Abstract
We conducted an open-label phase 1 study exploring the feasibility, safety, and biologic activity of epigenetic priming with decitabine before standard induction chemotherapy in patients with less-than-favorable risk of acute myelogenous leukemia (AML). We directly compared the clinical and DNA-hypomethylating activity of decitabine delivered at 20 mg/m2 by either a 1-hour infusion (Arm A) or a continuous infusion (Arm B) for 3, 5, or 7 days before a single, standard induction with infusional cytarabine (100 mg/m2 for 7 days) and daunorubicin (60 mg/m2 × 3 doses). Toxicity was similar to that of standard induction chemotherapy alone. Although we did not identify a maximum tolerated dose, there was more gastro-intestinal toxicity with 7 days of decitabine priming. Decitabine induced DNA hypomethylation at all dose levels and there was a trend toward greater hypomethylation in CD34+ bone marrow cells when decitabine was delivered by a short pulse (Arm A). Twenty-seven subjects (90%) responded to therapy: 17 with complete remission (57%) and 10 with partial remission (33%). Of the patients with partial remission to protocol treatment, 8 achieved remission to their next therapy, bringing the overall complete remission rate to 83%. We conclude that epigenetic priming of intensive chemotherapy can be safely delivered in an attempt to improve response rates. This trial was registered at www.clinicaltrials.gov as NCT00538876.
Introduction
Complete remission (CR) is required for the cure of patients with acute myelogenous leukemia (AML). Modern treatment has improved the survival of patients with AML and favorable molecular features, but the majority of patients do not have these favorable features. Standard therapy for younger patients with suboptimal-risk AML is associated with only 19%-37% long-term survival, and 17%-38% of these patients have persistent AML after standard induction chemotherapy.1-3 Accordingly, new treatment strategies to improve induction success rates are needed to increase survival.
Aberrant DNA methylation is a common means by which tumor suppressor genes (TSGs) are inactivated during carcinogenesis.4,5 Unlike genetic mechanisms of inactivation such as gene deletion or mutation, the epigenetic silencing of TSGs by DNA methylation is potentially reversible. This has led to broad interest by cancer biologists and oncologists in the study of DNA methylation and provides the rationale for incorporating DNA-hypomethylating agents into cancer therapy.
Acquired abnormalities of DNA methylation are frequently observed in AML and are particularly prevalent among patients with adverse risk features.6,7 Decitabine has single-agent activity in patients with poor-risk, relapsed, and resistant AML.8-12 This antileukemic activity is believed to result from the epigenetic induction of differentiation and cytotoxicity,13,14 but a link between clinical outcomes and the decitabine-induced hypomethylation of specific genomic loci has not yet been established.10,15-19 Furthermore, the absence of useful biomarkers of decitabine activity has hampered the identification of an optimal decitabine dose and delivery schedule.
Preclinical studies have shown that the mode and duration of decitabine delivery affects its antileukemic activity.20 In vitro, the growth of human leukemia–derived cell lines is inhibited to a greater degree if decitabine is administered over 24 hours rather than as a 1-hour pulse, even when the pulsed dose is 100-fold greater.21 Similarly, L1210 leukemia cells can be completely eradicated from mice when decitabine is administered by continuous infusion.22 This suggests that continuous exposure to decitabine may improve efficacy, perhaps as a result of improved penetration of the drug in sanctuary sites such as the central nervous system when decitabine is administered by long infusion23,24 or it may relate to fundamental aspects of DNA hypomethylation. Nonetheless, there is far greater experience with pulse administration of decitabine, and this mode of delivery appears to be effective as a single agent in AML and other myeloid malignancies.
The mechanistic basis for the clinical activity of decitabine has not been precisely defined, but reactivated expression of TSGs is commonly implicated because these genes often have roles in cellular differentiation, apoptosis, DNA repair, and checkpoint control.13,14 DNA-hypomethylating agents can sensitize resistant cancer cells to cytotoxic agents,25-28 but this approach has been sparsely studied in clinical trials. Nonetheless, the use of an epigenetic modifier for chemosensitization appears to be safe,29,30 and was reported to have restored cytarabine sensitivity to a cohort of younger patients with refractory acute lymphoblastic leukemia.26 Based on these observations, we hypothesized that pretreatment (ie, priming) with a hypomethylating agent would sensitize the leukemic cells to cytotoxic chemotherapeutics and increase the efficacy of induction chemotherapy for AML. To test this hypothesis, we conducted an open-label phase 1 evaluation of the feasibility, safety, and biologic activity of epigenetic priming using the hypomethylating agent decitabine before full-dose cytarabine and daunorubicin induction chemotherapy in patients with less-than-favorable risk AML.
Methods
Study group eligibility
Subjects ≤ 60 years of age with untreated AML and the absence of a favorable karyotype—APL, t(8;21) or inv(16)—were eligible to be enrolled. All subjects had an Eastern Cooperative Oncology Group performance status ≤ 2 and adequate hepatic (total bilirubin ≤ 2 mg/dL and aspartate transaminase and alanine aminotransferase < 1.5× the upper limit of normal), renal (serum creatinine ≤ 2 mg/dL or creatinine clearance > 50/mL/min/1.73 m2), and cardiac function (echocardiogram or multiple-gated acquisition scan demonstrating an ejection fraction within normal limits). Subjects must not have received chemotherapy (other than hydroxyurea) or radiation within the 2 weeks before enrollment in this study. Subjects with significant leukocytosis were enrolled once the circulating blast count was controlled (blast count ≤ 50 000/μL) with hydroxyurea. All subjects provided written informed consent. All study procedures and informed consent forms were approved by the Weill-Cornell Medical College institutional review board in accordance with the Declaration of Helsinki. This trial was registered at www.clinicaltrials.gov as NCT00538876.
