation by phosphorylation of tyrosine 701 as demonstrated by impaired antimicrobial immunity of patients and model organisms with defective STAT1 activation.2-6 Several additional posttranslational modifications have been proposed to modulate the transcriptional activity of STAT1,7 one modification of which is phosphorylation of serine residue 727 in the transactivation domain,8,9 which is required for full-fledged IFNγ-dependent innate immunity.10 In contrast, whether posttranslational modifications of STAT1 contribute to its negative regulation is less clear. The proposed inhibition of IFNγ signaling by acetylation of STAT1 on lysines 410 and 413 has recently been falsified,11 and the physiologic significance of another covalent modification of STAT1, the conjugation of small ubiquitin-like modifier (SUMO), has not been fully explored.12-15 STAT1 harbors a functional sumoylation consensus sequence (ΨKxE; Ψ, large hydrophobic residue; x, any residue) with the SUMO acceptor Lys703 in position +2 relative to the Tyr701 phosphorylation site.12 The SUMO consensus is evolutionarily conserved in STAT1 but mutated in the other STAT family members.15 SUMO modification is a dynamic and reversible process with generally repressive effects on the transactivation of transcription factors, but exceptions have been reported previously.16 We and others have recently shown that phosphorylation of STAT1 at Tyr701 and sumoylation at the adjacent Lys703 are mutually exclusive, such that transcriptionally active, that is, Tyr701-phosphorylated STAT1, remains unsumoylated.17,18 This phenomenon constitutes 1 of 2 mechanisms by which sumoylation diminishes the pool of activated STAT1, that is, reducing Tyr701 phosphorylation at the cytokine receptors. It was further demonstrated that activated STATs can polymerize into dynamic paracrystalline arrays in the nucleus of cytokine-stimulated cells and that localization in paracrystals protects the activated STATs from phosphatase attack.18 However, STAT1's unique ability among the STATs to SUMO conjugate triggers dimers that are semiphosphorylated at Tyr701, which dimers function as competitive polymerization inhibitors and hence preclude paracrystal assembly. Thus, sumoylation, although affecting only a disproportionately small fraction of STAT1 molecules (∼ 2% at steady state), dramatically increases the solubility of the activated STAT1. This, in turn, constitutes the second SUMO-dependent mechanism to diminish the activity of STAT1, that is, increased dephosphorylation. Together, these 2 additive mechanisms profoundly curtail the presence of activated STAT1 in the cell nucleus.18 But, it was not resolved how these consequences of STAT1 SUMO conjugation affect IFNγ signaling. Here, we explore the impact for cellular phenotype, and we report that SUMO conjugation of STAT1 raises the threshold of IFNγ responsiveness essential to protect cells against hyperresponsiveness to this cytokine.
Methods
Animal experimentation and cell culture
Mice expressing SUMO-free STAT1 (Glu705Gln) were generated using knockin approach and maintained on a mixed 129/C57Bl/6 background (M.D., A.B., U.V., K.-P.K., and Ronald Naumann, manuscript in preparation). Embryonic fibroblasts were prepared from 13.5-day-old embryos by standard methods and were genotyped by restriction fragment length polymorphism analysis. Bone marrow from 17-week-old wild-type or homozygous mutant mice was cultured in L cell–conditioned medium to obtain macrophages,19 which were used for experiments between days 8 and 22. Culture of cell lines and transfections were described previously.20
NO assay and cell viability
NO production was assessed by nitrite determination using Griess assay; cellular ATP content was measured with CellTiter-Glo assay to score metabolic activity. Both assays were performed as described by the manufacturer (Promega).
