Abstract
The t(12;21) translocation that generates the ETV6-RUNX1 (TEL-AML1) fusion gene, is the most common chromosomal rearrangement in childhood cancer and is exclusively associated with B-cell precursor acute lymphoblastic leukemia (BCP-ALL). The translocation arises in utero and is necessary but insufficient for the development of leukemia. Single-nucleotide polymorphism array analysis of ETV6-RUNX1 patient samples has identified multiple additional genetic alterations; however, the role of these lesions in leukemogenesis remains undetermined. Moreover, murine models of ETV6-RUNX1 ALL that faithfully recapitulate the human disease are lacking. To identify novel genes that cooperate with ETV6-RUNX1 in leukemogenesis, we generated a mouse model that uses the endogenous Etv6 locus to coexpress the Etv6-RUNX1 fusion and Sleeping Beauty transposase. An insertional mutagenesis screen was performed by intercrossing these mice with those carrying a Sleeping Beauty transposon array. In contrast to previous models, a substantial proportion (20%) of the offspring developed BCP-ALL. Isolation of the transposon insertion sites identified genes known to be associated with BCP-ALL, including Ebf1 and Epor, in addition to other novel candidates. This is the first mouse model of ETV6-RUNX1 to develop BCP-ALL and provides important insight into the cooperating genetic alterations in ETV6-RUNX1 leukemia.
Introduction
The ETV6-RUNX1 fusion gene generated by the t(12;21)(p13;q22) chromosomal translocation,1 is the most prevalent fusion gene in childhood acute lymphoblastic leukemias (ALLs), the most common malignancy of childhood. It occurs in ∼ 20% of cases and is almost exclusively associated with the common B-cell precursor (BCP) subset of ALL (also known as common ALL).2 The fusion gene arises during fetal hematopoiesis in a BCP,3 giving rise to a preleukemic common ALL-propagating cell that was recently identified by the surface phenotype CD34+CD38–/lowCD19+.4,5 However, the frequency of individuals carrying the ETV6-RUNX1 fusion gene at birth considerably exceeds the number of patients presenting with clinically overt ALL,6 and twin studies and retrospective analysis of neonatal blood spots from ALL patients indicate that ETV6-RUNX1–expressing fetal clones expand and can persist in a clinically covert state for more than a decade.5 Together, this indicates the requirement for additional secondary (postnatal) genetic events (“hits”) for the development of ALL.
ETV6-RUNX1+ ALL at diagnosis is characterized by multiple copy number alterations (4-7 per case). These commonly include deletions in genes regulating B-cell differentiation or cell cycling,7 and studies on identical twins with concordant ALL indicate that these genetic events are secondary to ETV6-RUNX1 fusion and probably occur postnatally.8 In agreement with these clinical observations, animal models of ETV6-RUNX1 in both mice and zebrafish have shown that expression of the ETV6-RUNX1 fusion alone is insufficient for leukemogenesis; yet, similarly to the RUNX1-RUNX1T1 fusion,9 leukemia may occur after the acquisition of cooperating mutations.10-14 However, these models of ETV6-RUNX1 leukemia have not been suitable for the identification of cooperating mutations for two reasons. First, they do not accurately recapitulate the precursor B-cell phenotype associated with expression of the ETV6-RUNX1 fusion. Second, the models either use mutations already known to co-occur in ETV6-RUNX1 expressing ALL (such as deletion of the CDKN2A [p16] and p19 genes10 ), which provide no further pathogenetic information, or use agents such as N-ethyl-N-nitrosourea to induce secondary mutations, which are difficult to identify. To overcome these limitations, we have developed a new mouse model of ETV6-RUNX1 ALL in which expression of the fusion gene is driven from the endogenous Etv6 promoter and is linked to expression of the Sleeping Beauty (SB) transposase. This not only allows for expression of the fusion gene at endogenous levels but also recapitulates expression of the fusion gene in the pattern of endogenous Etv6 and allows forward mutagenesis to occur in the same cellular compartment.
Our model demonstrates that in the presence of mutations induced by the SB transposon, mice carrying the Etv6-RUNX1 allele can develop BCP-ALL. Furthermore, transposon insertions can be used to identify gene mutations that cooperate with Etv6-RUNX1 in leukemogenesis. This model therefore represents a unique tool for both studying the biology of this common disease and for identifying mutations that mediate development of ALL in cooperation with ETV6-RUNX1.
Methods
Generation of Etv6-RUNX1 mice
A 10.1-kb fragment of 129S5/SvEVBrd genomic DNA (NCBI m37: Chr6; 134200205-134210356) from bacterial artificial chromosome clone bMQ-66f22 was captured into pBluescript (Stratagene) by recombineering. This fragment contained most of introns 5 and 6, based on the longest protein-coding transcript of Etv6 (Ensembl ID: ENSMUST00000081028). Into the captured genomic fragment a cassette was inserted containing a splice acceptor, exons 1 to 6 of human RUNX1, an encephalomyocarditis virus internal ribosomal entry site (IRES), a hyperactive variant of the SB transposase (HSB5),15 and a flippase recognition target (FRT)-flanked phosphoglucokinase gene (PGK)–PuroΔTK drug selection marker.16 This entire cassette was synthesized by GENEART to ensure fidelity and was flanked by Lox66/Lox71 sites (to allow Cre-mediated inactivation of the Etv6-RUNX1 fusion gene) and HpaI sites (to allow cloning into the two StuI sites of the captured genomic fragment). The final targeting construct was termed pAML-IRES-SB-puro. Targeting was performed in E14Tg2a ES cells (129P2Ola/Hsd) and correctly targeted clones identified by Southern blotting on StuI-digested genomic DNA (gDNA) using a 5′ probe A (generated by PCR on gDNA using forward (FOR: 5′-GGG CAC CTA CGA TGC ACA GAT AAA TAC ATC CGC-3′ and reverse (REV): 5′-GAG ATA GGG TCA GTC TGG AAC TGG TCG CAG GGA-3′) and 3′ probe B (generated by PCR on gDNA using FOR: 5′-TCC TCC TTG GTC CTC ATA TAA AGG TGG TA-3′ and REV: 5′-TGA GTT CTG CTG AGT TAA TGG GCC CCG GA-3′). Targeted embryonic stem (ES) cells were injected into C57BL/6J blastocysts to generate chimaeras that were bred to 129S5/SvEvBrd or C57BL/6J mice for germ line transmission of the allele (Etv6+/RUNX1). Mice were housed in accordance with Home Office regulations (United Kingdom), and all procedures were carried out with Home Office approval. ES cell targeting of the RUNX1-FLAG was performed in the same way (Etv6+/RUNX1-FLAG).
Immunoprecipitation of FLAG-tagged proteins and Western blotting
FLAG-tagged proteins were immunoprecipitated from whole-cell ES cell lysates using Dynabead protein G (Invitrogen) according to the manufacturer's instructions, and Western blot analysis was performed using monoclonal anti-FLAG M2 antibody (Sigma-Aldrich). Western blot was performed on whole-cell mouse bone marrow lysates using anti-SB transposase antibody (R&D Systems) and actin (C-11) polyclonal antibody (Santa Cruz Biotechnology). The ETV6-RUNX1-Flag expression plasmid,17 which was used as a positive control for immunoprecipitation and Western blotting, was kindly provided by Dr O. Williams (University College London).
qPCR
Total RNA from mouse tissue (spleen, thymus, and bone marrow) was isolated using TRIzol reagent (Invitrogen), and cDNA was reverse transcribed using SuperScript First-Strand RT-PCR kit (Invitrogen), according to the manufacturer's instructions. Quantitative PCR (qPCR) was performed using ABsolute qPCR ROX Mix kit on an ABI PRISM 7900HT sequence detection system (Applied Biosystems). qPCR probes with 5′ 5-carboxyfluorescein and 3′ 5-carboxytetramethylrhodamine modifications (MWG Operon) were as follows: Etv6 probe: 5′-CAC GCC ATG CCC ATT GGG AGA A-3′ (FWD primer: 5′-TCT CTA TGT CCC CAC CGG AAG-3′; REV primer: 5′-CAT AAT CCC AAA GCA GTC TAC AGT CT-3′), Etv6-RUNX1 probe: 5′-AGC ACG CCA TGC CCA TTG GG-3′ (FWD primer: 5′-CTT GAA CCA CAT CAT GGT CTC TAT G-3′; REV primer: 5′-TCG TGC TGG CAT CTG CTAT T-3′), and β-actin probe: 5′-TTT GAG ACC TTC AAC ACC CCA GCC A-3′ (FWD primer: 5′-CGT GAA AAG ATG ACC CAG ATC A-3′ and REV primer: 5′-CAC AGC CTG GAT GGC TAC GT-3′).