Treatment
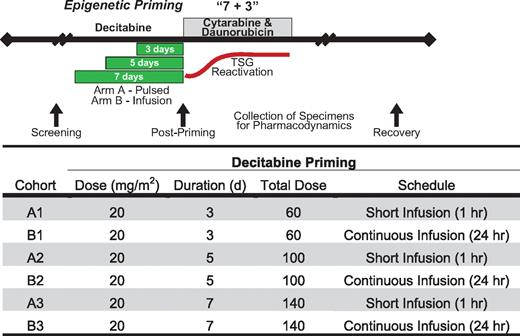
Cohorts of sequential subjects were treated with escalating doses of decitabine before a standard, “7 + 3” induction chemotherapy using infusional cytarabine (100 mg/m2/d continuous IV infusion for 7 consecutive days) with daunorubicin (60 mg/m2 daily for 3 days, generally the first 3 days of cytarabine). Because prior studies suggested that the hypomethylating activity of decitabine plateaus at 20 mg/m2/d,19 we escalated the total decitabine dose delivered by increasing the treatment duration rather than the daily dose. Decitabine was administered at 1 of 3 total dose levels: Level 1, 3 days of decitabine priming (total dose delivered, 60 mg/m2); Level 2, 5 days of decitabine priming to 100 mg/m2 (total dose delivered, 100 mg/m2); and Level 3, 7 days of priming (total dose delivered, 140 mg/m2; Figure 1). To determine whether the delivery schedule of decitabine affects its clinical and hypomethylating activity, we administered decitabine either as a daily 1-hour infusion (Arm A) or as a continuous infusion (Arm B). Decitabine has limited stability at ambient temperature but is stable for at least 8 hours at 4-10°C.31 To ensure stability, we chilled the decitabine infusate and changed bags every 8 hours during the continuous infusion. Standard 7 + 3 induction chemotherapy was initiated ∼ 24 hours after starting the last day of decitabine priming. We refer to the first day of 7 + 3 induction as Day +1 for all cohorts. Subjects received supportive care measures as clinically indicated, including standard antiemetic and antimicrobial therapy. Growth factor support was permitted but was only administered to one subject. After a single induction cycle, subjects received subsequent therapy at the discretion of their treating physician.
Dose escalation schema. Decitabine (20 mg/m2) was administered either as a short infusion (Arm A) or by continuous infusion (Arm B) for 3, 5, or 7 days (dose levels 1, 2, and 3, respectively) before standard infusional cytarabine, daunorubicin induction chemotherapy (7 + 3). Bone marrow and blood specimens were collected for pharmacodynamic analyses before and immediately after decitabine administration. The red line represents the hypothesized reactivation of chemosensitizing tumor suppressor genes (TSGs) resulting from epigenetic priming.
Dose escalation schema. Decitabine (20 mg/m2) was administered either as a short infusion (Arm A) or by continuous infusion (Arm B) for 3, 5, or 7 days (dose levels 1, 2, and 3, respectively) before standard infusional cytarabine, daunorubicin induction chemotherapy (7 + 3). Bone marrow and blood specimens were collected for pharmacodynamic analyses before and immediately after decitabine administration. The red line represents the hypothesized reactivation of chemosensitizing tumor suppressor genes (TSGs) resulting from epigenetic priming.
Response criteria and statistical methods
Responses were assessed using International Working Group criteria.32 A CR required normalization of peripheral blood counts with neutrophils > 103/μL and platelets > 105/μL and bone marrow with < 5% blasts. A partial remission (PR) required all of the hematologic values for a CR but with a decrease of at least 50% in the percentage of blasts to 5%-25% in the bone marrow aspirate
Cohorts of 5 subjects were sequentially enrolled starting with the lowest dose level of Arm A. Each cohort was closed when all patients reached one of 3 potential end points: (1) CR, (2) persistent AML, or (3) dose-limiting toxicity (DLT). Once 3 subjects within a cohort reached a study end point, accrual to the next available arm and dose level was permitted. Accrual alternated between Arm A and Arm B, with the planned dose escalation schema proceeding as follows: Arm A Level 1 (5 patients) → Arm B Level 1 (5 patients) → Arm A Level 2 (5 patients) → Arm B Level 2 (5 patients) → Arm A Level 3 (5 patients) → Arm B Level 3 (5 patients).
Toxicities were assessed using the National Cancer Institute Common Toxicity Criterion Version 3. DLT was defined as any grade 4 or higher nonhematologic toxicity or a grade 3 nonhematologic toxicity lasting > 7 days. Persistent bone marrow aplasia lasting > 59 days was also considered a DLT. The decitabine dose was escalated within an arm if no more than 1 subject per cohort sustained a DLT. The maximum tolerated dose (MTD) was defined as the dose level below the one that caused more than one DLT. An arm was closed once the MTD was identified, and dose escalation proceeded in the remaining treatment arm until all cohorts were filled or an MTD was reached in both arms.
Demographic and baseline disease characteristics were summarized descriptively for all subjects. Severe adverse events and efficacy were summarized for the entire population and within dosing cohorts. Statistical significance of differences between the treatment arms and individual cohorts was assessed using the t test or the χ2 test.
Analysis of DNA methylation
At the dose levels used in this protocol, decitabine primarily acts to induce DNA hypomethylation, and the site and extent of demethylation is thought to underlie its clinical activity. To measure the effect of decitabine treatment on DNA methylation, research bone marrow and blood specimens were obtained from each patient before initiating therapy (screening), after the completion of the final dose of decitabine (postpriming), and after the patient achieved remission or had documentation of persistent leukemia after treatment (recovery, ∼ day +28 to +60). We used bisulfite quantitative PCR to assess DNA methylation at selected loci that are constitutively methylated (LINE1 consensus promoter, HIST1H2AA) or methylated in the blasts of a subset of AML subjects (CDKN2B).33 We used 2 sets of bisulfite PCR primers insensitive to DNA methylation as normalizing controls (ALUC4 and GAPDH). Quantitation was performed using standard curves made from plasmid stocks of known copy number.
Peripheral blood cells were fractionated using Ficoll-Paque into a buoyant mononuclear cell fraction (PBMCs) and a dense granulocyte fraction (GRAN). Ficoll-buoyant bone marrow mononuclear cells (BMMCs) were further separated into CD34-expressing (CD34P) and CD34-depleted (CD34N) cells using immunomagnetic beads (Miltenyi Biotec). Genomic DNA was extracted from each cell fraction, quantified with PicoGreen (Life Tech), and bisulfite converted (Zymo Research). We compared the normalized methylation of each patient specimen obtained before and immediately after decitabine treatment to assess the pharmacodynamic activity of decitabine delivered at different dose levels and schedules. Differences in the decitabine-induced methylation changes were assessed across the dose levels, treatment arms, cohorts, and treatment responses using ANOVA and pairwise t tests of statistical significance.