End point PCR and real-time PCR
Total RNA extraction and end point RT-PCR were done as described previously.20 Real-time PCR was performed in a final volume of 25 μL containing 0.2 μg of each primer and 0.5 μg of template cDNA. Results for real-time PCR were obtained using the QantiTect SYBR Green PCR kit (QIAGEN) and the iCycler instrument (Bio-Rad Laboratories). The following real-time PCR protocol was used: 2-minute denaturation at 95°C, followed by 45 cycles of 95°C for 30 seconds, primer-specific annealing for 30 seconds and 72°C for 1 minute; the melting curve program was 55°C for 1 minute and 80 cycles of 55°C + 0.5°C/cycle (10 seconds). For each gene, the relative quantification of its expression in comparison with the reference gene (Gapdh) was determined 3 times as described previously.21 The primer sequences used in PCR assays are in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Quantitative immunoblotting
Whole cell extraction, 7% and 10% SDS-PAGE, quantitative immunoblotting, and membrane stripping were as described previously.18 Primary antibody decoration was detected with IRdye800-conjugated secondary immunoglobulin; signals were quantified using the Odyssey system (Li-Cor Biosciences). Nickel-nitrilotriacetic acid (Ni-NTA; QIAGEN) chromatography was done as described previously.18
Fluorescence microscopy
Microscopy was done using a TCP-SP2 confocal microscope (Leica) equipped with automated shutter and motorized X, Y, and Z stack controller together with a Q-Imaging charge-coupled device camera with 12-bit gray scale resolution.18 For immunofluorescence experiments, we used polyclonal anti-Tyr701–phosphorylated STAT1-specifc first antibody (Cell Signaling Technology), and Cy3-coupled anti–rabbit IgG secondary antibody (Stratech Scientific). Images were acquired with a 63 × /1.4 oil Ph3 CS objective (Leica) using immersion oil (Leica). Fluorescence signal intensities were obtained with LCS Lite 2.61 software (Leica).
Results and discussion
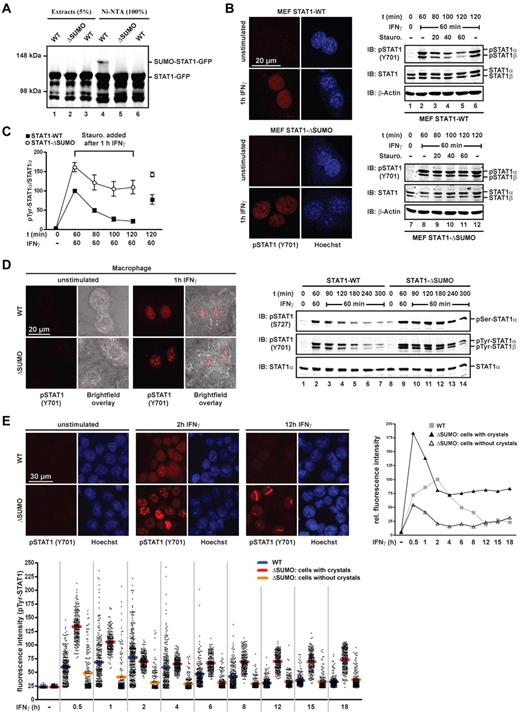
The structurally conservative SUMO consensus mutation Glu705Gln has minimal SUMO-independent effects on STAT1 activity,18 yet suffices to preclude SUMO conjugation (Figure 1A). To study the physiologic consequences, we therefore generated STAT1-Glu705Gln knockin mice that exclusively express STAT1 from the endogenous gene locus and that can no longer be SUMO conjugated. As shown in Figure 1B, primary fibroblasts (mouse embryonic fibroblasts) extracted from embryos expressing SUMO-free STAT1 did not present Tyr701-phosphorylated STAT1 irrespective of the genotype. On IFNγ stimulation, however, small STAT1 paracrystals containing Tyr701-phosphorylated STAT1 appeared in the nucleus of cells expressing SUMO-free STAT1, which were absent for activated wild-type STAT1. Comparison of Tyr701-phosphorylation revealed increased and prolonged activation of the mutant STAT1 (Figure 1B). In addition, mouse embryonic fibroblasts were treated with IFNγ for 60 minutes followed by a pulse chase with the tyrosine kinase inhibitor staurosporine to block continued Tyr701-phosphorylation.22 Immunoblotting revealed that increased activation of SUMO-free STAT1 resulted not solely from facilitated Tyr701-phosphorylation but also from markedly reduced dephosphorylation (Figure 1B-C). These results were confirmed and extended with primary BMMs, where SUMO-free STAT1 assembled very large paracrystals on IFNγ stimulation, whereas wild-type STAT1 remained soluble (Figure 1D). Paracrystal incorporation protects activated STAT1 molecules from phosphatase attack;18 however, thus far only Tyr701 modification has been considered. We therefore examined modification at the second confirmed STAT1 phosphorylation site, namely, Ser727 in the transactivation domain, which is required for increased transcription activation and innate immune responses.10 As shown in Figure 1D, phosphorylation of Ser727 like that of Tyr701 was markedly increased and thus persisted longer in the paracrystal-containing marrow-derived macrophages expressing SUMO-free STAT1. This finding agrees with a protective role of paracrystals for phosphorylated STAT1 and provides another mechanistic explanation for increased transcriptional activity of STAT1 in the nucleus.