Embryonic lethality and leukemogenesis studies
Embryonic lethality studies were performed by timed matings of Etv6+/RUNX1 mice, and the embryos were collected at day 10.5 of gestation. The embryos were genotyped by PCR using primers to detect the wild-type Etv6 allele (FOR: 5′-AGG CAT TGT GCA AAG AAT GAG AGA C-3′; REV: 5′-GAC CAA CCA AAC CAA ACA AAC AAA A-3′, with a 65°C annealing temperature to produce a 360-bp band) and the Etv6+/RUNX1 allele (FOR: 5′-AAG AAG CCA CTG CTC CAA AA-3′ and REV: 5′-GCA CTT GCT CTC CCA AAG TC-3′, with a 60°C annealing temperature to produce a 376-bp band). Mice carrying the SB transposon array (T2Onc)18 were kindly provided by Lara Collier (University of Wisconsin). Genotyping for the T2Onc allele and the excision PCR was performed as described previously.18 Mice carrying the Etv6+/RUNX1 and T2Onc+/Tg alleles were intercrossed, and their offspring were placed on tumor watch. These mice were examined twice daily for signs of disease, at which time they were killed, and a full necropsy was performed.
Histology and immunohistochemistry
Tissues and tumors were fixed in 10% neutral-buffered formalin at room temperature overnight. Samples were then transferred to 50% ethanol, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. For samples where no bone marrow was available for flow cytometric analysis of CD antigen expression, immunophenotyping was performed by immunohistochemical on formalin-fixed, paraffin-embedded tissue sections that had undergone antigen retrieval (microwaving in citrate buffer pH 6.0 for 20 minutes) using antibodies for CD3 (clone SP7; Abcam), B220 (clone RA3-6B2; R&D systems), and myeloperoxidase (Dako UK). Immunohistochemical signal was detected by secondary biotinylated goat anti–rabbit antibody (Vector Laboratories), followed by VECTASTAIN Elite ABC kit (Vector Laboratories) according to the manufacturer's instructions.
Flow cytometric analysis
For analysis of CD antigen expression, single-cell suspensions from spleen or bone marrow were stained with combinations of Mac-1-FITC, Gr-1-PE/Cy7, Ter-119-PE, B220-AlexaFluor 647, CD4-PE/Cy7, CD19-PE, B220-PE/Cy7, CD43-PE, IgM-FITC, IgD-AlexaFluor 647, c-Kit-PE, AA4.1-PE, CD24-FITC, BP1-PE, and IL7Ra-PE/Cy7 anti–mouse antibodies, obtained from BioLegend, according to the supplier's recommendations. FACS analysis was performed with a CyAn ADP analyzer (Beckman Coulter). For the IgH rearrangement analysis, cells were sorted on a MoFlo cell sorter (Beckman Coulter).
IgH rearrangement analysis
PCR analysis was performed using Phusion High Fidelity DNA polymerase (Finnzymes PCR Products, Thermo Fisher Scientific), amplifying from upstream of DQ52 (5′-DQ52 FOR: TTG GGT CAC TTT CCT GCT GT) to JH4 (J4 REV: AGA CC TGG AGA GGC CAT TCT) or from upstream of DFL/DSP genes (5′-D FOR: GCA TGT CTC AAA GCA CAA TG) to JH4 (J4 REV2: CTG AGG AGA CGG TGA CTG AG). PCR conditions used a 65°C annealing temperature and 90-second extension time for 40 cycles. PCR products were purified (QIAGEN Gel Extraction kit; QIAGEN), cloned into pGEM-T Easy plasmid (Promega), and transformed into subcloning efficiency DH5α chemically competent cells (Invitrogen); clones were sequenced using either the appropriate D primer or a primer in the vector. For some samples, it was necessary to perform whole-genome amplification before IgH rearrangement analysis, and this was performed using the GenomiPhi DNA Amplification kit (GE Healthcare), according to the manufacturer's instructions.
Whole-genome expression profiling
Total RNA extracted from the spleens of 15 Etv6-RUNX1 BCP-ALL cases and 3 Etv6-RUNX1 nondiseased mice using TRIzol (Invitrogen), according to the manufacturer's instructions. Expression profiling of the RNA was performed using a MouseWG-6 v2.0 expression beadchip kit (Illumina), according to the manufacturer's instructions. Data were quantile normalized19 and analyzed using the bioconductor packages limma and lumi (http://www.bioconductor.org/). Data were P values adjusted to yield a sorted list of differentially expressed genes.20 Gene annotations were further refined using ReMOAT (http://remoat.sysbiol.cam.ac.uk/). Gene set enrichment analysis was performed on the differentially expressed genes using Ingenuity Pathway Analysis software (Ingenuity Systems).
Isolation and statistical analysis of transposon insertion sites
Isolation of the transposon insertion sites from the leukemias was performed using splinkerette PCR to produce barcoded PCR products that were pooled and sequenced as described previously.21 The pooled PCRs were sequenced on 454 GS-FLX sequencers (Roche Diagnostics platform) over 4 separate lanes, with 1 lane per restriction enzyme and a maximum of 48 leukemias per lane. Processing of 454 reads, identification of insertion sites, and Gaussian kernel convolution (GKC) statistical methods used to identify common insertion sites (CISs) have been described previously.21 The P value for each CIS was adjusted by chromosome and a cut-off of P < .05 was used. Any CISs on mouse chromosome 1 are not reported because this is the “donor chromosome” where the transposon array is located. Data from this chromosome are automatically excluded from analysis because of the phenomenon of “local hopping.”18 The insertion sites have been made publically available by submission to both the Retrovirus Tagged Cancer Gene Database (RTCGD; http://rtcgd.ncifcrf.gov/) and the Insertional Mutagenesis Database (http://imdb.nki.nl/).
Results
Generation of Etv6-RUNX1 mice
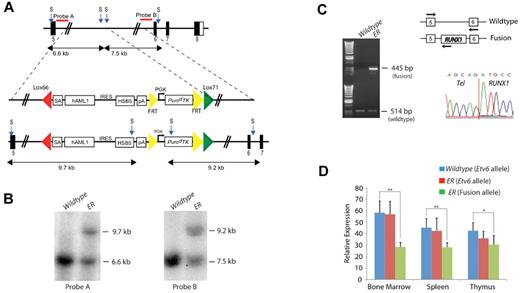
The endogenous Etv6 locus was targeted to introduce a Lox66/Lox71-flanked cassette containing a splice acceptor (SA), exons 1 to 6 of human RUNX1, an IRES, a hyperactive variant of the SB transposase (HSB5), and an FRT-flanked PGK-PuroΔTK drug selection marker into Etv6 intron 5, as shown in Figure 1A. The targeting vector was introduced into ES cells and correctly targeted clones, Etv6+/RUNX1 (ER), were identified by Southern blotting using a 5′ and 3′ external probe on StuI-digested genomic DNA (Figure 1B). These ES cell clones were used to generate chimeric mice that transmitted the mutated allele through the germ line. All offspring were genotyped by PCR to detect the presence of the transposase. The PuroΔTK drug marker was removed by breeding the mice to a Flpe-deleter strain (FLPeR mice)22 before tumor watch experiments.
Generation of the Etv6-RUNX1 transposase knockin allele (Etv6+/RUNX1). (A) The endogenous Etv6 locus was targeted to introduce a SA, exons 1 to 6 of human RUNX1, an IRES followed by a hyperactive variant of the SB transposase (HSB5), and an FRT-flanked PGK-PuroΔTK drug selection marker between exons 5 and 6. The entire cassette was flanked by Lox66 and Lox71 sites for potential Cre-mediated inactivation of the Etv6-RUNX1 fusion gene. The PuroΔTK drug marker was removed by breeding mice to an Flpe deleter strain before tumor watch experiments. Probe A (5′) and probe B (3′) are shown. S, StuI. (B) Southern blot analysis on StuI-digested tail DNA confirmed germ line transmission and homologous recombination of the 5′ and 3′ targeting vector arms. (C) RT-PCR showing expression of Etv6-RUNX1 fusion transcripts in bone marrow from Etv6+/RUNX1 mice, using primers located as indicated by the arrows on the schematic of the Etv6 locus. Bottom panel shows a sequence trace showing splicing of the Etv6 and RUNX1 transcripts to produce an in-frame fusion transcript. (D) qPCR of Etv6 and Etv6-RUNX1 fusion transcripts (using primers as mentioned herein). RNA was extracted from bone marrow, spleen, and thymus. Relative Etv6 gene expression in wild-type (blue) and Etv6+/RUNX1 tissues (red), and relative Etv6-RUNX1 fusion transcript expression in Etv6+/RUNX1 tissues (green) is shown. The data were collected from the analysis of 5 littermate mice of each genotype (± SD). **P < .005, *P < .05.