Results
Study group
A total of 30 subjects with newly diagnosed AML were enrolled in the study. Baseline characteristics are shown in Table 1. Subjects were evenly split between men and women and had a median age of 55 years. Almost half of the subjects had a baseline karyotype associated with adverse outcome and 8 subjects (27%) had an antecedent hematologic disorder. An FMS-related tyrosine kinase 3 internal tandem duplication (FLT3-ITD) was detected in 4 subjects (13%) and 5 subjects (17%) presented with a white blood cell count (WBC) > 105/μL. The majority (83%) of those treated had at least 1 characteristic associated with poor outcome.
Patient characteristics
| Characteristic . | . |
|---|---|
| Median age, y (range) | 54.5 (23-60) |
| Sex, M:F | 16:14 |
| Median WBC, ×109/L (range) | 14.4 (1.5-211) |
| Median platelet count, ×109/L (range) | 57 (9-175) |
| Cytogenetics, no. subjects (%)* | |
| Intermediate, diploid, abn(12p),† +8,† +21 | 15 (50) |
| Adverse complex, −7/7q, 11q23,‡ t(6;9) | 13 (43) |
| Unknown add(18p), t(2:11)(q31;p15) | 2 (7) |
| FLT3-ITD, no. subjects (%) | 4 (13) |
| Antecedent hematologic disorder, no. subjects (%) | 8 (27) |
| WBC, no. subjects (%) | |
| > 100 000 | 5 (17) |
| > 50 000 | 8 (27) |
| ≥ 1 Adverse molecular feature, no. subjects (%)§ | 17 (57) |
| ≥ 1 Adverse feature, no. subjects (%)¶ | 25 (83) |
| Characteristic . | . |
|---|---|
| Median age, y (range) | 54.5 (23-60) |
| Sex, M:F | 16:14 |
| Median WBC, ×109/L (range) | 14.4 (1.5-211) |
| Median platelet count, ×109/L (range) | 57 (9-175) |
| Cytogenetics, no. subjects (%)* | |
| Intermediate, diploid, abn(12p),† +8,† +21 | 15 (50) |
| Adverse complex, −7/7q, 11q23,‡ t(6;9) | 13 (43) |
| Unknown add(18p), t(2:11)(q31;p15) | 2 (7) |
| FLT3-ITD, no. subjects (%) | 4 (13) |
| Antecedent hematologic disorder, no. subjects (%) | 8 (27) |
| WBC, no. subjects (%) | |
| > 100 000 | 5 (17) |
| > 50 000 | 8 (27) |
| ≥ 1 Adverse molecular feature, no. subjects (%)§ | 17 (57) |
| ≥ 1 Adverse feature, no. subjects (%)¶ | 25 (83) |
The majority of subjects had at least 1 adverse prognostic feature.
ECOG/SWOG criterion.2
abn(12p), +8 are considered adverse risk by CALGB.1
t(11;19)(q23;p13.1), t(10;11)(p12;q23).
Adverse karyotype or FLT3-ITD.
Adverse molecular features or antecedent hematologic disorder, age ≥ 55, WBC > 50 000.
Assessment of safety
The toxicity of epigenetic primed induction was similar to that of standard induction chemotherapy alone. An MTD was not identified. The most common nonhematologic toxicities were related to infection or colitis/mucositis (Tables 2–3). Subjects receiving 7 days of decitabine priming experienced more toxicity than those treated at either of the lower dose levels. Two subjects treated at the highest dose level (Cohorts A3 and B3) had grade 3 mucositis that resolved before count recovery but met criterion for DLT. There was no decline in cardiac ejection fraction after decitabine-primed induction. There was no induction mortality and no subject required transfer to intensive care. All subjects completed study treatment and were successfully discharged from the hospital within 35 days of induction chemotherapy. As expected, grade 4 neutropenia and thrombocytopenia were universal, but we observed no excess hematologic toxicity and peripheral blood counts recovered within 40 days (Table 4). All subjects went on to receive additional AML therapy and 20 received consolidation with an allogeneic hematopoietic stem cell transplantation (allo-HSCT). We did not identify late toxicities that delayed the delivery of subsequent AML therapy. We also did not observe excess toxicity when subsequent therapy was delivered, whether it was chemotherapy or allo-HSCT.
Adverse events possibly related to decitabine priming
| Adverse event . | Grade 1 . | Grade 2 . | Grade 3 . | Total . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort . | A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | |
| GI | 5 | 3 | 5 | 5 | 5 | 5 | 3 | 4 | 4 | 2 | 1 | 5 | 1 | 3 | 5 | 56 | |||
| Infection | 1 | 2 | 1 | 2 | 1 | 5 | 5 | 3 | 5 | 5 | 5 | 35 | |||||||
| Cardiac | 3 | 2 | 4 | 2 | 3 | 2 | 1 | 2 | 1 | 1 | 21 | ||||||||
| Dermatology | 2 | 4 | 1 | 2 | 2 | 2 | 1 | 3 | 1 | 1 | 1 | 1 | 21 | ||||||
| Constitutional | 1 | 3 | 2 | 5 | 2 | 2 | 3 | 1 | 1 | 20 | |||||||||
| Musculoskeletal | 3 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 14 | ||||||||||
| Respiratory | 1 | 4 | 2 | 3 | 1 | 11 | |||||||||||||
| Neurology | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 10 | |||||||||||
| Hepatobiliary | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 9 | |||||||||||
| Bleeding | 4 | 2 | 1 | 1 | 1 | 9 | |||||||||||||
| Adverse event . | Grade 1 . | Grade 2 . | Grade 3 . | Total . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort . | A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | |
| GI | 5 | 3 | 5 | 5 | 5 | 5 | 3 | 4 | 4 | 2 | 1 | 5 | 1 | 3 | 5 | 56 | |||
| Infection | 1 | 2 | 1 | 2 | 1 | 5 | 5 | 3 | 5 | 5 | 5 | 35 | |||||||
| Cardiac | 3 | 2 | 4 | 2 | 3 | 2 | 1 | 2 | 1 | 1 | 21 | ||||||||
| Dermatology | 2 | 4 | 1 | 2 | 2 | 2 | 1 | 3 | 1 | 1 | 1 | 1 | 21 | ||||||
| Constitutional | 1 | 3 | 2 | 5 | 2 | 2 | 3 | 1 | 1 | 20 | |||||||||
| Musculoskeletal | 3 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 14 | ||||||||||
| Respiratory | 1 | 4 | 2 | 3 | 1 | 11 | |||||||||||||
| Neurology | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 10 | |||||||||||
| Hepatobiliary | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 9 | |||||||||||
| Bleeding | 4 | 2 | 1 | 1 | 1 | 9 | |||||||||||||
The most common side effects according to system included: GI tract—diarrhea, nausea, constipation, and mucositis; infectious—bacteremia; cardiac—dizziness, hypotension, and chest pain; dermatology—macular erythematous rash; and constitutional—fatigue, insomnia, anxiety, and chills. Biological systems with less than 9 adverse effects, all grade ≤ 2, are not shown. No grade 4 nonhematologic toxicities were observed.