SUMO-mediated paracrystal dispersal curtails STAT1 activity in the nucleus. (A) Green fluorescent protein (GFP)–tagged wild-type STAT1 or mutant Glu705Gln (ΔSUMO) was coexpressed with Ubc9 (all lanes) and His-tagged SUMO1 (lanes 1, 2, 4, and 5) in HEK293T cells. Whole cell extracts were prepared in buffer containing 120mM N-ethyl-maleimide (Sigma-Aldrich) and subjected to native affinity chromatography on Ni-NTA agarose (QIAGEN) to enrich His-tagged SUMO1 conjugates. Shown are the results of a representative immunoblot analysis of cell extracts and bound Ni-NTA chromatography fraction using anti STAT1-specific antibody (C24; Santa Cruz Biotechnology). (B) Left: Immunofluorescence confocal micrographs of unstimulated or 1-hour IFNγ-stimulated (50 U/mL mouse IFNγ; Calbiochem) mouse embryonic fibroblasts derived from SUMO-free STAT1-Glu705Gln (ΔSUMO) knockin mice or wild-type littermates using anti–Tyr701-phosphorylated STAT1 antibody. Nuclei were stained with Hoechst dye. Right: immunoblot analyses of corresponding whole cell extracts depicting STAT1 activation kinetics. Cells were left untreated or were treated with IFNγ for 60 minutes, after which time the medium was replaced by growth medium without or with 0.5μM tyrosine kinase inhibitor staurosporine. The cells were incubated for the indicated times before cell extraction and consecutive Western blotting on the same membrane using anti-Tyr701–phosphorylated STAT1-specific antibody (Cell Signaling Technology), anti-STAT1–specific antibody (E23; Santa Cruz Biotechnology) and then anti–β-actin specific antibody (Sigma-Aldrich). Note that antibody E23 recognizes both STAT1 splice variants, full-length STAT1α, and truncated STAT1β. (C) Diagram depicting specific Tyr701-phosphorylation of SUMO-free and wild-type STAT1 using fibroblast extracts as shown in panel B. IFNγ-stimulated wild type was set as 100. Data are presented as the mean ± SD of 3 independent immunoblot analyses for each STAT1 variant. (D) Left: Immunofluorescence confocal micrographs of unstimulated or 1-hour IFNγ-stimulated (50 U/mL mouse IFNγ) BMMs from SUMO-free STAT1 (ΔSUMO) knockin mice or wild-type littermates using anti Tyr701-phosphorylated STAT1-specific antibody. Cell dimensions are shown using bright-field microscopy. Right: Representative immunoblot analyses of corresponding whole cell extracts depicting STAT1 phosphorylation kinetics at residues Ser727 and Tyr701. Anti-Ser727–phosphorylated STAT1-specific antibody (44-382G; Invitrogen), anti-Tyr701–phosphorylated STAT1-specific antibody (Cell Signaling Technology), and then anti-STAT1–specific antibody (M23; Santa Cruz Biotechnology; note that this antibody recognizes full-length STAT1α splice variant only) were used. (E) Time course of soluble activated STAT1 in the nucleus of HEK293T cells expressing wild-type or SUMO-free STAT1, as determined by quantitative immunofluorescence confocal microscopy. Top left: Representative immunofluorescence micrographs of cells before and after treatment with 5 ng/mL human IFNγ (Calbiochem) using anti-Tyr701–phosphorylated STAT1-specific antibody. Bottom: Scatter plot depicting unprocessed fluorescence signal intensities recorded outside paracrystals and nucleoli in the nucleoplasm of 170-250 randomly selected cells per time point for each STAT1 variant. For SUMO-free STAT1, cells with paracrystals and without are grouped separately; horizontal bars indicate average fluorescence signal intensity. Top right: Graph depicting the time course of average fluorescence signal intensities. Data were background subtracted; in addition, to correct for the ∼ 15% of cells that were unresponsive to IFNγ (not applicable to dataset ΔSUMO with paracrystals), the bottom 15% intensities of each time point were excluded in this representation. Data are shown relative to the wild-type maximum (t = 2 h), which was set as 100.