Generation of the Etv6-RUNX1 transposase knockin allele (Etv6+/RUNX1). (A) The endogenous Etv6 locus was targeted to introduce a SA, exons 1 to 6 of human RUNX1, an IRES followed by a hyperactive variant of the SB transposase (HSB5), and an FRT-flanked PGK-PuroΔTK drug selection marker between exons 5 and 6. The entire cassette was flanked by Lox66 and Lox71 sites for potential Cre-mediated inactivation of the Etv6-RUNX1 fusion gene. The PuroΔTK drug marker was removed by breeding mice to an Flpe deleter strain before tumor watch experiments. Probe A (5′) and probe B (3′) are shown. S, StuI. (B) Southern blot analysis on StuI-digested tail DNA confirmed germ line transmission and homologous recombination of the 5′ and 3′ targeting vector arms. (C) RT-PCR showing expression of Etv6-RUNX1 fusion transcripts in bone marrow from Etv6+/RUNX1 mice, using primers located as indicated by the arrows on the schematic of the Etv6 locus. Bottom panel shows a sequence trace showing splicing of the Etv6 and RUNX1 transcripts to produce an in-frame fusion transcript. (D) qPCR of Etv6 and Etv6-RUNX1 fusion transcripts (using primers as mentioned herein). RNA was extracted from bone marrow, spleen, and thymus. Relative Etv6 gene expression in wild-type (blue) and Etv6+/RUNX1 tissues (red), and relative Etv6-RUNX1 fusion transcript expression in Etv6+/RUNX1 tissues (green) is shown. The data were collected from the analysis of 5 littermate mice of each genotype (± SD). **P < .005, *P < .05.
RT-PCR performed on ER bone marrow cDNA confirmed the presence of the fusion transcript (not seen in Etv6+/+ [wild-type]; bone marrow cDNA; Figure 1C). Sequencing the RT-PCR product confirmed faithful splicing of the Etv6 and RUNX1 transcripts to produce an in-frame fusion transcript (Figure 1C). Quantitative RT-PCR performed on RNA extracted from bone marrow, spleen, and thymus showed that the relative expression level of the wild-type Etv6 gene to be comparable between wild-type and ER mice, whereas the relative expression level of the Etv6-RUNX1 fusion transcript was less than that of the wild-type Etv6 transcript in ER tissues (Figure 1D). Immunoprecipitation followed by Western blot analysis confirmed the presence of the fusion protein (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Etv6-RUNX1 in embryonic development
Mice heterozygous for the Etv6-RUNX1 allele (ER) were grossly normal at birth and fertile (data not shown). However, homozygosity resulted in embryonic lethality at embryonic day 10.5 (supplemental Figure 2A), similar to that seen in homozygous null Etv6 mice (Etv6−/−).23 This result confirms that our Etv6-RUNX1 allele does not produce a functional Etv6 transcript. This is important when modeling human leukemia, because loss of the second ETV6 allele is one of the most common genetic alterations in ETV6-RUNX1 ALL patients.7,24
These studies were performed using ER mice on a mixed genetic background (129;C57), and intercrossing of ER mice resulted in 54 of the 94 (57%) offspring being ER, which is within normal Mendelian ratios for homozygous lethal alleles (P = .2285; χ2 test, 2-tailed). This suggests that ETV6-RUNX1 does not have a dominant-negative function over the remaining fusion partner alleles during embryogenesis, unlike the RUNX1-RUNX1T1 fusion.9 However, when the proportion of C57 genetic background was increased by breeding, ER mice were produced at sub-Mendelian ratios (supplemental Figure 2B)—a phenotype similar to that observed in a recently described conditional knockin ETV6-RUNX1 mouse generated on a C57 background,14 suggesting the C57 strain may contain a genetic modifier of the function of ETV6-RUNX1 in embryonic development.
Etv6-RUNX1 expression predisposes to hematologic malignancies
Consistent with previous reports that ETV6-RUNX1 expression does not grossly affect hematopoiesis in the postnatal period,14 we did not observe significant gross perturbation of hematopoietic homeostasis in Etv6-RUNX1 mice (as assessed by immunophenotyping of bone marrow at 3, 6, and 12 months of age; supplemental Figure 3). To model the “second hit” event(s) that occurs in ETV6-RUNX1 patients facilitating the development of overt leukemia, we intercrossed Etv6-RUNX1 (Etv6+/RUNX1; ER) mice with mice carrying the T2Onc transposon array (T2Onc+/Tg).18 To confirm that the SB system was functioning in these mice, we first analyzed bone marrow cell lysates for the presence of SB transposase protein. Western blotting of these lysates using an anti-SB transposase antibody showed the presence of a 40-kDa band in mice carrying the Etv6-RUNX1 allele, consistent with the molecular weight of the SB transposase (Figure 2A). Second, we performed “excision PCR” on genomic tail DNA from the offspring (Figure 2B). A small PCR product (∼ 200 bp) is only present in mice carrying both the transposon and transposase (Etv6+/RUNX1; T2Onc+/Tg, hereafter referred to as EROnc mice), indicative of “jumping” of single transposons out of the array (into the genome), whereas mice carrying the transposon array alone (Etv6+/+; T2Onc+/Tg, hereafter referred to as Onc mice) show only a large PCR product (∼ 2 kb) representing the transposon concatamer.
Activation of transposon mutagenesis in Etv6+/RUNX1 mice. (A) Western blot showing expression of the SB transposase in the bone marrow of Etv6+/RUNX1 (ER) mice but not mice carrying the T2Onc transposon (Onc). (B) “Excision” PCR showing mobilization of the transposon in bone marrow from Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice, as evidenced by the 200-bp “excision” band (instead of the 2-kb band seen in mice carrying the unmobilized transposon array, ie, Etv6+/+; T2Onc+/Tg [Onc] mice). (C) Kaplan-Meier curves showing the tumor latency of “jumping” Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice and “non-jumping” control mice (ER, Onc, and wild-type). (D) Categorization of the malignancies developed by mice shown in the Kaplan-Meier curve according to the Bethesda criteria for lymphoid and nonlymphoid murine malignancies.25,26
Activation of transposon mutagenesis in Etv6+/RUNX1 mice. (A) Western blot showing expression of the SB transposase in the bone marrow of Etv6+/RUNX1 (ER) mice but not mice carrying the T2Onc transposon (Onc). (B) “Excision” PCR showing mobilization of the transposon in bone marrow from Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice, as evidenced by the 200-bp “excision” band (instead of the 2-kb band seen in mice carrying the unmobilized transposon array, ie, Etv6+/+; T2Onc+/Tg [Onc] mice). (C) Kaplan-Meier curves showing the tumor latency of “jumping” Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice and “non-jumping” control mice (ER, Onc, and wild-type). (D) Categorization of the malignancies developed by mice shown in the Kaplan-Meier curve according to the Bethesda criteria for lymphoid and nonlymphoid murine malignancies.25,26
Mice of all 4 genotypes that arose from the intercrossing of Etv6+/RUNX1 mice with T2Onc+/Tg mice: EROnc (n = 90), Etv6+/RUNX1; T2Onc+/+ (hereafter referred to as ER; n = 54), Onc (n = 50), and Etv6+/+; T2Onc+/+ (hereafter referred to as wild-type; n = 22) were placed on tumor watch and observed twice daily for signs of illness. Although mice of all genotypes developed hematologic malignancies, EROnc mice showed a statistically significant increased incidence of leukemia and decreased survival compared with the other genotypes (P < .0001; Figure 2C). ER mice (which carry the Etv6RUNX1 allele without the T2Onc transposon allele, hence no mutagenesis) also showed decreased survival compared with Onc mice carrying just the T2Onc allele (ie, no Etv6-RUNX1 or transposase expression, hence no mutagenesis).
This decreased survival in both cohorts was because of the development of hematologic malignancies that were classified according to the Bethesda criteria for murine malignancies using standard morphologic, histologic, and immunophenotypic analysis (Figure 2D).25,26 Occasional mice (11/72 mice, 14%) in the Onc and WT cohorts presented with long latency predominantly T-cell acute leukemias. However, the incidence of leukemia was significantly increased in the cohorts expressing Etv6-RUNX1 (73/90 mice, 81% for the EROnc cohort and 16/54, 29% for ER). The presence of Etv6-RUNX1 expression not only increased the incidence of acute leukemias but also modulated the phenotype. In the EROnc cohort, acute myeloid leukemia (AML) predominated (34/73 cases, 46%), with T-cell ALL also seen (21/73, 28%). T-cell ALL predominated in the ER mice (11/16 cases, 68%), with an occasional case of AML seen. Importantly, 15 of 73 (21%) of EROnc mice developed BCP (BCP-ALL) as did a slightly smaller proportion of the ER cohort (2/16 cases, 13%). Most importantly, BCP-ALL was only seen in mice carrying the ETV6-RUNX1 allele. This is the only mouse model of ETV6-RUNX1 to develop BCP-ALL, either through spontaneous or mutagenesis-driven acquisition of “second hits.”