Significant nonhematologic toxicities
| Adverse event . | Cohort . | ||||||
|---|---|---|---|---|---|---|---|
| A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | Total . | |
| Febrile neutropenia | 5 | 4 | 3 | 5 | 5 | 5 | 27 |
| Infection and neutropenia | 3 | 1 | 2 | 5 | 2 | 2 | 15 |
| LFT abnormality | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Rash | 0 | 0 | 1 | 0 | 1 | 0 | 2 |
| GI toxicity | 0 | 1 | 0 | 0 | 3 | 5 | 9 |
| Mucositis | 1 | 3 | 2 | 6 | |||
| Enteritis | 1 | 1 | |||||
| Colitis | 4 | 4 | |||||
| Adverse event . | Cohort . | ||||||
|---|---|---|---|---|---|---|---|
| A1 . | B1 . | A2 . | B2 . | A3 . | B3 . | Total . | |
| Febrile neutropenia | 5 | 4 | 3 | 5 | 5 | 5 | 27 |
| Infection and neutropenia | 3 | 1 | 2 | 5 | 2 | 2 | 15 |
| LFT abnormality | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Rash | 0 | 0 | 1 | 0 | 1 | 0 | 2 |
| GI toxicity | 0 | 1 | 0 | 0 | 3 | 5 | 9 |
| Mucositis | 1 | 3 | 2 | 6 | |||
| Enteritis | 1 | 1 | |||||
| Colitis | 4 | 4 | |||||
The most common grade 3 nonhematologic toxicities are shown (there were no grade 4 nonhematologic toxicities). The number of subjects with toxicity is shown for each cohort. In cohort B3, two patients experienced both mucositis and colitis—one had a history of Crohn disease. LFT abnormality indicates liver function test abnormality (serum transaminitis or hyperbilirubinemia).
Time to blood count recovery
| Cohort . | CR . | PR . | ||||
|---|---|---|---|---|---|---|
| Subjects . | Platelets . | ANC . | Subjects . | Platelets . | ANC . | |
| No. . | Days, median (range) . | No. . | Days, median (range) . | |||
| A1 | 3 | 22 (21-23) | 21 (21-39) | 2 | 22 (20-24) | 34 (27-41) |
| B1 | 3 | 22 (20-26) | 30 (24-35) | 1 | 23 (23-23) | 30 (30-30) |
| A2 | 2 | 24 (22-26) | 26 (24-28) | 2 | 23 (21-24) | 32 (28-35) |
| B2 | 2 | 29 (22-36) | 33 (30-36) | 1 | 23 (23-23) | 22 (22-22) |
| A3 | 3 | 22 (20-26) | 23 (23-27) | 2 | 27 (25-28) | 27 (21-33) |
| B3 | 4 | 24 (21-27) | 23 (19-29) | 1 | 28 (28-28) | 37 (37-37) |
| All | 17 | 22 (20-36) | 24 (19-39) | 9 | 24 (20-28) | 30 (21-41) |
| Cohort . | CR . | PR . | ||||
|---|---|---|---|---|---|---|
| Subjects . | Platelets . | ANC . | Subjects . | Platelets . | ANC . | |
| No. . | Days, median (range) . | No. . | Days, median (range) . | |||
| A1 | 3 | 22 (21-23) | 21 (21-39) | 2 | 22 (20-24) | 34 (27-41) |
| B1 | 3 | 22 (20-26) | 30 (24-35) | 1 | 23 (23-23) | 30 (30-30) |
| A2 | 2 | 24 (22-26) | 26 (24-28) | 2 | 23 (21-24) | 32 (28-35) |
| B2 | 2 | 29 (22-36) | 33 (30-36) | 1 | 23 (23-23) | 22 (22-22) |
| A3 | 3 | 22 (20-26) | 23 (23-27) | 2 | 27 (25-28) | 27 (21-33) |
| B3 | 4 | 24 (21-27) | 23 (19-29) | 1 | 28 (28-28) | 37 (37-37) |
| All | 17 | 22 (20-36) | 24 (19-39) | 9 | 24 (20-28) | 30 (21-41) |
Data are shown for responders. By definition, all responders had a complete hematologic response. There were no statistically significant differences in the time of nadir between treatment arms, dose levels, or cohorts.
Assessment of efficacy
Of the 30 subjects enrolled, 27 (90%) responded to the single induction cycle of study therapy: 17 CR (57%) and 10 PR (33%). All subjects with a PR had a complete hematologic response with a median bone marrow blast count of 7% before starting a second induction. Of the patients with PR, 8 achieved remission with a second induction, bringing the overall CR rate to 83% (Table 5). The high conversion rate of subjects with PR (8 of 10), suggests that the PRs were not merely cosmetic. Selection of a second induction regimen was made by the treating physician and for subjects with a PR, therapies included “5 + 2” infusional cytarabine (100 mg/m2 for 5 days) with daunorubicin (60 mg/m2 × 2 doses; 5 + 2, 5 subjects), mitoxantrone, etoposide, cytarabine34 (MEC, 2 subjects), high-dose cytarabine (3000 mg/m2 q12 hours every other day for a total of 6 doses; HiDAC, 1 subject), a phase 1 clinical trial of an oral multikinase inhibitor (1 subject) and one subject with 5% bone marrow blasts went directly to allo-HSCT. Subjects with no response (NR) to the single study induction received a second induction with MEC (3 subjects), but none resulted in remission.