SUMO-mediated paracrystal dispersal curtails STAT1 activity in the nucleus. (A) Green fluorescent protein (GFP)–tagged wild-type STAT1 or mutant Glu705Gln (ΔSUMO) was coexpressed with Ubc9 (all lanes) and His-tagged SUMO1 (lanes 1, 2, 4, and 5) in HEK293T cells. Whole cell extracts were prepared in buffer containing 120mM N-ethyl-maleimide (Sigma-Aldrich) and subjected to native affinity chromatography on Ni-NTA agarose (QIAGEN) to enrich His-tagged SUMO1 conjugates. Shown are the results of a representative immunoblot analysis of cell extracts and bound Ni-NTA chromatography fraction using anti STAT1-specific antibody (C24; Santa Cruz Biotechnology). (B) Left: Immunofluorescence confocal micrographs of unstimulated or 1-hour IFNγ-stimulated (50 U/mL mouse IFNγ; Calbiochem) mouse embryonic fibroblasts derived from SUMO-free STAT1-Glu705Gln (ΔSUMO) knockin mice or wild-type littermates using anti–Tyr701-phosphorylated STAT1 antibody. Nuclei were stained with Hoechst dye. Right: immunoblot analyses of corresponding whole cell extracts depicting STAT1 activation kinetics. Cells were left untreated or were treated with IFNγ for 60 minutes, after which time the medium was replaced by growth medium without or with 0.5μM tyrosine kinase inhibitor staurosporine. The cells were incubated for the indicated times before cell extraction and consecutive Western blotting on the same membrane using anti-Tyr701–phosphorylated STAT1-specific antibody (Cell Signaling Technology), anti-STAT1–specific antibody (E23; Santa Cruz Biotechnology) and then anti–β-actin specific antibody (Sigma-Aldrich). Note that antibody E23 recognizes both STAT1 splice variants, full-length STAT1α, and truncated STAT1β. (C) Diagram depicting specific Tyr701-phosphorylation of SUMO-free and wild-type STAT1 using fibroblast extracts as shown in panel B. IFNγ-stimulated wild type was set as 100. Data are presented as the mean ± SD of 3 independent immunoblot analyses for each STAT1 variant. (D) Left: Immunofluorescence confocal micrographs of unstimulated or 1-hour IFNγ-stimulated (50 U/mL mouse IFNγ) BMMs from SUMO-free STAT1 (ΔSUMO) knockin mice or wild-type littermates using anti Tyr701-phosphorylated STAT1-specific antibody. Cell dimensions are shown using bright-field microscopy. Right: Representative immunoblot analyses of corresponding whole cell extracts depicting STAT1 phosphorylation kinetics at residues Ser727 and Tyr701. Anti-Ser727–phosphorylated STAT1-specific antibody (44-382G; Invitrogen), anti-Tyr701–phosphorylated STAT1-specific antibody (Cell Signaling Technology), and then anti-STAT1–specific antibody (M23; Santa Cruz Biotechnology; note that this antibody recognizes full-length STAT1α splice variant only) were used. (E) Time course of soluble activated STAT1 in the nucleus of HEK293T cells expressing wild-type or SUMO-free STAT1, as determined by quantitative immunofluorescence confocal microscopy. Top left: Representative immunofluorescence micrographs of cells before and after treatment with 5 ng/mL human IFNγ (Calbiochem) using anti-Tyr701–phosphorylated STAT1-specific antibody. Bottom: Scatter plot depicting unprocessed fluorescence signal intensities recorded outside paracrystals and nucleoli in the nucleoplasm of 170-250 randomly selected cells per time point for each STAT1 variant. For SUMO-free STAT1, cells with paracrystals and without are grouped separately; horizontal bars indicate average fluorescence signal intensity. Top right: Graph depicting the time course of average fluorescence signal intensities. Data were background subtracted; in addition, to correct for the ∼ 15% of cells that were unresponsive to IFNγ (not applicable to dataset ΔSUMO with paracrystals), the bottom 15% intensities of each time point were excluded in this representation. Data are shown relative to the wild-type maximum (t = 2 h), which was set as 100.