Etv6-RUNX1–expressing mice develop a BCP-ALL
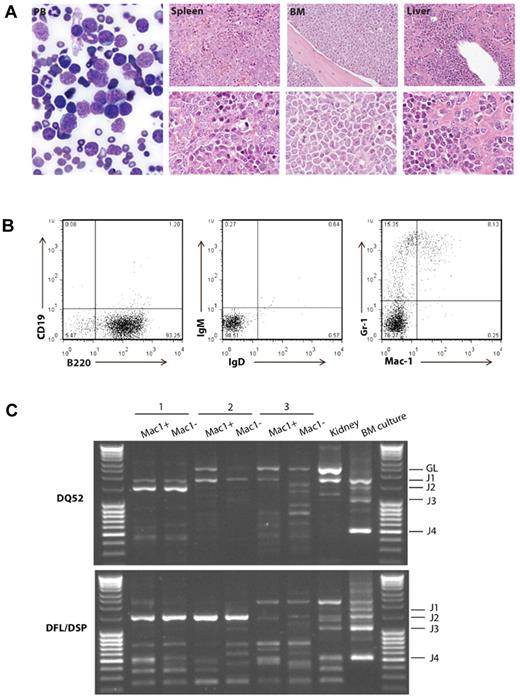
Mice with the BCP-ALL phenotype demonstrated the obvious presence of lymphoblasts in the peripheral blood and bone marrow, and heavy infiltration of lymphoblasts in the spleen and liver, resulting in effacement of the normal cellular architecture and replacement with predominantly nucleolated blasts (Figure 3A). Immunophenotypic analysis of the bone marrow by FACS from the 17 cases of BCP-ALL showed the majority of tumors to be of early B-cell progenitor immunophenotype (B220+ CD19−, IgM−, IgD−; Figure 3B; supplemental Figure 4). ETV6-RUNX1 patients typically have a surface phenotype reminiscent of pre-B cells (equivalent to Hardy fraction C).27 More detailed immunophenotyping of a subset of 7 cases predominantly showed a phenotypic profile (AA4.1+, CD24+, CD43+, BP1−, IL7Ra−; supplemental Figure 4) consistent with early pro-B cells (Hardy fraction B), suggesting transformation of a slightly earlier lymphoid progenitor. In addition, several cases also showed coexpression of myeloid (Mac-1+) markers on lymphoblasts (supplemental Figure 4). Coexpression of myeloid antigens (particularly CD13 and CD33) is well described for patients with ETV6-RUNX1 BCP-ALL.27
Etv6+/RUNX1 mice with “second hits” develop a pre-B ALL recapitulating features of the human disease. (A) Peripheral blood (PB; original magnification ×2000), spleen, bone marrow, and liver (original magnification ×400 in top panels and original magnification ×1000 in bottom panels) from a representative mouse with BCP-ALL. The presence of lymphoblasts in the PB is obvious, as is the infiltration of the spleen, bone marrow and liver, with effacement of the normal cellular architecture and replacement by nucleolated blasts. (B) FACS plots from the bone marrow of a representative mouse demonstrate only background Gr-1/Mac1 myeloid cells, with the majority of cells having a B220+/CD19−/sIg− phenotype in keeping with BCP-ALL. (C) D-J PCR rearrangement studies (involving DQ52 and DFL/DSP genes) were performed on spleen or bone marrow gDNA of 3 mice with phenotypic B-ALL. Negative control was gDNA from C57BL/6J mouse kidney (known to be unrearranged), and positive control was gDNA from a C57BL/6J primary pro-B cell bone marrow culture (known to contain a lot of DJ-rearranged alleles). Sample 1 shows 2 different rearrangement events (DQ52-J2 and DFL/DSP-J2), in keeping with a clonal B-cell population with both alleles D-J recombined. Sample 2 shows both a germ line band and an identical DFL/DSP-J2 band, in keeping with a clonal B-cell population with rearrangement of one allele and germ line configuration of the other. Sample 3 shows an absence of recombination.
Etv6+/RUNX1 mice with “second hits” develop a pre-B ALL recapitulating features of the human disease. (A) Peripheral blood (PB; original magnification ×2000), spleen, bone marrow, and liver (original magnification ×400 in top panels and original magnification ×1000 in bottom panels) from a representative mouse with BCP-ALL. The presence of lymphoblasts in the PB is obvious, as is the infiltration of the spleen, bone marrow and liver, with effacement of the normal cellular architecture and replacement by nucleolated blasts. (B) FACS plots from the bone marrow of a representative mouse demonstrate only background Gr-1/Mac1 myeloid cells, with the majority of cells having a B220+/CD19−/sIg− phenotype in keeping with BCP-ALL. (C) D-J PCR rearrangement studies (involving DQ52 and DFL/DSP genes) were performed on spleen or bone marrow gDNA of 3 mice with phenotypic B-ALL. Negative control was gDNA from C57BL/6J mouse kidney (known to be unrearranged), and positive control was gDNA from a C57BL/6J primary pro-B cell bone marrow culture (known to contain a lot of DJ-rearranged alleles). Sample 1 shows 2 different rearrangement events (DQ52-J2 and DFL/DSP-J2), in keeping with a clonal B-cell population with both alleles D-J recombined. Sample 2 shows both a germ line band and an identical DFL/DSP-J2 band, in keeping with a clonal B-cell population with rearrangement of one allele and germ line configuration of the other. Sample 3 shows an absence of recombination.
To further demonstrate clonality and determine the differentiation stage of the tumor, the VDJH recombination status of 3 BCP-ALLs (B220+/CD19−/sIg− phenotype) was analyzed. Clonal DJH rearrangements were detected in 2 samples; however, both alleles were in germ line configuration in the third (Figure 3C). Thus, taking into account the results of the IgH recombination and immunophenotyping analysis, these murine leukemias are BCP in origin, however, show an earlier arrest in B-cell ontogeny than typically found in ETV6-RUNX1 BCP-ALL patients (CD10+/CD19+ and evidence of VDJH recombination).
Gene expression analysis to look at transcripts differentially expressed between BCP-ALL spleen and normal splenic tissue (EROnc mice with no disease; n = 3) found statistically significant differential expression (typically down-regulation) of 125 genes (P < .05; supplemental Table 1), several of which are known to be important for transition through pro-B and pre-B cell stages of development (including Blk, Blnk, CD19, Ebf1, Foxo1, Irf4, Spib, Vpreb3i, and Zhx2). Gene set enrichment analysis performed on these differentially expressed genes found the top 5 canonical pathways involved B-cell signaling or development (supplemental Table 2). There was also decreased expression of the chemokine receptors Ccr6 (CD196) and Cxcr5 (CD185), reported to be down-regulated in BCP-ALL patients.28 Importantly, we also found decreased expression in genes found in deleted regions of human ETV6-RUNX1 patients (including Ebf1, Blnk, and Btg)7,24 and decreased expression of Ms4a1 (CD20), a B-lymphocyte surface antigen reported to be of lower expression in t(12;21)+ than t(12;21)− BCP-ALL cases.29 Thus, the molecular profiling of these leukemias in part mirrors human ETV6-RUNX1–positive BCP-ALL.
Identification of cooperating mutations in ETV6-RUNX1–expressing mice using insertional mutagenesis
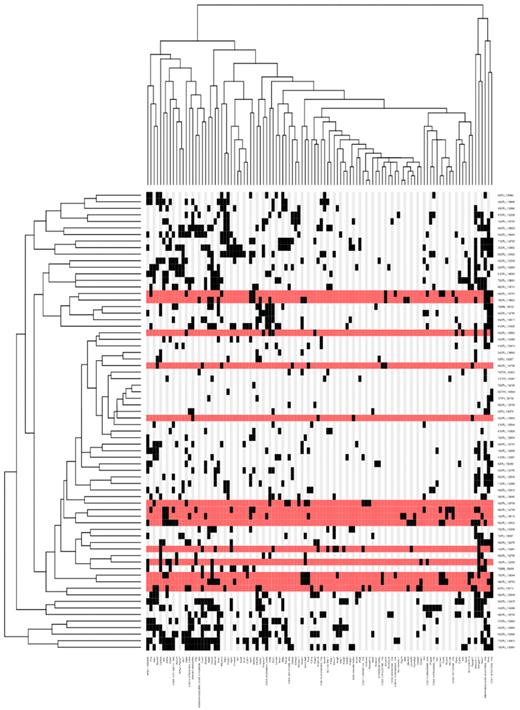
Genomic DNA extracted from leukemic tissue of 73 ETV6+/RUNX1; T2Onc+/Tg (EROnc) mice was used in a splinkerette PCR reaction to produce barcoded PCR products that were subsequently pooled and directly sequenced on the 454 GS-FLX platform.21 This generated 695,504 sequence reads, of which 51.5% unambiguously aligned to the mouse genome (supplemental Figure 5). Using a previously developed computational pipeline to trim, map, and annotate each sequence read,21 we were able to identify 23 529 unique (nonredundant) insertion sites. We used the GKC algorithm to determine statistically significant CISs, genomic regions with a higher density of insertion sites than expected by chance,30 and CISs were assigned to genes as described previously.21 In total, 71 leukemias (34 AML, 15 BCP-ALL, 19 T-lineage ALL [T-ALL], 2 unclassified, and 1 myeloproliferative) contributed to generate a total of 101 unique GKC CIS regions/genes, with an average of 15 ± 7 CIS/leukemia (listed in Figure 4; supplemental Table 3). To identify potential co-occurring CISs and common patterns of mutation between each of the leukemias, a matrix of the leukemias against the unique CIS regions was constructed, and hierarchical clustering was performed to aggregate those with similar patterns of insertions (Figure 4). Interestingly, many of the CISs that were found in the BCP-ALLs also were found in the AML and T-ALLs (supplemental Figure 6), suggesting that these CIS/genes may predispose to tumorigenesis in multiple hematopoietic lineages or that they may affect hematopoietic stem cell function before individual lineages are specified. However, the possibility cannot be excluded that because many of our BCP-ALLs also showed a population of cells with aberrant expression of myeloid markers (Mac-1 and Gr-1; Figure 3B; supplemental Figure 4), mutation of genes involved in the myeloid pathway would be expected or that these tumors were polyclonal and contained a separate myeloid clone.