Treatment response
| Response . | CR . | PR . | NR . | CR% . |
|---|---|---|---|---|
| First induction | 17 | 10 | 3 | 57% |
| Next therapy | 25 | 0 | 5 | 83% |
| Response . | CR . | PR . | NR . | CR% . |
|---|---|---|---|---|
| First induction | 17 | 10 | 3 | 57% |
| Next therapy | 25 | 0 | 5 | 83% |
Thirty subjects were treated and 17 met the criteria for CR after the first induction. Of the 10 subjects with PR after the first induction, 8 met the criteria for CR after their next treatment. Subjects who did not respond to the first induction were also refractory to a second induction attempt.
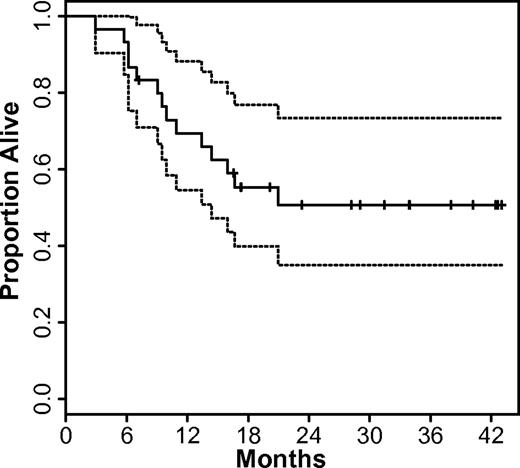
Because of the low power of our phase 1 design, no significant difference in response rate was identified among the treatment arms, cohorts, or dose levels. CR occurred in 13 of 15 patients (87%) treated with the 1-hour infusion schedule, including 8 patients (53%) who had a CR after 1 induction cycle. When decitabine was administered as a continuous infusion, 12 of 15 patients (80%) achieved CR, including 9 patients (60%) who had a CR after 1 induction cycle. With a median follow-up of 32 months, 16 of 30 subjects (53%) are alive in CR. Of the 14 subjects who died, 3 (10%) subjects died in remission from complications of allo-HSCT and the remainder (37%) died with relapsed or refractory AML. The median survival for the study group has not been reached, but the lower bound of the 95% confidence interval is 15 months (Figure 2).
Overall survival of study subjects. The Kaplan-Meier estimate of overall survival (solid line) and 95% confidence bounds (dashed lines) are shown for all subjects.
Overall survival of study subjects. The Kaplan-Meier estimate of overall survival (solid line) and 95% confidence bounds (dashed lines) are shown for all subjects.
Accelerated platelet recovery kinetics
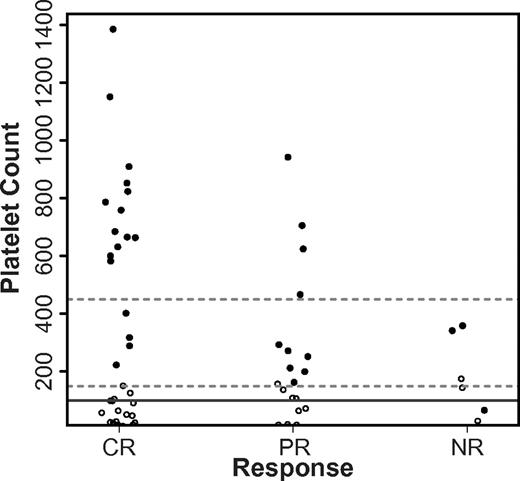
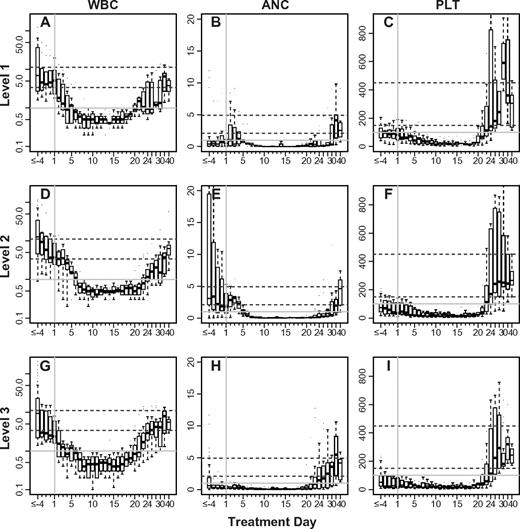
The platelet count for 29 of the 30 subjects treated returned to the normal range (≥ 150 × 103/μL) after a single induction (Figure 3). The pace of platelet recovery was generally brisk, requiring a median of 22 days for the platelet level to rise above 105/μL (Table 4). Strikingly, the peak platelet count for 17 subjects (all responders) rose above the normal range during their convalescence from decitabine-primed induction. Although the platelets rose above 750 000/μL in 8 subjects and as high as 1.4 million/μL in one subject, all counts returned to the normal range within a week and there were no thrombotic complications. Neutrophil recovery (≥ 103/μL) typically precedes platelet recovery after induction chemotherapy for AML, but we found that 22 of the 30 (73%) subjects we treated recovered their platelet count first.35 This was true both for responders (11 of 17 subjects with CR, 8 of 10 subjects with PR) and nonresponders (3 of 3 subjects with NR). Overall, 28 subjects normalized both their ANC (≥ 1500/μL) and platelet count, whereas only 3 subjects met these criteria before starting decitabine-primed induction. We did not identify differences in the blood count recovery between subjects treated within the different arms or cohorts (Figure 4).
Platelet count before and after decitabine-primed induction. Shown is a stripchart of pretreatment platelet counts (○) and the maximal platelet count observed at time of hematopoietic recovery (●) for subjects achieving CR, PR, or NR (NR). Dashed lines represent the extents of the normal range and the solid line is the platelet count required for CR or PR.