These results confirmed earlier work demonstrating paracrystals to function as reservoirs for the activated STATs. In conjunction with the dynamic exchange of paracrystal-incorporated STAT1 with the diffusible pool in the nucleoplasm,18 paracrystals thus appeared to have buffer-property for activated STAT1 rather than being static depots. To directly test this possibility, we used quantitative confocal immunofluorescence microscopy to determine the impact of paracrystals on the IFN-induced concentration change of Tyr701-phosphorylated STAT1 in the soluble phase, that is, the nucleoplasm (Figure 1E). We show results obtained with transfected human embryonic kidney (HEK293T) cells, which express low endogenous STAT1, because the large-sized paracrystals found in the nucleus of macrophages minimized the remaining area suitable for optical analyses, which often precluded meaningful measurements of soluble STAT1. In the nucleus of HEK293T cells expressing wild-type STAT1, Tyr701-phosphorylation of soluble STAT1 peaked between 1 and 4 hours after the addition of IFNγ, whereas it reached approximately double that value already after 30 minutes in nuclei containing SUMO-free STAT1. After this time, which coincided with the appearance of paracrystals, the nuclear concentration of diffusible Tyr701-phosphorylated STAT1 fell sharply in these cells to below wild-type level within ∼ 90 minutes, resulting in a much narrower activation peak (Figure 1E). Importantly, over the following 16 hours, the concentration of diffusible activated STAT1 remained constant, at ∼ 70% of the wild-type maximum. Of note, this behavior required assembly of paracrystals, because in their absence SUMO-free STAT1 resembled the wild-type situation, in which the nuclear concentration of activated STAT1 dwindled to background levels within < 10 hours (Figure 1D). These data demonstrated a buffer property of paracrystals that profoundly altered the kinetics of STAT1 activation in the nucleus.
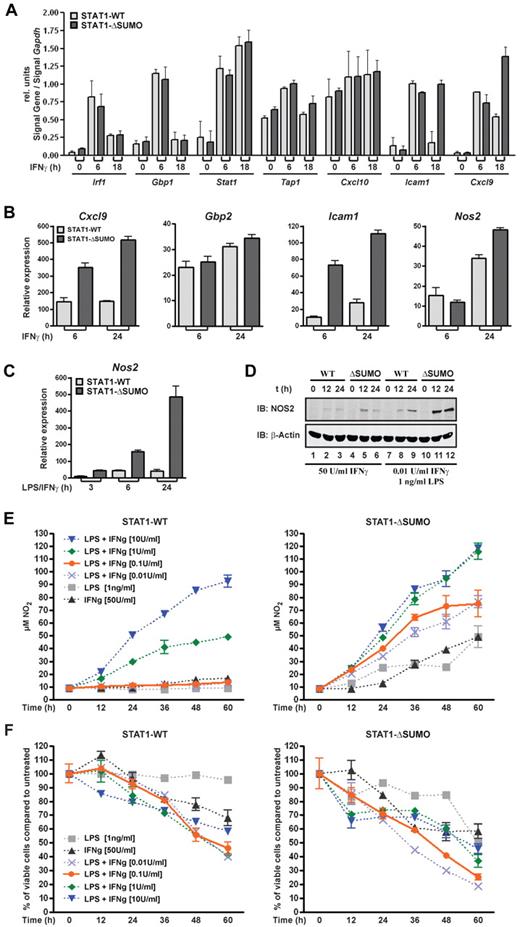
Next, we examined the consequences of prolonged STAT1 activity for the expression of IFNγ-regulated genes. Transcription kinetics was determined with embryonic fibroblasts from wild-type and mutant STAT1 knockin littermates using end point RT-PCR. Figure 2A shows that expression of IFNγ-responsive genes was selectively increased in fibroblasts from mice expressing SUMO-free STAT1, namely, intercellular adhesion molecule-1 (Icam1) and CXC chemokine ligand-9 (Cxcl9, or Mig). Other genes were not responsive to IFNγ in fibroblasts (inducible nitric-oxide synthase, Nos2; data not shown), or their expression was not affected by SUMO modification of STAT1, namely, guanylate binding protein 1 (Gbp1); interferon regulatory protein 1 (Irf1); CXC chemokine ligand-10, known also as Ip10; transporter 1 (Tap1); and Stat1. This outcome was largely confirmed for human fibrosarcoma cells stably reconstituted with wild-type or SUMO-free STAT1 (supplemental Figure 1). We then examined transcription in BMMs, which are highly responsive to IFNγ. We tested expression of Cxcl9, Gbp2, Icam1, and Nos2, and we found that their expression was increased up to 5-fold if STAT1 was not SUMO modified (Figure 2B). Consistent with the curtailed nuclear STAT1 activation profile, sumoylation diminished STAT1-dependent transcription not only in magnitude but also profoundly shortened its duration. The most potent increase in transcriptional activation associated with SUMO-free STAT1 was seen for Nos2 when IFNγ treatment was combined with