Hierarchical clustering of the leukemias and their mutated genes based on common insertion sites. A matrix of 71 leukemias against the 102 CIS genes identified by analysis of transposon insertions by the GKC method was constructed. Although 73 leukemias were subjected to ligation-mediated PCR, sample 120992 did not have any insertions that were mapped and sample 138641 had only 44 insertions sites that were mapped, but none of these contributed to any CISs; thus, these 2 samples do not appear in the heatmap because the heatmap is based on the co-occurrence of insertions in CISs. Hierarchical clustering was performed on a binarized version of the matrix using the Hamming distance metric along both the rows and columns, to aggregate leukemias and CIS genes with similar insertions patterns. The 2 resulting dendrograms were visualized as a heatmap, where black squares indicate the presence of at least one insertion in the tumor, and gray no insertion site. Leukemia rows highlighted in red indicate a BCP-ALL phenotype (n = 15; only 14 rows highlighted in red because no insertions were found in the called CIS for sample 138895). The clustering and visualization were performed using the R programming language (Version 2.10.0; http://www.r-project.org) and Bioconductor (Version 2.5; http://www.bioconductor.org).
Hierarchical clustering of the leukemias and their mutated genes based on common insertion sites. A matrix of 71 leukemias against the 102 CIS genes identified by analysis of transposon insertions by the GKC method was constructed. Although 73 leukemias were subjected to ligation-mediated PCR, sample 120992 did not have any insertions that were mapped and sample 138641 had only 44 insertions sites that were mapped, but none of these contributed to any CISs; thus, these 2 samples do not appear in the heatmap because the heatmap is based on the co-occurrence of insertions in CISs. Hierarchical clustering was performed on a binarized version of the matrix using the Hamming distance metric along both the rows and columns, to aggregate leukemias and CIS genes with similar insertions patterns. The 2 resulting dendrograms were visualized as a heatmap, where black squares indicate the presence of at least one insertion in the tumor, and gray no insertion site. Leukemia rows highlighted in red indicate a BCP-ALL phenotype (n = 15; only 14 rows highlighted in red because no insertions were found in the called CIS for sample 138895). The clustering and visualization were performed using the R programming language (Version 2.10.0; http://www.r-project.org) and Bioconductor (Version 2.5; http://www.bioconductor.org).
Restricting our analysis to the BCP-ALL cases (Table 1; supplemental Figure 7), the most frequently targeted genes/loci were an intragenic region on mouse chromosome 7 (CIS peak location at 37320759), translation initiation factor 2 (Eif2a), erythropoietin receptor (Epor), Ikaros family zinc finger 1 (Ikzf1), CCAAT/enhancer binding protein, α (Cebpa), and cannabinoid receptor 2 (Cnr2). In some cases, the transposons had inserted themselves in the forward orientation and were clustered in the noncoding introns upstream of the ATG start site, suggesting they act as activating mutations to induce overexpression of the gene (such as Raf1 and Mef2c). In other cases, the transposons had inserted themselves in both the forward and reverse orientations and were scattered throughout the gene, suggesting they act as inactivating mutations, to produce a truncated transcript (such as Ikzf1). Although some of these genes/loci also were found as CIS in AML and T-ALL cases, 66% (22/33) of these CIS were exclusive to the BCP-ALL cases (including Eif2a and Ikzf1). Intriguingly, these B cell–specific genes did not seem to cluster together (Figure 4), suggesting that there is significant heterogeneity in the genes that can contribute to the development of acute lymphoblastic leukemia in this model.
GKC CISs identified in all BCP-ALL cases
| Gene . | Chromosome . | CIS leak location . | GKC scale (kbp) . | Tumors with CISs . | Unique insertions . | Other genes in the CIS interval . |
|---|---|---|---|---|---|---|
| CIS7:37320759 | 7 | 37320759 | 15 | 6 | 12 | |
| Eif2a | 3 | 58350745 | 240 | 6 | 10 | 2810407C02Rik, AC111080.1, AC119873.1, AC132302.1, AC142228.1, Commd2, Gm410, Pfn2, Rnf13, Serp1, Siah2, Tsc22d2, U6, Wwtr1 |
| Ikzf1 | 11 | 11652893 | 15 | 4 | 9 | |
| Epor | 9 | 21769506 | 15 | 2 | 9 | |
| Cebpa | 7 | 35899198 | 15 | 2 | 8 | 5S_rRNA |
| Cnr2 | 4 | 135467435 | 15 | 4 | 7 | |
| Prr8 | 5 | 28716988 | 75 | 5 | 6 | AC121843.1, AC134530.1, Shh |
| Rcan2 | 17 | 44101905 | 75 | 3 | 6 | 7SK, CT025668.2 |
| CIS6:31099937 | 6 | 31099937 | 30 | 5 | 5 | AB041803, AC153820.1/4, RP23–459L15.3 |
| Rasgrf1 | 9 | 89867811 | 15 | 5 | 5 | |
| Slc10a1 | 12 | 82095071 | 15 | 5 | 5 | Sfrs5 |
| Gm1968 | 16 | 29983555 | 30 | 5 | 5 | CT025592.1, CT030736.1 |
| Mecom | 3 | 30047667 | 15 | 4 | 5 | |
| Psd3 | 8 | 70258943 | 30 | 4 | 5 | AC109142.1, AC109142.2 |
| Sfi1 | 11 | 3065855 | 50 | 4 | 5 | AL671968.1/2/4/5, Drg1, Eif4enif1 |
| CIS5:75770426 | 5 | 75770426 | 15 | 3 | 5 | U6 |
| Raf1 | 6 | 115605310 | 120 | 2 | 5 | |
| Kctd2 | 11 | 115288407 | 15 | 1 | 5 | |
| CIS11:54066112 | 11 | 54066112 | 15 | 3 | 4 | AL596103.1, Il3 |
| Asap1 | 15 | 64063966 | 30 | 3 | 4 | - |
| Met | 6 | 17424888 | 15 | 2 | 4 | - |
| Fam46d | X | 105018631 | 15 | 2 | 4 | 2610002M06Rik |
| Csf2rb2 | 15 | 78135922 | 15 | 1 | 4 | |
| CIS2:131423786 | 2 | 131423786 | 15 | 3 | 3 | |
| Zmym4 | 4 | 126594535 | 15 | 3 | 3 | |
| Zfp423 | 8 | 90421110 | 15 | 3 | 3 | |
| Mef2c | 13 | 83672435 | 15 | 3 | 3 | |
| Il2rb | 15 | 78328305 | 15 | 3 | 3 | |
| Rasgef1a | 6 | 118007445 | 15 | 2 | 3 | |
| L3mbtl3 | 10 | 26098605 | 15 | 2 | 3 | |
| CIS10:116095924 | 10 | 116095924 | 15 | 2 | 3 | |
| Thg1l | 11 | 45761354 | 15 | 1 | 3 | |
| Gata1 | X | 7537541 | 15 | 2 | 2 |
| Gene . | Chromosome . | CIS leak location . | GKC scale (kbp) . | Tumors with CISs . | Unique insertions . | Other genes in the CIS interval . |
|---|---|---|---|---|---|---|
| CIS7:37320759 | 7 | 37320759 | 15 | 6 | 12 | |
| Eif2a | 3 | 58350745 | 240 | 6 | 10 | 2810407C02Rik, AC111080.1, AC119873.1, AC132302.1, AC142228.1, Commd2, Gm410, Pfn2, Rnf13, Serp1, Siah2, Tsc22d2, U6, Wwtr1 |
| Ikzf1 | 11 | 11652893 | 15 | 4 | 9 | |
| Epor | 9 | 21769506 | 15 | 2 | 9 | |
| Cebpa | 7 | 35899198 | 15 | 2 | 8 | 5S_rRNA |
| Cnr2 | 4 | 135467435 | 15 | 4 | 7 | |
| Prr8 | 5 | 28716988 | 75 | 5 | 6 | AC121843.1, AC134530.1, Shh |
| Rcan2 | 17 | 44101905 | 75 | 3 | 6 | 7SK, CT025668.2 |
| CIS6:31099937 | 6 | 31099937 | 30 | 5 | 5 | AB041803, AC153820.1/4, RP23–459L15.3 |
| Rasgrf1 | 9 | 89867811 | 15 | 5 | 5 | |
| Slc10a1 | 12 | 82095071 | 15 | 5 | 5 | Sfrs5 |
| Gm1968 | 16 | 29983555 | 30 | 5 | 5 | CT025592.1, CT030736.1 |
| Mecom | 3 | 30047667 | 15 | 4 | 5 | |
| Psd3 | 8 | 70258943 | 30 | 4 | 5 | AC109142.1, AC109142.2 |
| Sfi1 | 11 | 3065855 | 50 | 4 | 5 | AL671968.1/2/4/5, Drg1, Eif4enif1 |
| CIS5:75770426 | 5 | 75770426 | 15 | 3 | 5 | U6 |
| Raf1 | 6 | 115605310 | 120 | 2 | 5 | |
| Kctd2 | 11 | 115288407 | 15 | 1 | 5 | |
| CIS11:54066112 | 11 | 54066112 | 15 | 3 | 4 | AL596103.1, Il3 |
| Asap1 | 15 | 64063966 | 30 | 3 | 4 | - |
| Met | 6 | 17424888 | 15 | 2 | 4 | - |
| Fam46d | X | 105018631 | 15 | 2 | 4 | 2610002M06Rik |
| Csf2rb2 | 15 | 78135922 | 15 | 1 | 4 | |
| CIS2:131423786 | 2 | 131423786 | 15 | 3 | 3 | |
| Zmym4 | 4 | 126594535 | 15 | 3 | 3 | |
| Zfp423 | 8 | 90421110 | 15 | 3 | 3 | |
| Mef2c | 13 | 83672435 | 15 | 3 | 3 | |
| Il2rb | 15 | 78328305 | 15 | 3 | 3 | |
| Rasgef1a | 6 | 118007445 | 15 | 2 | 3 | |
| L3mbtl3 | 10 | 26098605 | 15 | 2 | 3 | |
| CIS10:116095924 | 10 | 116095924 | 15 | 2 | 3 | |
| Thg1l | 11 | 45761354 | 15 | 1 | 3 | |
| Gata1 | X | 7537541 | 15 | 2 | 2 |
CISs shown in bold are only found in the BCP-ALLs (n = 15), not the other ALL cases. CISs that are not located within ± 150 kbp of a gene are given the label CIS followed by the chromosome and the peak location of the Gaussian kernel.