Platelet count before and after decitabine-primed induction. Shown is a stripchart of pretreatment platelet counts (○) and the maximal platelet count observed at time of hematopoietic recovery (●) for subjects achieving CR, PR, or NR (NR). Dashed lines represent the extents of the normal range and the solid line is the platelet count required for CR or PR.
Blood count recovery after decitabine-primed induction. Box plots showing the median (line), interquartile (box extents), and range (whiskers) of WBC (A,D,G), ANC (B,E,H), and platelet count (C,F,I) for responding subjects (CR or PR) enrolled on the phase 1 study. Each row of panels represent subjects treated at the dose level labeled on left: Level 1, 3 days of decitabine (A-C); Level 2, 5 days of decitabine (D-F); and Level 3, 7 days of decitabine (G-I). The dashed lines represent the extents of the normal range. Blood count data are summarized daily between days −3 and +21. Days earlier than day 3 are binned. Between day 22 through day 30, data were binned every 2 days and after day 30, 5-day bins were used. Day +1 is indicated with a vertical gray line.
Blood count recovery after decitabine-primed induction. Box plots showing the median (line), interquartile (box extents), and range (whiskers) of WBC (A,D,G), ANC (B,E,H), and platelet count (C,F,I) for responding subjects (CR or PR) enrolled on the phase 1 study. Each row of panels represent subjects treated at the dose level labeled on left: Level 1, 3 days of decitabine (A-C); Level 2, 5 days of decitabine (D-F); and Level 3, 7 days of decitabine (G-I). The dashed lines represent the extents of the normal range. Blood count data are summarized daily between days −3 and +21. Days earlier than day 3 are binned. Between day 22 through day 30, data were binned every 2 days and after day 30, 5-day bins were used. Day +1 is indicated with a vertical gray line.
Analysis of DNA methylation
We assessed decitabine pharmacodynamics by comparing DNA methylation in bone marrow and blood specimens obtained before initiating therapy (screening) and after the final dose of decitabine (postpriming). We analyzed several different cell populations to assess how cell source affected our measures. After bisulfite conversion of the DNA, we used quantitative PCR to measure methylation at genomic loci that are pathologically methylated in AML (CDKN2B) or constitutively methylated in hematopoietic tissues (LINE1 and HIST1H2AA).33 We used plasmid standards to convert the qPCR threshold cycle to copy number and then normalized for the total copies of template DNA using methylation insensitive primer sets to amplify repetitive and single-copy genomic regions (ALUC4 and GAPDH). We observed only low-level methylation of CDKN2B in a subset of our specimens, so we restricted our analyses to HIST1H2AA and LINE1.
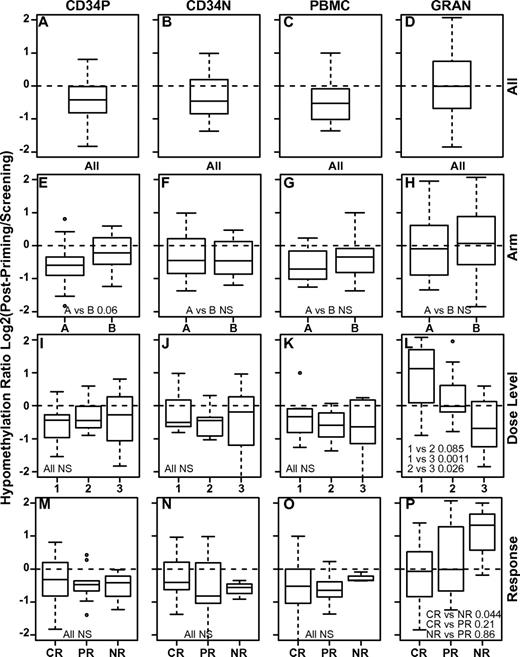
To compare the extent of DNA hypomethylation across subjects and cell types, we first calculated a hypomethylation ratio by dividing the normalized methylation observed after decitabine priming (postpriming) with that found at the time of screening (screening). All of the mononuclear cell populations (PBMCs, CD34P, and CD34N) underwent a similar degree of DNA hypomethylation, but granulocytes as a group were significantly less hypomethylated by decitabine (P ≤ .002, t test; Table 6 and Figure 5A-D). We observed DNA hypomethylation at all dose levels (Figure 5I-L), but found a clear dose response only within granulocytes. This result was partially due to a paradoxical increase in methylation within the granulocytes of subjects treated at the lowest dose level (Figure 5L). Although we presume that the increased methylation results from variation in the granulocyte cell population rather than a true change in DNA methylation, we could find no association between this effect and the pre- or postpriming blood counts.
Decitabine-induced change in DNA methylation
| Cell population locus . | BMCD34P . | BMCD34N . | BMMC Change in methylation (%), significance . | PBMC . | GRANS . |
|---|---|---|---|---|---|
| HIST1H2AA | −32, P = .001 | −16, P = .075 | −30, P = .005 | −25, P = .020 | 8, P = .67 |
| LINE1 | −19, P = .068 | −24, P = .012 | −46, P = .012 | −28, P = .001 | −14, P = .11 |
| Cell population locus . | BMCD34P . | BMCD34N . | BMMC Change in methylation (%), significance . | PBMC . | GRANS . |
|---|---|---|---|---|---|
| HIST1H2AA | −32, P = .001 | −16, P = .075 | −30, P = .005 | −25, P = .020 | 8, P = .67 |
| LINE1 | −19, P = .068 | −24, P = .012 | −46, P = .012 | −28, P = .001 | −14, P = .11 |
For each specimen type, DNA methylation at the HIST1H2AA locus and LINE1 repeat class was compared before (screening) and after (post-priming) decitabine priming. The negative -fold change reflects the reduction in DNA methylation induced by decitabine. Statistical significance (P) of the -fold change was calculated using a one-sided Student t test for the paired specimens.