lipopolysaccharide (LPS) administration, which is known to synergistically induce Nos2.23 In wild-type macrophages, Nos2 expression reached a plateau between 6 and 24 h of IFNγ/LPS treatment (Figure 2C). Macrophages from littermates expressing SUMO-free STAT1, in stark contrast, showed elevated and rising Nos2 transcription, which was ∼ 10-fold higher than wild type at the end of the 24-hour observation period. We subsequently used Western blotting to determine the corresponding protein expressing of NOS2 enzyme in the macrophages (Figure 2D), which showed moderately increased protein expression on treatment with IFNγ alone but dramatically increased enzyme expression on LPS costimulation, thus faithfully mirroring the gene expression data. Given the influence of sumoylation on STAT1 activation and IFNγ-induced gene expression, this posttranslational modification thus resembles another inhibitor of STAT1 activation, namely, suppressor of cytokine signaling 1, which too is required for the timely attenuation of IFNγ signaling. However, suppressor of cytokine signaling 1 inhibits STAT1 temporarily by an IFNγ-induced feedback mechanism,24 whereas SUMO conjugation occurs constitutively,12 and therefore could function like a rheostat that permanently diminishes cellular responsiveness to IFNγ.
Sumoylation of STAT1 desensitizes cells to IFNγ. (A) End point RT-PCR analyses of IFNγ-induced genes in mouse embryonic fibroblasts derived from SUMO-free STAT1 (ΔSUMO) knockin mice or wild-type littermates. Cells were left untreated or treated with 50 U/mL mouse IFNγ for the indicated times, followed by RNA extraction, reverse transcription, and PCR. Shown are the Gapdh-normalized signal intensities of ethidium bromide–stained PCR products. Data are the mean and SEM of 3 independent experiments. (B) Real-time PCR analyses using BMMs from mice expressing SUMO-free STAT1 (ΔSUMO) or wild-type littermates. Cells were left untreated or were treated with mouse IFNγ (50 U/mL) for 6 or 24 hours. Shown are Gapdh-normalized gene expression data (mean ± SEM) of 3 independent experiments. (C) As in panel B, but BMMs were left untreated or were cotreated with IFNγ (50 U/mL) and LPS (1 ng/mL) for 3, 6, or 24 hours. Shown is the Gapdh-normalized expression of Nos2; values are mean ± SEM of 3 independent experiments. (D) Consecutive immunoblot analysis using NOS2-specific antibody (iNOS; BD Biosciences) and then β-actin antibodies with whole cell extracts from macrophages derived from SUMO-free STAT1 (ΔSUMO) knockin mice or wild-type littermates. The cells were treated for the indicated times with IFNγ alone or in combination with LPS. Data are representative of 2 independent experiments. (E) Nitric oxide production of BMMS derived from wild-type (left) or SUMO-free STAT1 knockin mice (ΔSUMO; right). Macrophages were seeded in 96-well plates at a density of 1 × 104 cells/well and kept for 60 hours in L cell–conditioned medium (10% L cell–supplemented DMEM with 10% calf serum) supplemented for the indicated times with IFNγ or LPS or combinations thereof. Half the culture supernatant (50 μL) and Griess reagent were subsequently used to determine absorption at 550 nm, before nitrite concentrations were calculated using a standard curve. Data are representative for 2 independent experiments carried out in duplicate. (F) Corresponding viability of the cells used in panel E, as determined by their ATP content. The value obtained for cells kept in L cell–conditioned medium for 60 hours was set to 100 and used as the reference point.