Insertional mutagenesis in Etv6-RUNX1–expressing mice models human ALL
By performing cross-species comparisons, we found an overlap between some of the CIS in our BCP-ALL cases and genes recurrently found to be altered in patients with ETV6-RUNX1–positive ALL (Table 1), specifically insertions in the coding region of Ikzf1 and Epor. IKZF1 has a central role in the pathogenesis of BCP-ALL. Deletions involving IKZF1 are common (30%) in high-risk/poor prognosis BCP-ALL31 (although not in ETV6-RUNX1 ALL) and EPOR is consistently highly expressed in ETV6-RUNX1 ALL.32 Compared with control spleens (n = 3), those spleens with insertions in Epor (n = 2) showed a log-fold change of 0.35 for Epor expression (which is ∼ 27% increase in expression),20 whereas those without insertions in Epor (n = 13) showed a log-fold change of −0.23 (which is ∼ 15% decrease in expression). This suggests that the insertions in the promoter region of Epor up-regulate Epor expression, a finding consistent with ETV6-RUNX1 ALL patients. In addition, we found Met to be a CIS, and c-MET activation specifically enhances FAS-mediated apoptosis in ETV6-RUNX1 cells. Indeed, the c-MET/FAS complex is only found in normal B lymphocytes and ETV6-RUNX1 leukemias and not B-ALLs that lack the t(12;21) translocation, an observation that may account for their high sensitivity to chemotherapeutic regimens.33 We also found CISs in genes mutated in AML, specifically Gata1 (Gata-1 mutation is found in acute megakaryoblastic leukemia associated with Down syndrome)34 and Cebpa (mutations in C/EBPA are seen in AML).35
Seven of the CIS genes in our BCP-ALL samples had genomic locations syntenic with human chromosomal locations known to be regions of recurrent copy number change in ETV6-RUNX1 patient samples, including Fam46 days, Gata1, and L3mbtl3 (n ≥ 3/50 patients; Table 2). Although the genomic size of the lesions was quite large in some cases, it is possible that our CIS genes can serve as a focal point for identification of the causative gene in these regions.
CIS identified in the BCP-ALL samples whose homologous human chromosomal location was found to be a recurrent region of copy number (CN) change in human ETV6-RUNX1 leukemias
| Mouse CIS (no. insertions) . | Homologous human region (chromosome: bp) . | No. patients . | CN changes in patient samples . | CN lesion size (Mb): mean ± SD (median) . |
|---|---|---|---|---|
| Fam46d (4) | X: 79589304–79616318 | 10 | 1.4, 1.34, 1.36, 1.28, 1.53, 1.11, 1, 2.99, 0.85, 1.23 | 124 ± 43 (150) |
| Gata1 (2) | X: 48645231–48659263 | 9 | 2.47, 1.34, 1.36, 1.28, 1.11, 1, 2.99, 0.85, 1.23 | 132 ± 34 (151) |
| L3mbtl3 (3) | 6: 130336865–130339827 | 5 | 0.88, 1.28, 0.84, 1.55, 3.17 | 110 ± 57 (87) |
| CIS7:37320759 (12) | 19: 31962394–32030766 | 3 | 1.68, 1.65, 1.06 | 23 ± 29 (7) |
| Cebpa (8) | 19: 33765960–33835715 | 3 | 1.68, 1.65, 1.06 | 23 ± 29 (7) |
| Gm1968 (5) | 3: 193728382–193781161 | 3 | 0.98, 0.94, 1.59 | 37 ± 42 (20) |
| Psd3 (5) | 8: 18414769–18459953 | 3 | 1.16, 0.93, 2.6 | 58 ± 37 (37) |
| Mouse CIS (no. insertions) . | Homologous human region (chromosome: bp) . | No. patients . | CN changes in patient samples . | CN lesion size (Mb): mean ± SD (median) . |
|---|---|---|---|---|
| Fam46d (4) | X: 79589304–79616318 | 10 | 1.4, 1.34, 1.36, 1.28, 1.53, 1.11, 1, 2.99, 0.85, 1.23 | 124 ± 43 (150) |
| Gata1 (2) | X: 48645231–48659263 | 9 | 2.47, 1.34, 1.36, 1.28, 1.11, 1, 2.99, 0.85, 1.23 | 132 ± 34 (151) |
| L3mbtl3 (3) | 6: 130336865–130339827 | 5 | 0.88, 1.28, 0.84, 1.55, 3.17 | 110 ± 57 (87) |
| CIS7:37320759 (12) | 19: 31962394–32030766 | 3 | 1.68, 1.65, 1.06 | 23 ± 29 (7) |
| Cebpa (8) | 19: 33765960–33835715 | 3 | 1.68, 1.65, 1.06 | 23 ± 29 (7) |
| Gm1968 (5) | 3: 193728382–193781161 | 3 | 0.98, 0.94, 1.59 | 37 ± 42 (20) |
| Psd3 (5) | 8: 18414769–18459953 | 3 | 1.16, 0.93, 2.6 | 58 ± 37 (37) |
In total, 50 ETV6-RUNX1 patient samples were analyzed by single-nucleotide polymorphism array, and the CN changes for the relevant lesions shown. Deletions correspond to CN < 2 and amplifications to CN > 2. The LiftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver) was used to find the associated human chromosomal region for each mouse CIS gene (using the GRCh37/hg19 assembly). Statistical significance for each CIS was calculated (using 1000 randomly selected Refseq genes) and a cut-off of P < 0.2 was used.