Early pharmacodynamic analyses of DNA hypomethylation. Shown is the hypomethylation ratio induced by decitabine in 4 different cell populations: CD34P indicates bone marrow CD34+ mononuclear cells (A,E,L,M); CD34N, bone marrow CD34-depleted mononuclear cells (B,F,J,N); PBMCs (C,G,K,O); and GRAN, peripheral blood granulocytes (D,H,L,P). The hypomethylation ratio is calculated as the ratio of DNA methylation detected just after completion of decitabine priming to that detected before beginning decitabine. The hypomethylation ratio is shown for all subjects (A-D), for subjects treated on each arm (E-H), at each dose level (I-L), and according to treatment response: CR, PR, or NR (M-P). Paired t test P values are indicated (NS indicates that the P value was nonsignificant).
Early pharmacodynamic analyses of DNA hypomethylation. Shown is the hypomethylation ratio induced by decitabine in 4 different cell populations: CD34P indicates bone marrow CD34+ mononuclear cells (A,E,L,M); CD34N, bone marrow CD34-depleted mononuclear cells (B,F,J,N); PBMCs (C,G,K,O); and GRAN, peripheral blood granulocytes (D,H,L,P). The hypomethylation ratio is calculated as the ratio of DNA methylation detected just after completion of decitabine priming to that detected before beginning decitabine. The hypomethylation ratio is shown for all subjects (A-D), for subjects treated on each arm (E-H), at each dose level (I-L), and according to treatment response: CR, PR, or NR (M-P). Paired t test P values are indicated (NS indicates that the P value was nonsignificant).
We next investigated the relative hypomethylating activity of decitabine delivered by 1-hour infusion (Arm A) or continuous infusion (Arm B). As a whole, the 2 arms had comparable activity (Figure 5E-H), but there was a trend toward more hypomethylation in the most immature CD34+ bone marrow population on Arm A (P = .06). This difference was significant (P = .042) when we restricted the analysis to subjects achieving a CR or PR to the study induction. The study was not sufficiently powered to identify significant differences in the hypomethylation ratio linked to other parameters such as karyotype, FLT3 status, pretreatment WBC, or the use of hydroxyurea (not shown).
Although DNA hypomethylation is believed to underlie the clinical activity of decitabine, prior studies have not shown that the early pharmacodynamics of hypomethylation predict decitabine response.19,36-38 We also found no significant linkage between DNA hypomethylation within the mononuclear cell subpopulations and treatment response. As a group, hypomethylation in granulocytes was greater for subjects who achieved CR compared with those who had NR (Figure 5P, P = .044), but we could not identify a threshold of hypomethylation that predicted response for individual subjects using the Fisher exact test (P > .075 for all cutoffs). Because the response rate was so unbalanced (only 3 subjects with NR), these results should be viewed as preliminary despite the statistical significance of the grouped analysis. Similar analyses of the late pharmacodynamics comparing DNA methylation at screening with that at the time of count recovery did not provide additional insights (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Discussion
CR is requisite for the cure of AML, and this study demonstrates that it is safe to combine the epigenetic modifier decitabine with full-dose, standard, cytotoxic chemotherapy as an approach to improving remission rates. We did not identify an MTD in the present study, although there was more GI toxicity at the highest dose level of decitabine. The overall CR rate of 83% is encouraging. The epigenetic mechanism of decitabine may complement the cytotoxic effects of standard induction chemotherapy, and we observed DNA hypomethylation within the most relevant cell population (bone marrow CD34+ cells) at all dose levels. Therefore, decitabine could act as a chemosensitizer by reactivating TSG expression during the window of exposure to cytotoxic induction chemotherapy. A larger trial will be required to definitively demonstrate the clinical utility of epigenetic priming.
One concern about combining decitabine with full-intensity induction chemotherapy was the risk of significantly prolonging the duration of myelosuppression. Instead, we found that the time to neutrophil recovery after decitabine-primed induction was similar (median 24 days) to that for standard induction chemotherapy alone.35 Surprisingly, the time to platelet recovery (median 22 days) was even shorter than we typically find with cytarabine-containing induction chemotherapy. Time to platelet recovery is associated with improved survival in ALL, and in AML, the time to CR may predict survival.39-41 Because the kinetics of hematopoietic recovery was brisk in our study group, the median time to CR was 28 days.
The most clinically significant nonhematologic toxicity was grade 3 GI toxicity related to enteritis/colitis with fever during the period of neutropenia. All grade 3 GI toxicities resolved with blood count recovery and lasted < 7 days for all but 2 subjects (DLTs, for whom it lasted 8 days). Although an MTD was not identified, the GI toxicity appeared to be dose related and was most significant for dose level 3 (7-day decitabine priming, 140 mg/m2) in both arms. For this reason, a subsequent phase 2 study will be conducted using the 5-day dosing schedule.
Although this was a phase 1 trial, the overall CR rate (83%) was encouraging considering that the study population was predominantly composed of older subjects (67% ≥ 50 years) with adverse molecular characteristics (57%) and at least one adverse risk feature (83%). This CR rate is comparable to a recent Eastern Cooperative Oncology Group study demonstrating the superiority of dose-intensive daunorubicin over standard induction chemotherapy in AML.42 Whereas daunorubicin intensification proved valuable for younger subjects (< 50 years) with an intermediate-risk karyotype, older subjects (≥ 50 years) and those with unfavorable karyotype or an FLT3-ITD did not benefit. Most responders (70%) on our phase 1 study had at least one of these adverse characteristics, suggesting that decitabine-primed induction should be explored as a complementary approach to anthracycline dose intensification (Table 7).