Sumoylation of STAT1 desensitizes cells to IFNγ. (A) End point RT-PCR analyses of IFNγ-induced genes in mouse embryonic fibroblasts derived from SUMO-free STAT1 (ΔSUMO) knockin mice or wild-type littermates. Cells were left untreated or treated with 50 U/mL mouse IFNγ for the indicated times, followed by RNA extraction, reverse transcription, and PCR. Shown are the Gapdh-normalized signal intensities of ethidium bromide–stained PCR products. Data are the mean and SEM of 3 independent experiments. (B) Real-time PCR analyses using BMMs from mice expressing SUMO-free STAT1 (ΔSUMO) or wild-type littermates. Cells were left untreated or were treated with mouse IFNγ (50 U/mL) for 6 or 24 hours. Shown are Gapdh-normalized gene expression data (mean ± SEM) of 3 independent experiments. (C) As in panel B, but BMMs were left untreated or were cotreated with IFNγ (50 U/mL) and LPS (1 ng/mL) for 3, 6, or 24 hours. Shown is the Gapdh-normalized expression of Nos2; values are mean ± SEM of 3 independent experiments. (D) Consecutive immunoblot analysis using NOS2-specific antibody (iNOS; BD Biosciences) and then β-actin antibodies with whole cell extracts from macrophages derived from SUMO-free STAT1 (ΔSUMO) knockin mice or wild-type littermates. The cells were treated for the indicated times with IFNγ alone or in combination with LPS. Data are representative of 2 independent experiments. (E) Nitric oxide production of BMMS derived from wild-type (left) or SUMO-free STAT1 knockin mice (ΔSUMO; right). Macrophages were seeded in 96-well plates at a density of 1 × 104 cells/well and kept for 60 hours in L cell–conditioned medium (10% L cell–supplemented DMEM with 10% calf serum) supplemented for the indicated times with IFNγ or LPS or combinations thereof. Half the culture supernatant (50 μL) and Griess reagent were subsequently used to determine absorption at 550 nm, before nitrite concentrations were calculated using a standard curve. Data are representative for 2 independent experiments carried out in duplicate. (F) Corresponding viability of the cells used in panel E, as determined by their ATP content. The value obtained for cells kept in L cell–conditioned medium for 60 hours was set to 100 and used as the reference point.
To assess whether this was the case, we researched the impact on cytotoxicity of BMMs, namely IFNγ and STAT1-dependent production of the enzymatic product of inducible NOS, that is, nitric oxide, which is critical for innate immunity and the control of infections.25 BMMs were exposed to IFNγ or bacterial molecular pattern, that is, LPS, and nitrite accumulation in the culture supernatant was determined, an established assay for NO production.3 As reported, treatment of wild-type macrophages either with IFNγ (50 U/mL) or LPS (1 ng/mL) did not cause detectable nitrite release (Figure 2E left panel).23 In contrast, SUMO-free STAT1-expressing macrophages showed robust NO production in response not only to IFNγ (Figure 2E right panel) but also to LPS, suggesting that lack of sumoylation renders STAT1 more sensitive to autocrine activation by LPS-induced type I IFN.26 The IFNγ hyperresponsiveness was strikingly confirmed by cotreatment with increasing IFNγ and LPS (1 ng/mL). Already at the lowest IFNγ concentration used (0.01 U/mL) a strong NO response was observed, with macrophages expressing SUMO-free STAT1. Wild-type macrophages, in contrast, remained unresponsive still at the 10 times higher dose of IFNγ (Figure 2C). Even when the dose was elevated 100-fold (1 U/mL), their NO production remained considerably lower. Indeed, mutant macrophages showed increased cytotoxicity at all interferon concentrations tested, although NO production seemed to reach saturation. Cellular ATP content was determined subsequently (Figure 2F), which demonstrated that viability of macrophage of both genotypes expectedly decreased in a time and NO concentration-dependent manner.27 Thus, the heightened NO production of BMMs expressing SUMO-free STAT1 agreed well with their demonstrated increase in both gene transcription and protein expression of inducible nitric oxide synthase shown in Figure 2C and D, respectively.
In sum, these results establish SUMO conjugation as an essential permanent negative regulator of STAT1. This posttranslational modification stands out from the known STAT1 inhibitors, tyrosine phosphatases, suppressor of cytokine signaling proteins, and protein inhibitor of activated STAT (PIAS) proteins,28 which all require activated, that is, Tyr701-phosphorylated STAT1. SUMO, in contrast, targets exclusively the unphosphorylated STAT1.17,18 As a result, SUMO diminishes the basic IFNγ sensitivity of cells and thus protects them against hyperresponsiveness to this cytokine with potent proinflammatory effects. This fundamental difference to the other STAT1 inhibitors, namely, permanent desensitization of STAT1 to IFNγ, however, seems at odds with the suggested need for macrophage sensitization to low concentrations of IFNγ during the early stages of immune responses.29 Future work will have to revisit this question. The PIAS proteins, another group of negative STAT1 regulators, can act as SUMO E3 ligases, but it has not been established whether this activity contributes to their STAT1 inhibition.28,30 Our results rather support the view that it does not.12,31 This is concluded from the largely nonoverlapping effects that PIAS proteins and SUMO have on STAT1. Aside from the fact that SUMO conjugates to unphosphorylated STAT1 and PIAS proteins associate with the phosphorylated STAT1,32 we note that SUMO-mediated STAT1 inhibition entails diminished tyrosine and serine phosphorylation. PIAS-mediated repression of STAT1-dependent gene activation, in contrast, does not seem to be associated with reduced STAT1 phosphorylation, because STAT1 phosphorylation is not increased in cells lacking PIAS133 or PIAS4,34 or in cells after removal of one allele of Pias4 or Pias1 in the Pias1−/− or Pias4−/− background, respectively.35 In addition, as demonstrated here, SUMO deficiency of STAT1 resulted in strongly increased macrophage cytotoxicity, whereas similar effects are not associated with PIAS1 and PIAS4 deficiencies,34,35 suggesting that PIAS proteins do not participate in STAT1 sumoylation in vivo. Nonetheless, results obtained with PIAS-deficient cells demonstrate cooperativity between the different PIAS proteins, so that at present a role for PIAS proteins in the augmentation of STAT1 sumoylation cannot be ruled out. However, irrespective of the actual SUMO E3 ligase activity responsible for STAT1 modification, SUMO-mediated STAT1 inhibition is distinguished by its exquisite target specificity, encoded in the defined single conjugation site on STAT1.12,18 In conjunction with its great significance for IFN signaling demonstrated here, the modulation of STAT1 SUMO conjugation thus becomes feasible for pharmacologic interventions with therapeutic potential in inflammatory and immune disorders.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ronald Naumann (Max-Planck-Institut für Zellbiologie, Dresden, Germany) for help in generating the STAT1ΔSUMO knockin mice.
This work was supported by the Deutsche Forschungsgemeinschaft (KFO 192) grants VI218/4 and 218/5 (to U.V.), and KN590/2-1 (to K.-P.K.), and a Wellcome Trust Value in People award (to A.B.).
National Institutes of Health
Authorship
Contribution: A.B., M.D., K.-P.K., and U.V. designed the experiments; A.B., M.D., and K.-P.K, performed the experiments; A.B., M.D., and U.V. analyzed the data and prepared the figures; and U.V. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Uwe Vinkemeier, School of Biomedical Sciences, Nottingham University Medical School, Nottingham NG7 2UH, United Kingdom; e-mail: uwe.vinkemeier@nottingham.ac.uk.
References
Author notes
A.B. and M.D. contributed equally to this work.