Given the small number of BCP-ALL cases in this cohort (n = 15), we were statistically underpowered to generate a large number of CISs. Therefore, for the BCP-ALL cases, we looked at the 4787 unique/nonredundant insertion sites in addition to the CISs, and we found that many of them were of interest because of their location relative to genes known to be dysregulated in ETV6-RUNX1–positive ALL patients. These genes included Lef1, Dpf3, Tbl1xr1, Ebf1, and Nr3c1 (Table 3). Although the insertion frequency was too low, or the genomic distribution of the insertions too broad to be classified as a CIS, a frequency of 1 to 3 cases from a total of 15 cases represents 6 to 20% and echoes the frequency reported in ETV6-RUNX1–positive ALL patients.7,24
Human-mouse comparison of the insertion sites in BCP-ALL cases
| Mouse gene with insertion . | Insertion location (relative to gene) . | No. unique insertions . | Dysregulation of gene in human ETV6-RUNX1 ALL lesions . | Reference . |
|---|---|---|---|---|
| Ikzf1 | Coding | 4 | In a deletion region | 36*,31† |
| C20orf94 | Coding | 3 | In a deletion region | 7 |
| Ebf1 | Coding | 3 | In a deletion region | 7,8 |
| Fhit | Coding | 3 | In a deletion region | 7 |
| Chn2 | Coding | 2 | Down-regulated expression | 37 |
| Epor | Promoter | 2 | Up-regulated expression | 32,37,38 |
| Flt3 | Promoter | 2 | Activating mutations | 39 |
| Mllt3 | Coding | 2 | In a deletion region | 7 |
| Nf1 | Coding | 2 | In a deletion region | 7 |
| Smarcc1 | Coding | 2 | In a deletion region | 24 |
| Tbl1xr1 | Coding | 2 | In a deletion region | 7,8 |
| Tcf4 (E2–2) | Coding | 2 | In a deletion region | 36,40 |
| Ankrd27 | Coding | 1 | In a deletion region | 24 |
| Arid1b | Coding | 1 | In a deletion region | 41 |
| Atf2 | Coding | 1 | Down-regulated expression | 37 |
| Atp10a | Coding | 1 | In a deletion region | 41 |
| B4galt1 | Coding | 1 | Down-regulated expression | 37 |
| Birc7 | Promoter | 1 | Up-regulated expression | 38 |
| Cd44 | Coding | 1 | Down-regulated expression | 32,37,38 |
| Clic5 | Coding | 1 | Up-regulated expression | 38 |
| Dpf3 | Coding | 1 | In a deletion region | 7 |
| Eif2s1 | Coding | 1 | Down-regulated expression | 37 |
| Fbn2 | Coding | 1 | Up-regulated expression | 38 |
| Flnb | Coding | 1 | Down-regulated expression | 37 |
| Fyn | Coding | 1 | In a deletion region | 36* |
| Grik2 | Coding | 1 | In a deletion region | 7 |
| Hmg20a | Coding | 1 | Up-regulated expression | 38 |
| Jak1 | Coding | 1 | Expression up-regulated | 37 |
| Lef1 | Coding | 1 | In a deletion region | 7 |
| Lztfl1 | Coding | 1 | In a deletion region | 24 |
| Myo10 | Coding | 1 | Up-regulated expression | 38 |
| Nr3c1 | Coding | 1 | In a deletion region | 7 |
| Nr3c2 | Coding | 1 | In a deletion region | 7 |
| Pbef1 (Nampt) | Coding | 1 | In a deletion region | 36 |
| Pde4b | Coding | 1 | In a deletion region | 40 |
| Pdlim1 | Coding | 1 | Down-regulated expression | 37 |
| Ptpn2 | Coding | 1 | Down-regulated expression | 37 |
| Ptprk | Coding | 1 | Up-regulated expression | 37,38 |
| Rap1b | Coding | 1 | In a deletion region | 41 |
| Rb1 | Coding | 1 | In a deletion region | 7,8 |
| Rhpn2 | Promoter | 1 | In a deletion region | 24 |
| Tcf3 (E2A) | Promoter | 1 | In a deletion region | 36* |
| Tcf12 | Coding | 1 | In a deletion region | 41 |
| Thada | Coding | 1 | In a deletion region | 7 |
| Usp9x | Coding | 1 | Down-regulated expression | 37 |
| Yipf7 | Coding | 1 | In a deletion region | 40 |
| Ywhaq | Coding | 1 | In a deletion region | 40 |
| Zcchc7 | Coding | 1 | In a deletion region | 8,24 |
| Mouse gene with insertion . | Insertion location (relative to gene) . | No. unique insertions . | Dysregulation of gene in human ETV6-RUNX1 ALL lesions . | Reference . |
|---|---|---|---|---|
| Ikzf1 | Coding | 4 | In a deletion region | 36*,31† |
| C20orf94 | Coding | 3 | In a deletion region | 7 |
| Ebf1 | Coding | 3 | In a deletion region | 7,8 |
| Fhit | Coding | 3 | In a deletion region | 7 |
| Chn2 | Coding | 2 | Down-regulated expression | 37 |
| Epor | Promoter | 2 | Up-regulated expression | 32,37,38 |
| Flt3 | Promoter | 2 | Activating mutations | 39 |
| Mllt3 | Coding | 2 | In a deletion region | 7 |
| Nf1 | Coding | 2 | In a deletion region | 7 |
| Smarcc1 | Coding | 2 | In a deletion region | 24 |
| Tbl1xr1 | Coding | 2 | In a deletion region | 7,8 |
| Tcf4 (E2–2) | Coding | 2 | In a deletion region | 36,40 |
| Ankrd27 | Coding | 1 | In a deletion region | 24 |
| Arid1b | Coding | 1 | In a deletion region | 41 |
| Atf2 | Coding | 1 | Down-regulated expression | 37 |
| Atp10a | Coding | 1 | In a deletion region | 41 |
| B4galt1 | Coding | 1 | Down-regulated expression | 37 |
| Birc7 | Promoter | 1 | Up-regulated expression | 38 |
| Cd44 | Coding | 1 | Down-regulated expression | 32,37,38 |
| Clic5 | Coding | 1 | Up-regulated expression | 38 |
| Dpf3 | Coding | 1 | In a deletion region | 7 |
| Eif2s1 | Coding | 1 | Down-regulated expression | 37 |
| Fbn2 | Coding | 1 | Up-regulated expression | 38 |
| Flnb | Coding | 1 | Down-regulated expression | 37 |
| Fyn | Coding | 1 | In a deletion region | 36* |
| Grik2 | Coding | 1 | In a deletion region | 7 |
| Hmg20a | Coding | 1 | Up-regulated expression | 38 |
| Jak1 | Coding | 1 | Expression up-regulated | 37 |
| Lef1 | Coding | 1 | In a deletion region | 7 |
| Lztfl1 | Coding | 1 | In a deletion region | 24 |
| Myo10 | Coding | 1 | Up-regulated expression | 38 |
| Nr3c1 | Coding | 1 | In a deletion region | 7 |
| Nr3c2 | Coding | 1 | In a deletion region | 7 |
| Pbef1 (Nampt) | Coding | 1 | In a deletion region | 36 |
| Pde4b | Coding | 1 | In a deletion region | 40 |
| Pdlim1 | Coding | 1 | Down-regulated expression | 37 |
| Ptpn2 | Coding | 1 | Down-regulated expression | 37 |
| Ptprk | Coding | 1 | Up-regulated expression | 37,38 |
| Rap1b | Coding | 1 | In a deletion region | 41 |
| Rb1 | Coding | 1 | In a deletion region | 7,8 |
| Rhpn2 | Promoter | 1 | In a deletion region | 24 |
| Tcf3 (E2A) | Promoter | 1 | In a deletion region | 36* |
| Tcf12 | Coding | 1 | In a deletion region | 41 |
| Thada | Coding | 1 | In a deletion region | 7 |
| Usp9x | Coding | 1 | Down-regulated expression | 37 |
| Yipf7 | Coding | 1 | In a deletion region | 40 |
| Ywhaq | Coding | 1 | In a deletion region | 40 |
| Zcchc7 | Coding | 1 | In a deletion region | 8,24 |
The Ensembl mouse genome annotation database (version 59) was used to find the associated mouse gene for the human genes relevant to ETV6-RUNX1 patients (genes that did not map were hand-curated). The location of insertion sites was scored as being in the coding region of the gene (“coding”) or ≤ 5K upstream of the gene (“promoter”).
Reported in childhood ALL (not necessarily ETV6-RUNX1–positive patients).
IKZF1 deletion outside of the BCR-ABL context.
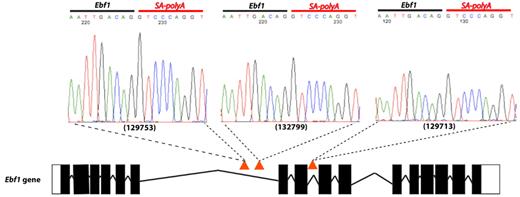
Of particular importance was Ebf1. EBF1 can activate expression of PAX5,42 and analysis of cDNA from the 3 cases that carried Ebf1 insertions showed splicing of Ebf1 directly onto the splice acceptor/polyA of the transposon (Figure 5). This resulted in premature truncation of the gene at exon 6 and thus presumably an inability to activate Pax5. As well as Ebf1, we also had insertions in many other key regulators of B-cell development, including Ikzf1 (supplemental Figure 8), Lef1, Tcf3, and Blnk, providing strong evidence that genetic alterations resulting in a block in B-lymphoid development are key events in the pathogenesis of B-ALL.
Insertions in the Ebf1 gene. Three Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice (129753, 132799, and 129713) that developed BCP-ALL carried insertions in the Ebf1 gene (indicated by the red triangles in introns 6 and 8). Sequencing of the insertion-genome junction from splenic cDNA of these mice showed the splicing of Ebf1 directly onto the SA-polyA from the transposon. Although sample 129713 carried an insertion in intron 8, we detected splicing onto the transposon directly from exon 6.
Insertions in the Ebf1 gene. Three Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice (129753, 132799, and 129713) that developed BCP-ALL carried insertions in the Ebf1 gene (indicated by the red triangles in introns 6 and 8). Sequencing of the insertion-genome junction from splenic cDNA of these mice showed the splicing of Ebf1 directly onto the SA-polyA from the transposon. Although sample 129713 carried an insertion in intron 8, we detected splicing onto the transposon directly from exon 6.
Discussion
ETV6-RUNX1 is the most common fusion oncogene associated with childhood ALL, and although generally associated with favorable prognosis, no faithful mouse models exist to provide mechanistic or potential therapeutic insight into this disease. Previous models have been hampered by potential overexpression of the fusion oncogene from transgenic or retroviral promoters, by a failure to generate leukemia, or by generation of a leukemia of a differing phenotype.10-14 In this report, we describe the generation of the first mouse model of Etv6-RUNX1 that develops BCP-ALL. In addition, through the use of the SB transposon system, we identify cooperating mutations that facilitate development of ALL, thereby providing an excellent resource to further dissect the biology of this disease.
We identified in total 102 unique statistically significant CIS regions/genes from the 71 leukemias analyzed (Figure 4; supplemental Table 3). Interestingly, several of the CIS that were found in the BCP-ALLs (Table 1) were also found in the AML and T-ALL cases. Because expression of the fusion in our model occurs in hematopoietic stem cells following the expression pattern of endogenous Etv6, it likely that the fusion “primes” cells to become leukemic with the actual phenotype driven by the nature of the cooperating mutations and the hematopoietic lineage/compartment in which they are acquired (before specification of the B-cell lineage). However, overlapping CISs from the BCP-ALL and AML cases may reflect the fact that several of these BCP-ALL cases also expressed myeloid markers (supplemental Figure 4), because the coexpression of myeloid antigens has been described in ETV6-RUNX1–positive ALL patients.27
Cross-species analysis of the BCP-ALL cases identified insertions/CISs in genes known to be dysregulated in human ETV6-RUNX1–positive ALL patients, including Btg1, Dpf3, Ebf1, Epor, Fhit, Grik2, Ikzf1, Lef1, Mllt3, Nf1, Nr3c1/2, Rap1b, Rb1, Tbl1xr1, Tcf12, and Zcchc7 (Table 3). Previous studies that have performed genome-wide profiling of genetic alterations in ETV6-RUNX1 BCP-ALL have shown that although a small number of mutations predominate, the mutational spectrum is highly heterogeneous, with numerous recurrent lesions generating mutations or copy number alterations in only a few patients.7,40 For example, in 47 cases of ETV6-RUNX1–positive ALL, deletions at chromosome 12p13.2 (ETV6) and 9p13.2 (PAX5) were found in as many as 33 and 13 patients (70% and 27% of cases), respectively. However, the majority of chromosomal gains/losses were found in < 8 (14%) patients, including deletions at 3q26.32 (TBL1XR1), 5q33.3 (EBF1), and 5q31.3 (NR3C1), with 8 of the genomic rearrangements only being found in 1 (2%) patient, including deletions at 4q25 (LEF1), 10q24.1 (BLNK), and 14q24.2 (DPF3).7 Thus, although our insertions in these genes do not classify as “CIS,” a “hit” frequency of 6% to 20% echoes the 2% to 14% frequency reported in ETV6-RUNX1–positive ALL patients.
In addition, it is interesting that some genes in recurrent regions of chromosomal imbalance were not involved, including Etv6, Cdkn2a/Cdkn2b (encoding Ink4a/Arf), and Pax5.7,24 This could be because of our small sample size of 15 BCP-ALL cases. Alternatively, it is possible that these genes were not available for insertion, through prior inactivation by other means such as focal deletion or point mutation (potentially through illegitimate recombinase-activating gene–mediated recombination). It is also recognized that the mutation mechanism itself can contribute to (bias) the selection of target genes. For example, AID expression in CML cells has been suggested to promote overall genetic instability by hypermutation of tumor suppressor and DNA repair genes.43 Moreover, malignancies of progenitor B cells also have been associated with aberrant recombinase-activating gene–mediated recombination.44 Both of these mechanisms are sequence specific and may contribute a different bias to our experimental system in the selection of specific targets. Finally, it is also possible that this reflects the fact that our disease involves an earlier cell type (pro-B) than typically seen in ETV6-RUNX1–positive ALL patients (which are typically pre-B).
However, although Pax5 was not involved in our screen, we identified mutations in Ebf1 and Tcf3 (E2A) that cooperatively regulate PAX545 and Tcf4, Lef1, and CD44 that are PAX5-activated genes.46,47 These “complementary” lesions therefore potentially having the same signaling impact as if we had directly mutated Pax5. Interestingly, a study recently found that many PAX5-regulated genes lie within 100 kb of an EBF1-bound region, suggesting coordinated regulation of common target genes by these transcription factors.48
Of the CIS regions/genes that were identified in the BCP-ALLs (Table 1), Epor and Met are known to show altered expression in ETV6-RUNX1–positive ALL patients (and Ikzf in some cases). Although the other regions/genes have not been associated with ETV6-RUNX1 ALL, some have been shown to play a role in lymphocyte biology. For example, Cebpa belongs to the CEBP family, which have recently been found to show dysregulated expression in BCPs and can contribute to their malignant transformation49 and Rasgrf1 has been shown to be overexpressed in B-cell chronic lymphocytic leukemia compared with normal B lymphocytes.50 Other regions/genes have been implicated previously as players in tumorigenesis, such as Cnr2, a proto-oncogene that has been postulated to offer a growth advantage in leukemia cells51 ; and Psd3, the reduced expression of which is believed to contribute to malignant progression.52 Thus, these genes are all attractive candidates to cooperate with ETV6-RUNX1 in promoting ALL, and as new technologies (such as RNA sequencing and exome sequencing) emerge that allow further interrogation of cancer biology, new genes will constantly be added to the list of those found to be dysregulated in ETV6-RUNX1 ALL.
In summary, we present the first mouse model of ETV6-RUNX1 that exhibits BCP-ALL. This model will prove a valuable resource to both further understanding of the disease and in the development of therapeutics in ETV6-RUNX1–positive ALL.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of the Research Support Facility at the Wellcome Trust Sanger Institute for looking after the mice and Mahrokh Nohadani for performing the immunohistochemistry.
L.v.d.W. is supported by a Fellowship from the Kay Kendall Leukaemia Fund. D.J.A. is supported by Cancer Research-UK and the Wellcome Trust. B.J.H. and G.G. are supported by an Medical Research Council-UK Senior Fellowship (B.J.H.) and Cancer Research-UK. F.W.v.D. and M.F.G. are supported by the Kay Kendall Leukaemia Fund and Leukaemia & Lymphoma Research UK. A.E.C. and L.S.M. are supported by the Biotechnology and Biological Sciences Research Council. C.G.M. is supported by the American Lebanese Syrian Associated Charities of St Jude Children's Research Hospital, and is a Pew Scholar in the Biomedical Sciences.
Wellcome Trust
Authorship
Contribution: L.v.d.W conceived and performed research; collected, analyzed, and interpreted data; and wrote the manuscript; G.G. performed research; collected, analyzed, and interpreted data; and assisted in writing the manuscript; A.G.R. performed statistical analysis and assisted in writing the manuscript; L.S.M. performed research, analyzed data, and edited the manuscript; F.W.v.D. analyzed data and edited the manuscript; J.K. performed research; A.E.C. analyzed data and edited the manuscript; M.F.G. analyzed data and edited the manuscript; C.G.M. performed research, analyzed data, and edited the manuscript; B.J.H. conceived and performed research; collected, analyzed, and interpreted data; and assisted in writing the manuscript; and D.J.A. conceived and performed research, interpreted data, and assisted in writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Louise van der Weyden, Experimental Cancer Genetics, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1HH, United Kingdom; e-mail: lvdw@sanger.ac.uk.
References
Author notes
B.J.H. and D.J.A. contributed equally to this work.

![Figure 2. Activation of transposon mutagenesis in Etv6+/RUNX1 mice. (A) Western blot showing expression of the SB transposase in the bone marrow of Etv6+/RUNX1 (ER) mice but not mice carrying the T2Onc transposon (Onc). (B) “Excision” PCR showing mobilization of the transposon in bone marrow from Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice, as evidenced by the 200-bp “excision” band (instead of the 2-kb band seen in mice carrying the unmobilized transposon array, ie, Etv6+/+; T2Onc+/Tg [Onc] mice). (C) Kaplan-Meier curves showing the tumor latency of “jumping” Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice and “non-jumping” control mice (ER, Onc, and wild-type). (D) Categorization of the malignancies developed by mice shown in the Kaplan-Meier curve according to the Bethesda criteria for lymphoid and nonlymphoid murine malignancies.25,26](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2011-02-338848/4/m_zh89991174930002.jpeg?Expires=1769132939&Signature=XLbPeonGblPjiuj8TlSphVA-tzIgNFOGnfe1XoULyTIh4NKVIOgYXxGnUX6PYNkQiDLguZ3kvL1cOfjd7~8Cot1IsQXLfrXDhzX0yrMpaYxxzbpXBMoGRIli5K7KqEgSYRNSF6bus9M7f4qwHY9Vq5yUAeeCly-VCmumpqNiQT1vRduIn4lDvu4CbpMmUZyDrNk6giQVmP8krygn3gT61U6zEbltA~wrCVXakuWNPMhjgdeIuVeQiulfxRdv4W2-7JXLjhjITCWI41MhjH12Dpjn22nREo6cFfWGi27svv~8UTsTQDQ~5ZcYXtvZDZI2j0B6HSsvrOl8jCuqv~7SkA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)