Subject characteristics by response to a single decitabine-primed induction chemotherapy cycle
| Characteristic . | Response . | ||
|---|---|---|---|
| CR . | PR . | NR . | |
| Median age, y (range) | 57 (37-60) | 50 (23-59) | 52 (50-58) |
| Sex, M:F | 8:9 | 7:3 | 1:2 |
| FLT3-mutated, no. subjects (%) | 2 (12) | 2 (20) | 0 (0) |
| Antecedent hematologic disorder, no. subjects (%) | 3 (18) | 4 (40) | 1 (33) |
| Prior chemotherapy, no. subjects (%) | 2 (12) | 2 (20) | 0 (0) |
| WBC, ×109/L | |||
| Median (range) | 16 (2.2-133) | 33 (1.5-211) | 2.6 (2.4-30) |
| > 50, no. subjects (%) | 4 (24) | 4 (40) | 0 (0) |
| > 100, no. subjects (%) | 2 (12) | 3 (30) | 0 (0) |
| Platelet recovery peak, ×109/L | |||
| Median (range) | 670 (223-1386) | 282 (163-943) | 342 (66-359) |
| > 450, no. subjects (%) | 13 (76) | 4 (40) | 0 (0) |
| Cytogenetics, no. subjects (%) | |||
| Normal | 7 (41) | 3 (30) | 2 (67) |
| −7/del(7q) | 4 (24) | 1 (10) | 1 (33) |
| Complex | 2 (12) | 3 (30) | 0 (0) |
| abn(12p) | 2 (12) | 0 (0) | 0 (0) |
| abn(11q23) | 4 (24) | 0 (0) | 0 (0) |
| +8 | 0 (0) | 3 (30) | 0 (0) |
| t(6;9)(p23;q34) | 1 (6) | 0 (0) | 0 (0) |
| t(2;11)(q31;p15) | 0 (0) | 1 (10) | 0 (0) |
| ≥ 1 Adverse feature, no. subjects (%) | 15 (88) | 8 (80) | 2 (67) |
| Characteristic . | Response . | ||
|---|---|---|---|
| CR . | PR . | NR . | |
| Median age, y (range) | 57 (37-60) | 50 (23-59) | 52 (50-58) |
| Sex, M:F | 8:9 | 7:3 | 1:2 |
| FLT3-mutated, no. subjects (%) | 2 (12) | 2 (20) | 0 (0) |
| Antecedent hematologic disorder, no. subjects (%) | 3 (18) | 4 (40) | 1 (33) |
| Prior chemotherapy, no. subjects (%) | 2 (12) | 2 (20) | 0 (0) |
| WBC, ×109/L | |||
| Median (range) | 16 (2.2-133) | 33 (1.5-211) | 2.6 (2.4-30) |
| > 50, no. subjects (%) | 4 (24) | 4 (40) | 0 (0) |
| > 100, no. subjects (%) | 2 (12) | 3 (30) | 0 (0) |
| Platelet recovery peak, ×109/L | |||
| Median (range) | 670 (223-1386) | 282 (163-943) | 342 (66-359) |
| > 450, no. subjects (%) | 13 (76) | 4 (40) | 0 (0) |
| Cytogenetics, no. subjects (%) | |||
| Normal | 7 (41) | 3 (30) | 2 (67) |
| −7/del(7q) | 4 (24) | 1 (10) | 1 (33) |
| Complex | 2 (12) | 3 (30) | 0 (0) |
| abn(12p) | 2 (12) | 0 (0) | 0 (0) |
| abn(11q23) | 4 (24) | 0 (0) | 0 (0) |
| +8 | 0 (0) | 3 (30) | 0 (0) |
| t(6;9)(p23;q34) | 1 (6) | 0 (0) | 0 (0) |
| t(2;11)(q31;p15) | 0 (0) | 1 (10) | 0 (0) |
| ≥ 1 Adverse feature, no. subjects (%) | 15 (88) | 8 (80) | 2 (67) |
The majority of responders had at least 1 adverse prognostic feature.
A more complete understanding of the tumor-specific pharmacodynamics of decitabine would facilitate its future incorporation into multidrug chemotherapeutic regimens. Because decitabine has S-phase activity, we postulated that delivering it by continuous infusion could expose a more quiescent cell population than pulsed administration. We directly compared the in vivo hypomethylating activity of infusional and pulsed decitabine and observed similar global and gene-specific DNA hypomethylation in all cell populations studied. Nonetheless, we did find a trend toward greater hypomethylation with pulsed decitabine (Arm A) in the most immature, and presumably the most quiescent, subpopulation studied (CD34P), which was contrary to our original hypothesis.
Although we found a statistical linkage between DNA hypomethylation in granulocytes and treatment response, we could not identify a threshold of hypomethylation that predicted response for any individual subject. Our pharmacodynamic analysis was confined to genomic loci (HIST1H2AA, LINE1 repetitive elements) that were unlikely to be involved in AML pathogenesis, and it is possible that a more comprehensive analysis of DNA methylation may identify DNA methylation biomarkers that predict response to decitabine therapy. Because of the variability between subjects, a definitive pharmacodynamic comparison of different decitabine dose and delivery schedules awaits a larger study.
This phase 1 study demonstrates that epigenetic priming of standard induction chemotherapy can be safely administered in an attempt to improve the response rates for patients with less-than-favorable risk AML. These findings provide the foundation for a subsequent phase 2 study to more clearly characterize the biologic and clinical activity of decitabine priming during AML induction chemotherapy. Although we did not identify an MTD, there was more GI toxicity at the 7-day dose level on both the continuous infusion and short infusion arms. Therefore, the 5-day priming schedule was chosen for a subsequent phase 2 study. Epigenetic silencing of TSG expression is a hallmark of cancer and is prototypically linked to aberrant DNA methylation. We hope that this study defining the feasibility of epigenetic priming in AML will translate to advances in the treatment of more common forms of cancer.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Tim Talbot and Elliot Coburn for data collection and to Emily McDonald for technical assistance.
This research support was provided by the Cornell Clinical and Translational Science Center (to J.M.S.) and by the Belfer Family (to J.M.S.).
National Institutes of Health
Authorship
Contribution: J.M.S. and E.J.F. conceived and designed the study; T.J.C., C.M.I., and L.V. provided administrative support; G.J.R., E.K.R., E.J.F., U.S.G., and S.A.M. provided study materials or patients; J.M.S., T.J.C., L.V., M.M., F.B., J.R.B., and E.W.M. collected and assembled data; J.M.S. analyzed and interpreted data; J.M.S., E.J.F., and E.W. wrote the manuscript; and all authors provided final approval of the manuscript.
Conflict-of-interest disclosure: MGI Pharma (Bloomington, MN) and subsequently Eisai (Woodcliff Lake, MN) provided decitabine. The authors declare no other competing financial interests.
Correspondence: Joseph M. Scandura, MD, PhD, Weill-Cornell Medical College, 1300 York Ave, C610D, New York, NY 10065; e-mail: jms2003@med.cornell.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal