Abstract
Systemic mastocytosis (SM) either presents as a malignant neoplasm with short survival or as an indolent disease with normal life expectancy. In both instances, neoplastic mast cells (MCs) harbor D816V-mutated KIT, suggesting that additional oncogenic mechanisms are involved in malignant transformation. We here describe that Lyn and Btk are phosphorylated in a KIT-independent manner in neoplastic MCs in advanced SM and in the MC leukemia cell line HMC-1. Lyn and Btk activation was not only detected in KIT D816V-positive HMC-1.2 cells, but also in the KIT D816V-negative HMC-1.1 subclone. Moreover, KIT D816V did not induce Lyn/Btk activation in Ba/F3 cells, and deactivation of KIT D816V by midostaurin did not alter Lyn/Btk activation. siRNAs against Btk and Lyn were found to block survival in neoplastic MCs and to cooperate with midostaurin in producing growth inhibition. Growth inhibitory effects were also obtained with 2 targeted drugs, dasatinib which blocks KIT, Lyn, and Btk activation in MCs, and bosutinib, a drug that deactivates Lyn and Btk without blocking KIT activity. Together, KIT-independent signaling via Lyn/Btk contributes to growth of neoplastic MCs in advanced SM. Dasatinib and bosutinib disrupt Lyn/Btk-driven oncogenic signaling in neoplastic MC, which may have clinical implications and explain synergistic drug interactions.
Introduction
Systemic mastocytosis (SM) is a myeloid neoplasm characterized by increased survival and accumulation of neoplastic mast cells (MCs) in one or more internal organs.1-6 Indolent and aggressive variants of SM have been described.1-9 Patients with indolent SM (ISM) may suffer from mediator-related symptoms and/or cosmetic problems, but otherwise have a normal or near-normal life expectancy.1-9 By contrast, patients with aggressive SM (ASM) or MC leukemia (MCL) have a grave prognosis with short survival.1-9 In these patients, the response to conventional cytoreductive drugs is poor. Therefore, these patients are candidates for experimental therapy. Indeed, several attempts have been made to identify novel therapeutic targets in neoplastic MCs.1-3,5,10-12
In a majority of all patients with SM including ISM, ASM, or MCL, KIT D816V is detectable.13-16 This mutation is associated with ligand-independent activation of KIT, and is considered to contribute to growth of MCs in SM.17 Therefore, attempts have been made to identify drugs interfering with the tyrosine kinase (TK) activity of KIT D816V.5,10-12,18-24 These drugs include midostaurin (PKC412), nilotinib, and dasatinib.18-24 However, despite impressive effects on cell lines,19 these drugs, when applied as single agents, may not be sufficient to induce long-lasting complete responses in patients with ASM or MCL.22 More recently, we have shown that combinations of various KIT TK-inhibitors (TKI) produce synergistic growth-inhibitory effects on neoplastic MCs.19,23,24 However, in neoplastic MCs bearing KIT D816V, only the combination “dasatinib + midostaurin” produced clear synergistic effects.24
Several observations suggest that KIT D816V alone is not a fully transforming oncoprotein. First, as mentioned above, the mutant is not only detectable in ASM or MCL, but also in the majority of patients with ISM, where the disease course remains stable over years or even decades.1-6,13-16,25 In addition, several cases of ASM or MCL present without KIT D816V, or the mutant even disappears during progression, sometimes after treatment with KIT-targeting drugs (clonal selection).22 Moreover, although inducing differentiation and maturation, KIT D816V per se is unable to promote oncogenic proliferation when inducibly expressed in Ba/F3 cells.25 Similarly, KIT D816V-transgenic mice usually develop slowly expanding, indolent accumulations of MCs in tissues, but rarely develop a malignant MC disease.26
All these observations suggest that apart from KIT D816V, other gene defects and pro-oncogenic hits may be responsible for disease evolution and SM progression to ASM or MCL. Therefore, we screened for additional KIT-independent oncogenic events and signaling molecules in neoplastic MCs. In the present study, we have identified 2 KIT D816V-independent signaling molecules, Lyn and Btk, which are expressed in activated form in neoplastic MCs in advanced SM. Both molecules were identified as targets of dasatinib and bosutinib in neoplastic MCs.
Methods
Reagents
Dasatinib was kindly provided by Dr F. Y. Lee (Bristol-Myers Squibb; Oncology Drug Discovery, Princeton, NJ). Midostaurin (PKC412) was kindly provided by Dr E. Buchdunger and Dr P. W. Manley (Novartis Pharma AG). Bosutinib (SKI-606) was kindly provided by Ordis Biomed or purchased from Selleck. Stock solutions (10mM) were prepared by dissolving in DMSO (Merck). RPMI 1640 medium and FCS were from PAA Laboratories, IMDM from Invitrogen Life Technologies, recombinant SCF from Strathmann Biotech, and 3H-thymidine from Amersham.
Cell lines and culture of primary cells
The MC leukemia cell line HMC-127 was kindly provided by Dr Joseph H. Butterfield (Mayo Clinic, Rochester, MN). Two subclones were used: HMC-1.1 harboring KIT V560G but not KIT D816V, and HMC-1.2 harboring KIT V560G as well as KIT D816V.19,28 HMC-1 were grown in IMDM plus 10% FCS, l-glutamine, and antibiotics (37°C, 5% CO2). Other cell lines used in this study were K562 and KU812, HL60, KG1, HEL, Raji, and TOM-1. These cell lines were grown in RPMI 1640 medium plus 10% FCS, l-glutamine, and antibiotics (37°C, 5% CO2). Ba/F3 cells stably expressing BCR/ABL T315I (Ba/F3p210T315I) were kindly provided by Dr M. Deininger (Oregon Health & Science University Cancer Institute, Portland, OR) and kept in RPMI 1640 plus 10% FCS. Ba/F3 cells with doxycycline-inducible expression of wt KIT (Ton.Kit.wt) or KIT D816V (Ton.Kit.D816V)19,25 were maintained in RPMI 1640 plus 10% FCS and IL-3 as described.19,25 Expression of KIT D816V or wt KIT was induced by exposure to doxycycline (1 μg/mL) for 12 hours.19,25
Design and application of siRNA
siRNAs directed against Lyn, Btk, Hck, or luciferase (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were synthesized in 2′-deprotected, duplexed, and desalted form by Dharmacon and transfected into HMC-1.2 using lipofectin (Invitrogen) as described.29 In brief, cells were exposed to lipofectin (75nM) and siRNA (100-200nM) in serum-free IMDM at 37°C for 4 hours. Cells were then washed and resuspended in IMDM and 10% FCS, and incubated for another 12-44 hours. In select experiments, shRNA against JAK2 was expressed in HMC-1.2 cells by lentivirus-mediated gene transfer, following a published protocol.30 For knockdown of JAK2, a pLKO.1 clone containing a shRNA targeting human JAK2 (5′-GCAGAATTAGCAAACCTTATA-3′) was used (Open Biosystems). Recombinant VSV-G pseudotyped lentiviruses were produced as described.30 HMC-1 cells were transduced in the presence of polybrene (7 μg/mL) and selected with puromycin (2 μg/mL, 48 hours). Knockdown of JAK2 was confirmed by immunoblotting using mAb D2E12 directed against JAK2 (supplemental Table 2).
Isolation of primary neoplastic cells
In 14 patients with SM, isolated MCs were examined (ISM, n = 7; smouldering SM [SSM], n = 1; ASM, n = 4; MCL, n = 2; Table 1). In 11 of 14 patients, KIT D816V was detectable. Neoplastic cells were isolated from BM aspirates as described.19,24 Control samples were obtained from patients with acute myeloid leukemia (AML, n = 5), chronic myeloid leukemia (CML, n = 5), chronic neutrophilic leukemia (CNL, n = 1), and normal BM (n = 3). Patients' characteristics are shown in supplemental Table 3. The study was approved by the institutional review board (Medical University of Vienna, Vienna, Austria) and conducted in accordance with the Declaration of Helsinki. Informed consent was obtained in each case.
Patients' characteristics
| No. . | Age, y . | Sex, M/F . | Diagnosis, SM variant . | KIT D816V . | Serum tryptase, ng/mL . | BM MC infiltration grade, %* . |
|---|---|---|---|---|---|---|
| 01 | 43 | M | ISM | − | 20 | 5 |
| 02 | 50 | M | ISM | + | 54 | 10 |
| 03 | 32 | M | ISM | + | 36 | 5 |
| 04 | 49 | F | ISM | + | 54 | 5 |
| 05 | 39 | F | ISM | + | 17 | 5 |
| 06 | 39 | M | ISM | + | 16 | 5 |
| 07 | 53 | M | ISM | + | 60 | 5 |
| 08 | 43 | M | ISM | + | 69 | 20 |
| 09 | 23 | F | ISM | + | 14 | 5 |
| 10 | 47 | F | ISM | + | 65 | 15 |
| 11 | 79 | M | ISM | + | 31 | 5 |
| 12 | 58 | F | ISM | − | 50 | 1 |
| 13 | 66 | M | ISM | + | 65 | 15 |
| 14 | 68 | F | ISM | + | 17 | 5 |
| 15 | 53 | F | ISM | + | 14 | 5 |
| 16 | 33 | M | ISM | + | 67 | 2 |
| 17 | 63 | F | ISM | + | 30 | 1 |
| 18 | 44 | F | ISM | + | 175 | 10 |
| 19 | 53 | M | ISM/CLL | + | 290 | 20 |
| 20 | 45 | F | ISM | + | 938 | 15 |
| 21 | 56 | M | ISM | + | 650 | 20 |
| 22 | 40 | M | SSM | + | nt | 35 |
| 23 | 42 | M | SSM | + | 630 | 40 |
| 24 | 60 | F | SSM | + | 188 | 40 |
| 25 | 55 | F | SSM | + | 133 | 20 |
| 26 | 64 | F | SSM | + | nt | 15 |
| 27 | 61 | F | ASM | − | nt | 10 |
| 28 | 48 | F | ASM | + | nt | 25 |
| 29 | 49 | F | ASM | + | nt | 30 |
| 30 | 43 | M | ASM | + | 100 | 40 |
| 31 | 46 | M | ASM/MDS | − | 421 | 30 |
| 32 | 73 | M | ASM/CMML | + | 654 | 40 |
| 33 | 71 | F | ASM/CMML | + | 295 | 60 |
| 34 | 44 | M | MCL | + | 308 | 30 |
| 35 | 66 | M | MCL/MDS | + | 763 | 70 |
| 36 | 35 | F | MCL | − | 489 | 21 |
| 37 | 26 | M | MCL | − | 2500 | 90 |
| 38 | 60 | M | MCL | + | 375 | 100 |
| No. . | Age, y . | Sex, M/F . | Diagnosis, SM variant . | KIT D816V . | Serum tryptase, ng/mL . | BM MC infiltration grade, %* . |
|---|---|---|---|---|---|---|
| 01 | 43 | M | ISM | − | 20 | 5 |
| 02 | 50 | M | ISM | + | 54 | 10 |
| 03 | 32 | M | ISM | + | 36 | 5 |
| 04 | 49 | F | ISM | + | 54 | 5 |
| 05 | 39 | F | ISM | + | 17 | 5 |
| 06 | 39 | M | ISM | + | 16 | 5 |
| 07 | 53 | M | ISM | + | 60 | 5 |
| 08 | 43 | M | ISM | + | 69 | 20 |
| 09 | 23 | F | ISM | + | 14 | 5 |
| 10 | 47 | F | ISM | + | 65 | 15 |
| 11 | 79 | M | ISM | + | 31 | 5 |
| 12 | 58 | F | ISM | − | 50 | 1 |
| 13 | 66 | M | ISM | + | 65 | 15 |
| 14 | 68 | F | ISM | + | 17 | 5 |
| 15 | 53 | F | ISM | + | 14 | 5 |
| 16 | 33 | M | ISM | + | 67 | 2 |
| 17 | 63 | F | ISM | + | 30 | 1 |
| 18 | 44 | F | ISM | + | 175 | 10 |
| 19 | 53 | M | ISM/CLL | + | 290 | 20 |
| 20 | 45 | F | ISM | + | 938 | 15 |
| 21 | 56 | M | ISM | + | 650 | 20 |
| 22 | 40 | M | SSM | + | nt | 35 |
| 23 | 42 | M | SSM | + | 630 | 40 |
| 24 | 60 | F | SSM | + | 188 | 40 |
| 25 | 55 | F | SSM | + | 133 | 20 |
| 26 | 64 | F | SSM | + | nt | 15 |
| 27 | 61 | F | ASM | − | nt | 10 |
| 28 | 48 | F | ASM | + | nt | 25 |
| 29 | 49 | F | ASM | + | nt | 30 |
| 30 | 43 | M | ASM | + | 100 | 40 |
| 31 | 46 | M | ASM/MDS | − | 421 | 30 |
| 32 | 73 | M | ASM/CMML | + | 654 | 40 |
| 33 | 71 | F | ASM/CMML | + | 295 | 60 |
| 34 | 44 | M | MCL | + | 308 | 30 |
| 35 | 66 | M | MCL/MDS | + | 763 | 70 |
| 36 | 35 | F | MCL | − | 489 | 21 |
| 37 | 26 | M | MCL | − | 2500 | 90 |
| 38 | 60 | M | MCL | + | 375 | 100 |
F indicates female; M, male; SM, systemic mastocytosis; MC, mast cell; ISM, indolent SM; CLL, chronic lymphatic leukemia; SSM smoldering SM; ASM, aggressive SM, MCL, mast cell leukemia; MDS, myelodysplastic syndrome; CMML, chronic myelomonocytic leukemia; and nt, not tested.
The percentage of infiltration of the BM by MCs was determined by tryptase IHC.
Western blot experiments
HMC-1 cells were incubated with dasatinib (0.01-1μM), midostaurin (1μM), bosutinib (0.01-5μM), LMF-A13 (50-100μM), or control medium at 37°C for 4-24 hours. In select experiments, HMC-1 cells were transfected with control siRNA (Luc) or siRNA directed against Btk, Lyn, or Hck. Ba/F3 cells inducibly expressing wt KIT (Ton.Kit.wt) or KIT D816V (Ton.Kit.D816V) were kept in control medium or in doxycycline (1 μg/mL) to induce KIT expression. In case of Ton.Kit.wt cells, KIT phosphorylation was induced by adding SCF (100 ng/mL). After incubation, cells were harvested and Western blotting was performed as described23,24 using Abs against total Lyn, p-LynTyr507, Btk, p-BtkTyr223, p-SrcTyr416, STAT5, p-STAT5, Hck, p-HckTyr411, Fyn, Fgr, KIT, JAK2, or β-actin (supplemental Table 2). Phosphorylation of Lyn, Hck, Fyn, Fgr, and KIT was confirmed by immunoprecipitation followed by immunoblotting with anti-phospho-Tyr Ab 4G10 (Upstate Biotechnology; work dilution 1:1000) as reported.23,24
IHC and ICC
In 29 patients with SM (ISM, n = 19; SSM, n = 4; ASM, n = 3; MCL, n = 3), expression of Lyn, p-LynTyr396, p-LynTyr507, Btk, and p-BtkTyr551 in MCs was examined on serial BM sections (paraffin-embedded, formalin-fixed, 2 μm) by IHC. The patients' characteristics are shown in Table 1. In addition, control cases (AML, CML, CNL, control BM) were examined. Indirect immunoperoxidase staining was performed as reported31-33 using Abs against tryptase (G3), p-LynTyr507, total Lyn, p-BtkTyr551, and total Btk (supplemental Table 2). Before staining, BM sections were pretreated by microwave oven. 3-amino-9-ethyl-carbazole was used as chromogen. Slides were counterstained in Mayer hemalaun. Immunocytochemistry (ICC) was performed using cytospin preparations of primary neoplastic MCs (n = 3) and HMC-1 cells as described.29 Abs against total Lyn, p-LynTyr507, p-LynTyr396, p-SrcTyr416, Btk, p-BtkTyr223, and p-BtkTyr551 (supplemental Table 2), as well as biotinylated anti–rabbit IgG (Biocare) were applied. As chromogen, streptavidin-alkaline-phosphatase complex (Biocare) was used. Ab reactivity was made visible using Neofuchsin (Nichirei). In select experiments, HMC-1 cells were incubated in control medium or dasatinib (1μM) for 4, 12, or 24 hours before Ab staining.
Measurement of 3H-thymidine uptake
Untransfected HMC-1 cells, HMC-1 cells transfected with siRNA (against Lyn, Btk, Hck, or Luc control siRNA), and primary cells isolated from patients with SM (n = 4) were incubated in control medium, midostaurin (0.01-1μM), or bosutinib (0.1-10μM) in 96-well culture plates (TPP) at 37°C for 24 or 48 hours. In a separate set of experiments, HMC-1 cells were cultured in the absence or presence of the Btk-inhibitor LFM-A13 (1-500μM; Calbiochem). After incubation of cells with various drugs, 0.5 μCi 3H-thymidine was added (37°C, 12 hours). Cells were then harvested on filter membranes (Packard Bioscience) in a Filtermate harvester (Packard Bioscience). Filters were then air-dried, and the bound radioactivity was counted in a β-counter (Top-Count NXT; Packard Bioscience). All experiments were performed in triplicate.
Evaluation of apoptosis by microscopy and TUNEL assay
In typical experiments, cells were incubated with various concentrations of Lyn-siRNA, Btk-siRNA, Hck-siRNA, control siRNA, or control medium at 37°C for 16 to 48 hours. Apoptotic cells were quantified on Wright-Giemsa–stained cytospin preparations. To further confirm apoptosis in HMC-1 cells exposed to siRNA, a TUNEL assay was performed using the In situ cell death detection kit–fluorescein (Roche) according to the recommendation of the manufacturer. Cells were analyzed with a Nikon Eclipse E-800 fluorescence microscope.
Evaluation of apoptosis by flow cytometry
For flow cytometric determination of apoptosis and viability, annexin V/propidium iodide staining was performed as described.19,23,24 In brief, HMC-1 cells were exposed to control medium or were transfected with control siRNA or siRNA against Lyn, Btk, or Hck. After 48 hours, cells were incubated with annexin V–FITC (Alexis Biochemicals) in binding buffer containing HEPES (10mM, pH 7.4), NaCl (140mM), and CaCl2 (2.5mM). Thereafter, propidium iodide (1 μg/mL) was added. Cells were then washed and analyzed by flow cytometry on a FACScan (BD Biosciences).
Drug affinity purification and MS
For drug-affinity binding and mass spectrometry (MS) experiments, HMC-1 cells and c-Dasatinib synthesized by WuXi PharmaTech, were used. Drug pulldown experiments were performed on total cell lysates from HMC-1 cells (both HMC-1.1 and HMC-1.2) according to a standard protocol.34,35 Tryptically digested proteins were analyzed by nanoLC tandem MS essentially as described.34 The acquired data were searched against the human IPI database with the MASCOT search engine (MatrixScience). The identified proteins were validated and clustered into protein groups using EpiCenter (Proxeon).
Sequencing of Btk, Lyn, and FcϵR1 β- and γ-chains in HMC-1 cells
The complete exons of Btk and Lyn and of the β- and γ-chains of the FcϵR1 were PCR-amplified from genomic DNA of HMC-1.2 cells using primer sequences shown in supplemental Table 4. PCR products were treated with ExoSAP-IT (USB Corporation) and sequenced on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems) using the BigDye Terminator Version 3.1 cycle Sequencing Kit (Applied Biosystems). All sequences were examined for point mutations (polymorphisms) using the Vector NTI software (Invitrogen). Multiplex RT-PCR (mDx Hema Vision; Bio-Rad Laboratories) was applied to seek for leukemia-related defects in HMC-1 cells: t(X;11), t(4;11), t(6;11), t(11;19), t(10;11), t(1;11), t(11;17), t(9;11), t(1;19), t(17;19), t(12;21), TAL1, t(8;21), t(3;21), t(16;21), t(15;17)(bcr1+bcr3), t(5;17), t(6;9), t(9;9), inv(16), t(9;22), t(9;12), t(5;12), t(12;22), t(3;5). The presence of JAK2 V617F and FLT3 ITD mutations was excluded by direct sequencing, and FIP1L1/PDGFRA and BCR/ABL by PCR.
Statistical analysis
To determine the significance in differences in proliferation and in the percentage of apoptotic cells after exposure to control medium, inhibitors, or siRNA, the Student t test for dependent samples was applied. Differences in expression of Lyn and Btk in neoplastic MCs (by IHC) in ISM versus advanced SM (SSM/ASM/MCL) were anaylzed using the χ2 test. P values were adjusted for multiple testing by Bonferroni-correction. Results were considered statistically significant when P was < .05. Drug interactions (additive/synergistic/antagonistic) were determined by calculating combination index (CI) values using Calcusyn software as reported.24 A CI value of 1 indicates an additive effect, whereas CI values < 1 indicates synergistic drug effects.
Results
Neoplastic MCs in advanced SM frequently express p-Lyn and p-Btk
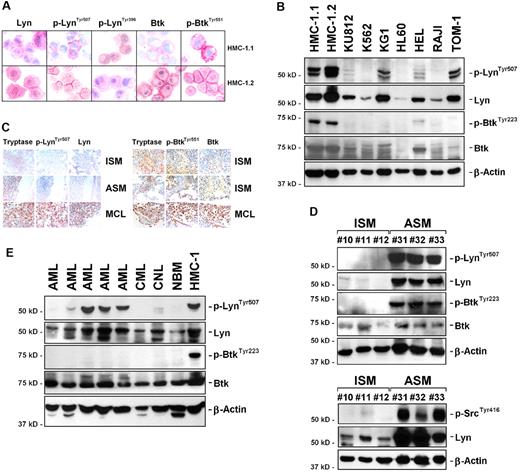
As assessed by immunocytochemistry, HMC-1.1 cells lacking KIT D816V and HMC-1.2 harboring KIT D816V were found to express p-Lyn and p-Btk (Figure 1A). Expression of p-Lyn and p-Btk in HMC-1 cells was confirmed by Western blotting (Figure 1B). In other leukemic cell lines, p-Lyn was also expressed, whereas p-Btk was weakly expressed or not detectable (Figure 1B, Table 2). We next examined p-Lyn and p-Btk expression in primary neoplastic MCs. As assessed by IHC, BM MCs in SM were found to express total Lyn and total Btk in most patients (Figure 1C, supplemental Figure 1; Table 3). However, p-Lyn and p-Btk were only expressed in MCs in a subset of patients (Table 3). Comparing subgroups of patients, we found that p-Btk is more frequently expressed in MCs in advanced SM (SSM, ASM, MCL; 80%) than in ISM (15.8%; P < .05; Tables 3–4). p-LynTyr396 and p-LynTyr507 were also found to be expressed more frequently in advanced SM than in ISM. This difference was found to be significant in case of p-LynTyr396, but not for p-LynTyr507 (Tables 3–4). Both p-Lyn (detected by mAb against p-SrcTyr416 and p-LynTyr507) and p-Btk were also detected in BM MCs in advanced SM by Western blotting (Figure 1D). Interestingly, p-Lyn was also detectable in some samples obtained from patients with AML or CML, whereas no substantial amounts of p-Btk were found (Figure 1E, Table 5) Normal BM cells expressed unphosphorylated Btk and Lyn, but did not express substantial amounts of p-Btk or p-Lyn (Figure 1E, Table 5). These data suggest that in advanced SM, both Lyn and Btk are constitutively activated in neoplastic MCs. Other BM cells including megakaryocytes were also found to express Lyn and p-Lyn, but did not express p-Btk (Table 6).
Expression and activation of Lyn and Btk in neoplastic MCs. (A) Immunocytochemical detection of p-LynTyr507, p-LynTyr396, and p-BtkTyr551 as well as total Lyn and total Btk in HMC-1.1 (top panels) cells and HMC-1.2 (bottom panels) cells. (B) Western blot analysis of Btk and Lyn expression in various cell lines using Abs against p-LynTyr507, Lyn, p-BtkTyr223, Btk, and β-actin as indicated. Molecular size markers are also shown. (C) Immunohistochemical detection of tryptase, p-LynTyr507, Lyn, p-BtkTyr551, and Btk in patients with indolent systemic mastocytosis (ISM), aggressive SM (ASM), and mast cell leukemia (MCL). The method of staining is provided in “IHC and ICC.” Figures shown in panels A and C were prepared using an Olympus DP11 camera connected to an Olympus BX50F4 microscope equipped with 100×/1.35 UPlan-Apo objective lens. Images were prepared using Adobe Photoshop CS2 Version 9.0 software (Adobe Systems) and processed with PowerPoint software (Microsoft). (D-E) Detection of p-LynTyr507, p-SrcTyr416, and p-BtkTyr223 in primary neoplastic cells by Western blotting. Neoplastic cells were obtained from patients with ISM (n = 3) and ASM (n = 3; D) as well as from patients with acute myeloid leukemia (AML, n = 5), chronic myeloid leukemia (CML, n = 1), and chronic neutrophilic leukemia (n = 1; E). In addition, normal BM cells (NBM, control) and HMC-1.2 cells (positive control) were examined (E). The β-actin control as well as size markers are also shown.
Expression and activation of Lyn and Btk in neoplastic MCs. (A) Immunocytochemical detection of p-LynTyr507, p-LynTyr396, and p-BtkTyr551 as well as total Lyn and total Btk in HMC-1.1 (top panels) cells and HMC-1.2 (bottom panels) cells. (B) Western blot analysis of Btk and Lyn expression in various cell lines using Abs against p-LynTyr507, Lyn, p-BtkTyr223, Btk, and β-actin as indicated. Molecular size markers are also shown. (C) Immunohistochemical detection of tryptase, p-LynTyr507, Lyn, p-BtkTyr551, and Btk in patients with indolent systemic mastocytosis (ISM), aggressive SM (ASM), and mast cell leukemia (MCL). The method of staining is provided in “IHC and ICC.” Figures shown in panels A and C were prepared using an Olympus DP11 camera connected to an Olympus BX50F4 microscope equipped with 100×/1.35 UPlan-Apo objective lens. Images were prepared using Adobe Photoshop CS2 Version 9.0 software (Adobe Systems) and processed with PowerPoint software (Microsoft). (D-E) Detection of p-LynTyr507, p-SrcTyr416, and p-BtkTyr223 in primary neoplastic cells by Western blotting. Neoplastic cells were obtained from patients with ISM (n = 3) and ASM (n = 3; D) as well as from patients with acute myeloid leukemia (AML, n = 5), chronic myeloid leukemia (CML, n = 1), and chronic neutrophilic leukemia (n = 1; E). In addition, normal BM cells (NBM, control) and HMC-1.2 cells (positive control) were examined (E). The β-actin control as well as size markers are also shown.
Expression of p-Btk and p-Lyn in various cell lines as assessed by ICC and Western blotting
| . | ICC . | Western blotting . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lyn . | p-Lyn Tyr507 . | p-Lyn Tyr396 . | p-Src Tyr416 . | Btk . | p-Btk Tyr223 . | Lyn . | p-Lyn Tyr507 . | p-Src Tyr416 . | Btk . | p-Btk Tyr223 . | |
| HMC-1.1 | + | + | + | + | + | + | + | + | + | + | + |
| HMC-1.2 | + | + | + | + | + | + | + | + | + | + | + |
| K562 | + | ± | nt | nt | + | ± | + | − | − | ± | − |
| KU812 | + | ± | nt | nt | + | ± | + | ± | ± | ± | − |
| KG1 | + | ± | nt | nt | + | ± | + | + | + | ± | − |
| HEL | nt | nt | nt | nt | nt | nt | + | ± | + | + | ± |
| Raji | + | ± | nt | nt | + | ± | + | − | + | ± | − |
| TOM-1 | + | ± | nt | nt | + | ± | + | + | + | ± | − |
| . | ICC . | Western blotting . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lyn . | p-Lyn Tyr507 . | p-Lyn Tyr396 . | p-Src Tyr416 . | Btk . | p-Btk Tyr223 . | Lyn . | p-Lyn Tyr507 . | p-Src Tyr416 . | Btk . | p-Btk Tyr223 . | |
| HMC-1.1 | + | + | + | + | + | + | + | + | + | + | + |
| HMC-1.2 | + | + | + | + | + | + | + | + | + | + | + |
| K562 | + | ± | nt | nt | + | ± | + | − | − | ± | − |
| KU812 | + | ± | nt | nt | + | ± | + | ± | ± | ± | − |
| KG1 | + | ± | nt | nt | + | ± | + | + | + | ± | − |
| HEL | nt | nt | nt | nt | nt | nt | + | ± | + | + | ± |
| Raji | + | ± | nt | nt | + | ± | + | − | + | ± | − |
| TOM-1 | + | ± | nt | nt | + | ± | + | + | + | ± | − |
Expression of total Lyn, p-LynTyr507, p-LynTyr396, p-SrcTyr416, total Btk, p-BtkTyr551, and p-BtkTyr223 in cell lines was assessed by ICC and Western blotting as described in “Western blot experiments” and “IHC and ICC.” Score of reactivity in ICC stains: + indicates positive; ±, weak reactivity; and −, negative stain. Score for Western blot analysis: + indicates positive;±, weak band; and −, negative.
ICC indicates immunocytochemistry; and nt, not tested.
Expression of p-Btk and p-Lyn in neoplastic MCs as assessed by IHC
| No. . | Diagnosis . | KIT D816V . | Lyn . | p-LynTyr507 . | p-LynTyr396 . | Btk . | p-BtkTyr551 . |
|---|---|---|---|---|---|---|---|
| 01 | ISM | − | + | + | ± | ± | − |
| 02 | ISM | + | + | − | nt | + | − |
| 03 | ISM | + | + | + | nt | + | − |
| 04 | ISM | + | nt | nt | nt | ± | − |
| 05 | ISM | + | + | − | nt | + | − |
| 06 | ISM | + | + | + | ± | + | − |
| 07 | ISM | + | ± | ± | − | + | − |
| 08 | ISM | + | + | − | ± | ± | − |
| 09 | ISM | + | − | − | nt | + | − |
| 10 | ISM | + | + | + | nt | + | + |
| 11 | ISM | + | − | − | nt | − | − |
| 12 | ISM | − | + | + | − | − | − |
| 13 | ISM | + | + | − | nt | + | − |
| 14 | ISM | + | ± | − | − | + | − |
| 15 | ISM | + | − | − | nt | + | − |
| 18 | ISM | + | − | − | nt | + | − |
| 19 | ISM/CLL | + | + | + | − | + | − |
| 20 | ISM | + | + | − | + | + | + |
| 21 | ISM | + | nt | nt | nt | + | + |
| 22 | SSM | + | − | − | + | + | + |
| 23 | SSM | + | ± | − | + | − | − |
| 24 | SSM | + | + | + | + | ± | |
| 26 | SSM | + | + | + | + | + | + |
| 27 | ASM | − | + | + | + | + | + |
| 28 | ASM | + | + | + | + | + | + |
| 29 | ASM | + | + | + | nt | + | − |
| 34 | MCL | + | + | − | + | + | ± |
| 37 | MCL | − | + | + | + | + | + |
| 38 | MCL | + | + | + | + | + | + |
| No. . | Diagnosis . | KIT D816V . | Lyn . | p-LynTyr507 . | p-LynTyr396 . | Btk . | p-BtkTyr551 . |
|---|---|---|---|---|---|---|---|
| 01 | ISM | − | + | + | ± | ± | − |
| 02 | ISM | + | + | − | nt | + | − |
| 03 | ISM | + | + | + | nt | + | − |
| 04 | ISM | + | nt | nt | nt | ± | − |
| 05 | ISM | + | + | − | nt | + | − |
| 06 | ISM | + | + | + | ± | + | − |
| 07 | ISM | + | ± | ± | − | + | − |
| 08 | ISM | + | + | − | ± | ± | − |
| 09 | ISM | + | − | − | nt | + | − |
| 10 | ISM | + | + | + | nt | + | + |
| 11 | ISM | + | − | − | nt | − | − |
| 12 | ISM | − | + | + | − | − | − |
| 13 | ISM | + | + | − | nt | + | − |
| 14 | ISM | + | ± | − | − | + | − |
| 15 | ISM | + | − | − | nt | + | − |
| 18 | ISM | + | − | − | nt | + | − |
| 19 | ISM/CLL | + | + | + | − | + | − |
| 20 | ISM | + | + | − | + | + | + |
| 21 | ISM | + | nt | nt | nt | + | + |
| 22 | SSM | + | − | − | + | + | + |
| 23 | SSM | + | ± | − | + | − | − |
| 24 | SSM | + | + | + | + | ± | |
| 26 | SSM | + | + | + | + | + | + |
| 27 | ASM | − | + | + | + | + | + |
| 28 | ASM | + | + | + | + | + | + |
| 29 | ASM | + | + | + | nt | + | − |
| 34 | MCL | + | + | − | + | + | ± |
| 37 | MCL | − | + | + | + | + | + |
| 38 | MCL | + | + | + | + | + | + |
MC indicates mast cell; ISM, indolent SM; CLL, chronic lymphatic leukemia; SSM smoldering SM; ASM, aggressive SM; and MCL, mast cell leukemia.
IHC staining results for Lyn, p-LynTyr507, p-LynTyr396, Btk, and p-BtkTyr551 were generated on serial sections stained with Abs directed against these signaling molecules and an Ab against tryptase. Score of reactivity in IHC stains: + indicates positive; ±, weak reactivity; −, negative stain; and nt, not tested.
Statistical analysis of the expression of p-Btk and p-Lyn as assessed by IHC in ISM and advanced SM
| Patient cohort . | Lyn . | p-LynTyr507 . | p-LynTyr396 . | Btk . | p-BtkTyr551 . |
|---|---|---|---|---|---|
| ISM (tryptase, < 200 ng/mL) | 11/15 (73.3%) | 6/15 (40%) | 3/6 (50%) | 14/16 (87.5%) | 1/16 (6.3%) |
| ISM (tryptase, > 200 ng/mL) | 2/2 (100%) | 1/2 (50%) | 1/2 (50%) | 3/3 (100%) | 2/3 (66.7%) |
| ISM (all) | 13/17 (76.5%) | 7/17 (41.2%) | 4/8 (50%) | 17/19 (89.5%) | 3/19 (15.8%) |
| SSM | 3/4 (75%) | 2/4 (50%) | 4/4 (100%) | 3/4 (75%) | 3/4 (75%) |
| ASM/MCL | 6/6 (100%) | 5/6 (83.3%) | 5/5 (100%) | 6/6 (100%) | 5/6 (83.3%) |
| Advanced SM* | 9/10 (90%) | 7/10 (70%) | 9/9 (100%) | 9/10 (90%) | 8/10 (80%) |
| Patient cohort . | Lyn . | p-LynTyr507 . | p-LynTyr396 . | Btk . | p-BtkTyr551 . |
|---|---|---|---|---|---|
| ISM (tryptase, < 200 ng/mL) | 11/15 (73.3%) | 6/15 (40%) | 3/6 (50%) | 14/16 (87.5%) | 1/16 (6.3%) |
| ISM (tryptase, > 200 ng/mL) | 2/2 (100%) | 1/2 (50%) | 1/2 (50%) | 3/3 (100%) | 2/3 (66.7%) |
| ISM (all) | 13/17 (76.5%) | 7/17 (41.2%) | 4/8 (50%) | 17/19 (89.5%) | 3/19 (15.8%) |
| SSM | 3/4 (75%) | 2/4 (50%) | 4/4 (100%) | 3/4 (75%) | 3/4 (75%) |
| ASM/MCL | 6/6 (100%) | 5/6 (83.3%) | 5/5 (100%) | 6/6 (100%) | 5/6 (83.3%) |
| Advanced SM* | 9/10 (90%) | 7/10 (70%) | 9/9 (100%) | 9/10 (90%) | 8/10 (80%) |
Based on the results obtained by IHC shown in Table 3, the percentages of patients expressing Lyn, p-LynTyr507, Btk, and p-BtkTyr551 were calculated.
ISM indicates indolent SM; SM, systemic mastocytosis; SSM: smoldering SM; ASM, aggressive SM; and MCL, mast cell leukemia.
Advanced SM includes SSM, ASM, and MCL. As assessed by statistical analysis (χ2 test), the difference in p-Btk and p-LynTyr396 expression in ISM (all ISM) and advanced SM (SSM/ASM/MCL) was significant (p-Btk: 15.8% vs 80%, P < .05; p-LynTyr396: 50% vs 100%, P < 0.05). Expression of p-LynTyr507 was also detected more frequently in advanced SM, but statistical significance was not reached.
Expression of p-Lyn and p-Btk in various primary BM cells (MNC) as determined by Western blotting
| No. . | Diagnosis . | KIT D816V . | Lyn . | p-LynTyr507 . | p-SrcTyr416 . | Btk . | p-BtkTyr223 . |
|---|---|---|---|---|---|---|---|
| 10 | ISM | + | ± | − | − | + | − |
| 11 | ISM | + | ± | − | − | + | − |
| 12 | ISM | − | − | − | − | ± | − |
| 13 | ISM | + | + | − | nt | + | − |
| 16 | ISM | + | + | − | nt | + | − |
| 25 | SSM | + | + | + | nt | nt | nt |
| 31 | ASM/MDS | − | + | + | + | + | + |
| 32 | ASM/CMML | + | + | + | + | + | + |
| 33 | ASM/CMML | + | + | + | + | + | + |
| 36 | MCL | − | nt | nt | nt | + | + |
| AML 1 | AML | − | + | − | nt | + | − |
| AML 2 | AML | − | + | ± | nt | + | − |
| AML 3 | AML | − | + | + | nt | + | − |
| AML 4 | AML | − | + | + | nt | + | − |
| AML 5 | AML | − | + | + | nt | + | − |
| CML 1 | CML | − | + | − | nt | + | − |
| CML 2 | CML | − | + | + | nt | + | − |
| CML 3 | CML | − | + | ± | nt | + | − |
| CML 4 | CML | − | + | ± | nt | + | − |
| CML 5 | CML | − | + | − | nt | + | − |
| CNL | CNL | − | + | ± | nt | + | − |
| NBM 1 | NBM | − | + | − | nt | + | − |
| NBM 2 | NBM | − | + | − | nt | + | − |
| NBM 3 | NBM | − | + | − | nt | + | − |
| No. . | Diagnosis . | KIT D816V . | Lyn . | p-LynTyr507 . | p-SrcTyr416 . | Btk . | p-BtkTyr223 . |
|---|---|---|---|---|---|---|---|
| 10 | ISM | + | ± | − | − | + | − |
| 11 | ISM | + | ± | − | − | + | − |
| 12 | ISM | − | − | − | − | ± | − |
| 13 | ISM | + | + | − | nt | + | − |
| 16 | ISM | + | + | − | nt | + | − |
| 25 | SSM | + | + | + | nt | nt | nt |
| 31 | ASM/MDS | − | + | + | + | + | + |
| 32 | ASM/CMML | + | + | + | + | + | + |
| 33 | ASM/CMML | + | + | + | + | + | + |
| 36 | MCL | − | nt | nt | nt | + | + |
| AML 1 | AML | − | + | − | nt | + | − |
| AML 2 | AML | − | + | ± | nt | + | − |
| AML 3 | AML | − | + | + | nt | + | − |
| AML 4 | AML | − | + | + | nt | + | − |
| AML 5 | AML | − | + | + | nt | + | − |
| CML 1 | CML | − | + | − | nt | + | − |
| CML 2 | CML | − | + | + | nt | + | − |
| CML 3 | CML | − | + | ± | nt | + | − |
| CML 4 | CML | − | + | ± | nt | + | − |
| CML 5 | CML | − | + | − | nt | + | − |
| CNL | CNL | − | + | ± | nt | + | − |
| NBM 1 | NBM | − | + | − | nt | + | − |
| NBM 2 | NBM | − | + | − | nt | + | − |
| NBM 3 | NBM | − | + | − | nt | + | − |
Expression of total Lyn, p-Lyn, total Btk, and p-Btk was assessed by Western blotting as described in “Western blot experiments.” Score: + indicates positive;±, weak band; and −, negative.
MNC indicates mononuclear cell; SM, systemic mastocytosis; ISM, indolent SM; SSM, smouldering SM; ASM, aggressive SM; MDS, myelodysplastic syndrome; CMML, chronic myelomonocytic leukemia; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; MCL, mast cell leukemia; CNL, chronic neutrophilic leukemia; NBM, normal BM; and nt, not tested.
Cellular distribution of Btk, p-Btk, Lyn, and p-Lyn in BM sections in patients with systemic mastocytosis* as determined by IHC
| . | Reactivity of BM cells with Abs against . | |||
|---|---|---|---|---|
| Lyn . | p-LynTyr507 . | Btk . | p-BtkTyr223 . | |
| Megakaryocytes | + | + | − | − |
| Myeloid progenitors | ± | + | − | − |
| Neutrophil granulocytes | ± | + | − | − |
| Eosinophil granulocytes | − | − | − | − |
| Erythroid progenitors | ± | ± | − | − |
| Lymphocytes | ± | ± | ± | ± |
| MCs | + | + | + | + |
| . | Reactivity of BM cells with Abs against . | |||
|---|---|---|---|---|
| Lyn . | p-LynTyr507 . | Btk . | p-BtkTyr223 . | |
| Megakaryocytes | + | + | − | − |
| Myeloid progenitors | ± | + | − | − |
| Neutrophil granulocytes | ± | + | − | − |
| Eosinophil granulocytes | − | − | − | − |
| Erythroid progenitors | ± | ± | − | − |
| Lymphocytes | ± | ± | ± | ± |
| MCs | + | + | + | + |
BM sections were stained with Abs against signaling molecules as described in “IHC and ICC.” Score: − indicates negative; +, positive; and ±, subset of cells positive.
MC indicates mast cell.
The same results were obtained with sections of normal BM as well as BM sections obtained from patients with CML (exception: MCs were only examined in mastocytosis).
Activated Lyn and Btk are expressed in neoplastic MCs independent of KIT
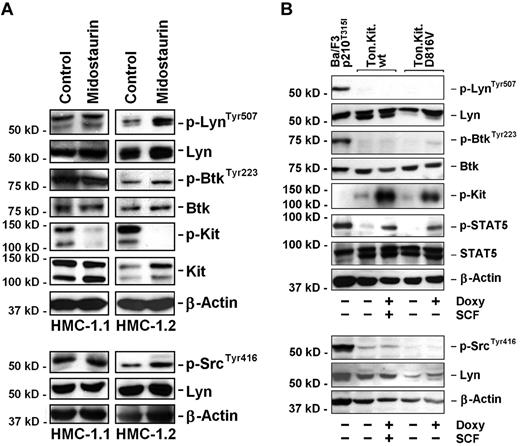
Midostaurin (PKC412) is a multikinase inhibitor that blocks the TK activity of wild-type KIT and KIT D816V. In this study, midostaurin (1μM) was found to inhibit the phosphorylation of KIT D816V in HMC-1.1 and HMC-1.2 cells, without down-regulating expression of p-Lyn or p-Btk (Figure 2A). These data suggest that p-Lyn and p-Btk are expressed in HMC-1 cells independent of KIT. We next examined Ton-Kit.D816V cells, a Ba/F3 clone in which expression of KIT D816V can be induced by addition of doxycycline. After induction of KIT D816V by doxycycline, Ton-Kit.D816V cells were found to express p-KIT and several KIT-downstream signaling molecules including p-STAT5, but did not express p-Btk (Figure 2B). Ba/F3 cells stably expressing BCR/ABL-T315I (Ba/F3p210T315I cells) were used as positive control and found to express p-STAT5, p-Lyn, and p-Btk (Figure 2B). Together, these data suggest that p-Lyn and p-Btk are expressed in neoplastic MCs independent of KIT D816V.
Effect of KIT D816V on Lyn and Btk activation in HMC-1 cells and Ba/F3 cells. (A) HMC-1.1 and HMC-1.2 cells were kept in control medium or in the presence of 1μM midostaurin for 4 hours. Thereafter, Western blot (WB) analysis was performed using Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and KIT. Moreover, the phosphorylation of KIT was analyzed by immunoprecipitation using an anti-KIT Ab followed by immunoblotting using an anti-phosphotyrosine Ab. The β-actin control is also shown. To confirm the specificity of this WB result, we also performed immunoprecipitation experiments using anti-Lyn Ab followed by immunoblotting with anti-pSrcTyr416 and anti-pLynTyr507 as well as anti-phospho-Ab 4G10. Again, midostaurin failed to suppress expression of p-Lyn (not shown). (B) Ba/F3 cells with doxycycline-inducible expression of wt KIT (Ton.Kit.wt) or KIT D816V (Ton.Kit.D816V) were kept in control medium or in the presence of doxycycline (doxy, 1 μg/mL) for 24 hours. In case of Ton.Kit.wt, SCF (100 ng/mL) was added to induce KIT phosporylation for 15 minutes. Thereafter, cells were harvested and subjected to Western blot analysis using an anti-phosphotyrosine Ab for detection of activated KIT and Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and phosphorylated STAT5 (p-STAT5) as well as total STAT5. Ba/F3 cells stably expressing BCR/ABL T315I (Ba/F3p210T315I) served as a positive control. The dual bands observed with Abs against Lyn and STAT5 in Ton.Kit.D816V cells may be explained by alternative splicing.
Effect of KIT D816V on Lyn and Btk activation in HMC-1 cells and Ba/F3 cells. (A) HMC-1.1 and HMC-1.2 cells were kept in control medium or in the presence of 1μM midostaurin for 4 hours. Thereafter, Western blot (WB) analysis was performed using Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and KIT. Moreover, the phosphorylation of KIT was analyzed by immunoprecipitation using an anti-KIT Ab followed by immunoblotting using an anti-phosphotyrosine Ab. The β-actin control is also shown. To confirm the specificity of this WB result, we also performed immunoprecipitation experiments using anti-Lyn Ab followed by immunoblotting with anti-pSrcTyr416 and anti-pLynTyr507 as well as anti-phospho-Ab 4G10. Again, midostaurin failed to suppress expression of p-Lyn (not shown). (B) Ba/F3 cells with doxycycline-inducible expression of wt KIT (Ton.Kit.wt) or KIT D816V (Ton.Kit.D816V) were kept in control medium or in the presence of doxycycline (doxy, 1 μg/mL) for 24 hours. In case of Ton.Kit.wt, SCF (100 ng/mL) was added to induce KIT phosporylation for 15 minutes. Thereafter, cells were harvested and subjected to Western blot analysis using an anti-phosphotyrosine Ab for detection of activated KIT and Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and phosphorylated STAT5 (p-STAT5) as well as total STAT5. Ba/F3 cells stably expressing BCR/ABL T315I (Ba/F3p210T315I) served as a positive control. The dual bands observed with Abs against Lyn and STAT5 in Ton.Kit.D816V cells may be explained by alternative splicing.
Role of Lyn and Btk in growth and survival of neoplastic MCs
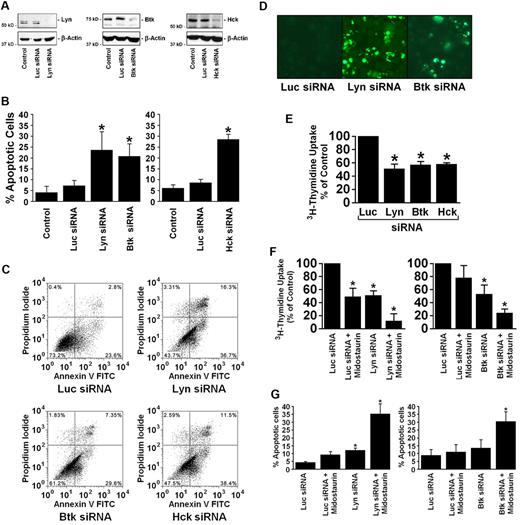
To define a functional role for Lyn and Btk in neoplastic MCs, we performed experiments using siRNAs. In addition, we applied siRNA against Hck. Transfection of HMC-1.2 cells with siRNA resulted in a knockdown of Lyn (by Lyn siRNA), Btk (by Btk siRNA), and Hck (by Hck siRNA) in these cells (Figure 3A). Interestingly, all 3 siRNAs produced growth inhibition and apoptosis in HMC-1.2 cells (Figure 3B-E). Notably, the number of apoptotic cells was significantly higher in HMC-1.2 cells transfected with Lyn siRNA, Btk siRNA, or Hck siRNA compared with cells transfected with control siRNA (Figure 3B). The apoptosis-inducing effects of the siRNAs were confirmed by combined annexin V/propidium iodide staining (Figure 3C) and by TUNEL assay (Figure 3D). Furthermore, all 3 siRNAs (Lyn, Btk, Hck) were found to inhibit 3H-thymidine uptake in HMC-1 cells (Figure 3E). We also applied the Btk inhibitor LFM-A13. At high concentrations (100-500μM), this drug was found to down-regulate p-Btk expression and growth in HMC-1 cells (supplemental Figure 2).
Effects of Lyn, Btk, and Hck siRNA of the viability and proliferation of HMC-1.2 cells. (A-E) HMC-1.2 cells were kept in control medium or transfected with siRNA directed against luciferase (Luc siRNA, 200nM) or siRNA against Lyn, Btk, or Hck (100-200nM each) using lipofectin as described in “Design and application of siRNA.” After 16 hours, cells were examined for Lyn, Btk, and Hck protein expression by Western blotting using Abs against Lyn, Btk, Hck, and β-actin (loading control; A). Cell viability and apoptosis in each condition were determined after 16 hours by light microscopy (B), combined annexin V/propidium iodide staining after 48 hours (C), or by TUNEL assay after 16 hours (D). Cell proliferation after 24 hours was determined by 3H-thymidine uptake (E). (F-G) HMC-1.2 cells were transfected with control siRNA (Luc siRNA) or 100nM Lyn or Btk siRNA as indicated. Four hours after transfection, cells were washed and resuspended in IMDM containing 10% FCS. Then, cells were kept for 24 hours in control medium or in the presence of 100nM midostaurin. Proliferation was determined by 3H-thymidine uptake (F), and the numbers of apoptotic cells by light microscopy (G). Results represent the mean ± SD of 3 independent experiments. *P < .05.
Effects of Lyn, Btk, and Hck siRNA of the viability and proliferation of HMC-1.2 cells. (A-E) HMC-1.2 cells were kept in control medium or transfected with siRNA directed against luciferase (Luc siRNA, 200nM) or siRNA against Lyn, Btk, or Hck (100-200nM each) using lipofectin as described in “Design and application of siRNA.” After 16 hours, cells were examined for Lyn, Btk, and Hck protein expression by Western blotting using Abs against Lyn, Btk, Hck, and β-actin (loading control; A). Cell viability and apoptosis in each condition were determined after 16 hours by light microscopy (B), combined annexin V/propidium iodide staining after 48 hours (C), or by TUNEL assay after 16 hours (D). Cell proliferation after 24 hours was determined by 3H-thymidine uptake (E). (F-G) HMC-1.2 cells were transfected with control siRNA (Luc siRNA) or 100nM Lyn or Btk siRNA as indicated. Four hours after transfection, cells were washed and resuspended in IMDM containing 10% FCS. Then, cells were kept for 24 hours in control medium or in the presence of 100nM midostaurin. Proliferation was determined by 3H-thymidine uptake (F), and the numbers of apoptotic cells by light microscopy (G). Results represent the mean ± SD of 3 independent experiments. *P < .05.
Lyn siRNA and Btk siRNA cooperate with midostaurin (PKC412) in inducing growth inhibition in neoplastic MCs
We next asked whether siRNAs against Lyn or Btk and midostaurin would produce cooperative antineoplastic effects. To address this question, we performed drug combination experiments, and found that transfection with 100nM siRNA against either Lyn or Btk, sensitize HMC-1 cells against growth-inhibitory and apoptosis-inducing effects of midostaurin (Figure 3F-G). These data suggest that Lyn/Btk-targeting drugs may represent optimal drug partners for midostaurin, resulting in synergistic growth-inhibitory effects in neoplastic MCs bearing KIT D816V. We therefore were interested to identify drugs that would interact with and would block Lyn- and Btk activation in neoplastic MCs.
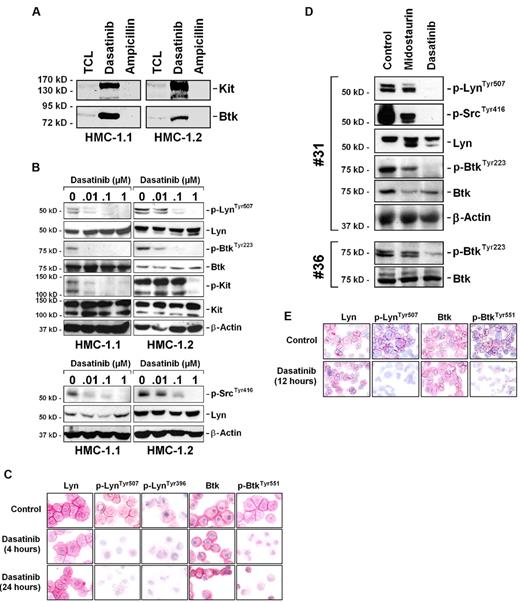
Identification of Lyn and Btk as major dasatinib binders and targets
Recent data suggest that dasatinib binds to and deactivates Lyn and Btk in CML cells.33-35 In this study, we examined whether dasatinib binds to Lyn and Btk in neoplastic MCs. Using drug-affinity chromatography followed by MS, we were able to show that dasatinib binds to several target kinases including Btk and Lyn, in HMC-1.1 and HMC-1.2 cells (Table 7). Western blotting confirmed that dasatinib binds to Btk in HMC-1 cell extracts (Figure 4A). In addition, we were able to show that dasatinib blocks activation/phosphorylation of Lyn and Btk in HMC-1.1 cells and HMC-1.2 cells (Figure 4B-C). Finally, we were able to show that dasatinib, but not midostaurin, inhibits the expression of p-Lyn and p-Btk in primary neoplastic MCs in patients with ASM or MCL (Figure 4D-E). These data suggest that Btk and Lyn are primary targets of dasatinib in neoplastic MCs. Other dasatinib binders in neoplastic MCs identified in this study were Fyn and Fgr (Table 7). As assessed by immunoprecipitation and Western blotting, Fyn and Fgr were found to be expressed in phosphorylated form in HMC-1 cells, and dasatinib (but not midostaurin) was found to counteract phosphorylation of Fyn and Fgr in these cells (supplemental Figure 3).
Proteomics profiling of HMC-1 cell subclones using dasatinib
| AC . | Entrez . | Description . | HMC-1.1 . | HMC1.2 . | ||
|---|---|---|---|---|---|---|
| PC . | SC . | PC . | SC . | |||
| IPI00221171 | ABL1 | Proto-oncogene tyrosine-protein kinase ABL1 (c-ABL) | 7 | 8 | 9 | 9 |
| IPI00719587 | ABL2 | Tyrosine-protein kinase ABL2 (ARG) | 7 | 7 | 2 | 2 |
| IPI00297220 | ACVR1B | Activin receptor type 1B | 2 | 4 | — | — |
| IPI00029132 | BTK | Tyrosine-protein kinase BTK | 47 | 71 | 48 | 66 |
| IPI00013212 | CSK | Tyrosine-protein kinase CSK | 25 | 63 | 26 | 63 |
| IPI00016613 | CSNK2A1 | Casein kinase 2, alpha subunit | 2 | 6 | 9 | 22 |
| IPI00219996 | DDR1 | Discoidin domain-containing receptor 1 | 16 | 19 | 8 | 9 |
| IPI00016645 | EPHA7 | Ephrin type-A receptor 7 | 2 | 2 | 4 | 4 |
| IPI00334334 | EPHB1 | Ephrin type-B receptor 1 | 12 | 17 | 23 | 29 |
| IPI00644408 | EPHB2 | Ephrin type-B receptor 2 | 4 | 6 | 11 | 13 |
| IPI00789482 | EPHB3 | Ephrin type-B receptor 3 | 2 | 15 | 2 | 14 |
| IPI00016871 | FGR | Proto-oncogene tyrosine-protein kinase FGR | 5 | 9 | 13 | 22 |
| IPI00219012 | FYN | Proto-oncogene tyrosine-protein kinase FYN, isoform 1 | 14 | 26 | 10 | 17 |
| IPI00640091 | FYN | Proto-oncogene tyrosine-protein kinase FYN, isoform 2 | 13 | 24 | 10 | 17 |
| IPI00298949 | GAK | Cyclin G-associated kinase | 5 | 5 | 26 | 19 |
| IPI00013219 | ILK | Integrin-linked kinase | — | — | 2 | 4 |
| IPI00784013 | JAK1 | Janus kinase 1 | — | — | 2 | 1 |
| IPI00022296 | KIT | Mast/stem cell growth factor receptor (c-KIT) | 39 | 42 | 41 | 40 |
| IPI00216433 | LIMK1 | Lim-domain kinase1 | — | — | 13 | 19 |
| IPI00456673 | LYK5 | STE20-related adapter protein | — | — | 3 | 8 |
| IPI00432416 | LYN | Tyrosine-protein kinase LYN, isoform LYN B | 25 | 57 | 24 | 53 |
| IPI00298625 | LYN | Tyrosine-protein kinase LYN, isoform LYN A | 25 | 55 | 24 | 51 |
| IPI00513803 | MAP3K2 | MAPK kinase kinase 2 | — | — | 8 | 9 |
| IPI00017801 | MAP3K3 | MAPK kinase kinase 3 | — | — | 4 | 6 |
| IPI00186536 | MAP3K4 | MAPK kinase kinase 4 | 7 | 5 | 17 | 12 |
| IPI00294842 | MAP4K5 | MAPK kinase kinase kinase 5 (KHS) | — | — | 19 | 23 |
| IPI00019473 | MAPK11 | MAPK 11 | 2 | 5 | — | — |
| IPI00002857 | MAPK14 | p38a, isoform 2 | 12 | 0.37 | 17 | 55 |
| IPI00221141 | MAPK14 | p38a, isoform 1 | 10 | 37 | 16 | 56 |
| IPI00742852 | PKMYT1 | 55-kDa protein | — | — | 3 | 6 |
| IPI00383291 | — | PRO3078 | 5 | 69 | 4 | 54 |
| IPI00015927 | PTK6 | Tyrosine-protein kinase 6 | — | — | 3 | 6 |
| IPI00021917 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | 5 | 12 | 12 | 23 |
| IPI00739386 | SGK223 | Tyrosine-protein kinase SGK223 | 4 | 0.04 | — | — |
| IPI00465291 | SNF1LK2 | Serine/threonine-protein kinase SNF1-like kinase 2 (QIK) | 5 | 4 | 5 | 6 |
| IPI00641230 | SRC | Proto-oncogene tyrosine-protein kinase SRC | 19 | 41 | 21 | 42 |
| IPI00000878 | TEC | Tyrosine-protein kinase TEC | 6 | 11 | 19 | 32 |
| IPI00102677 | TESK2 | Dual-specificity testis-specific protein kinase 2 | 11 | 18 | 19 | 36 |
| IPI00218905 | TNIK | TRAF2 and NCK-interacting protein kinase | — | — | 7 | 4 |
| IPI00022353 | TYK2 | Non–receptor tyrosine-protein kinase TYK2 | — | — | 34 | 28 |
| IPI00013981 | YES1 | Proto-oncogene tyrosine-protein kinase YES | 12 | 22 | 13 | 21 |
| IPI00029643 | ZAK | MAPK kinase kinase MLT, isoform 2 | 9 | 21 | 12 | 26 |
| IPI00329638 | ZAK | MAPK kinase kinase MLT, isoform 1 | 7 | 1 | 17 | 22 |
| AC . | Entrez . | Description . | HMC-1.1 . | HMC1.2 . | ||
|---|---|---|---|---|---|---|
| PC . | SC . | PC . | SC . | |||
| IPI00221171 | ABL1 | Proto-oncogene tyrosine-protein kinase ABL1 (c-ABL) | 7 | 8 | 9 | 9 |
| IPI00719587 | ABL2 | Tyrosine-protein kinase ABL2 (ARG) | 7 | 7 | 2 | 2 |
| IPI00297220 | ACVR1B | Activin receptor type 1B | 2 | 4 | — | — |
| IPI00029132 | BTK | Tyrosine-protein kinase BTK | 47 | 71 | 48 | 66 |
| IPI00013212 | CSK | Tyrosine-protein kinase CSK | 25 | 63 | 26 | 63 |
| IPI00016613 | CSNK2A1 | Casein kinase 2, alpha subunit | 2 | 6 | 9 | 22 |
| IPI00219996 | DDR1 | Discoidin domain-containing receptor 1 | 16 | 19 | 8 | 9 |
| IPI00016645 | EPHA7 | Ephrin type-A receptor 7 | 2 | 2 | 4 | 4 |
| IPI00334334 | EPHB1 | Ephrin type-B receptor 1 | 12 | 17 | 23 | 29 |
| IPI00644408 | EPHB2 | Ephrin type-B receptor 2 | 4 | 6 | 11 | 13 |
| IPI00789482 | EPHB3 | Ephrin type-B receptor 3 | 2 | 15 | 2 | 14 |
| IPI00016871 | FGR | Proto-oncogene tyrosine-protein kinase FGR | 5 | 9 | 13 | 22 |
| IPI00219012 | FYN | Proto-oncogene tyrosine-protein kinase FYN, isoform 1 | 14 | 26 | 10 | 17 |
| IPI00640091 | FYN | Proto-oncogene tyrosine-protein kinase FYN, isoform 2 | 13 | 24 | 10 | 17 |
| IPI00298949 | GAK | Cyclin G-associated kinase | 5 | 5 | 26 | 19 |
| IPI00013219 | ILK | Integrin-linked kinase | — | — | 2 | 4 |
| IPI00784013 | JAK1 | Janus kinase 1 | — | — | 2 | 1 |
| IPI00022296 | KIT | Mast/stem cell growth factor receptor (c-KIT) | 39 | 42 | 41 | 40 |
| IPI00216433 | LIMK1 | Lim-domain kinase1 | — | — | 13 | 19 |
| IPI00456673 | LYK5 | STE20-related adapter protein | — | — | 3 | 8 |
| IPI00432416 | LYN | Tyrosine-protein kinase LYN, isoform LYN B | 25 | 57 | 24 | 53 |
| IPI00298625 | LYN | Tyrosine-protein kinase LYN, isoform LYN A | 25 | 55 | 24 | 51 |
| IPI00513803 | MAP3K2 | MAPK kinase kinase 2 | — | — | 8 | 9 |
| IPI00017801 | MAP3K3 | MAPK kinase kinase 3 | — | — | 4 | 6 |
| IPI00186536 | MAP3K4 | MAPK kinase kinase 4 | 7 | 5 | 17 | 12 |
| IPI00294842 | MAP4K5 | MAPK kinase kinase kinase 5 (KHS) | — | — | 19 | 23 |
| IPI00019473 | MAPK11 | MAPK 11 | 2 | 5 | — | — |
| IPI00002857 | MAPK14 | p38a, isoform 2 | 12 | 0.37 | 17 | 55 |
| IPI00221141 | MAPK14 | p38a, isoform 1 | 10 | 37 | 16 | 56 |
| IPI00742852 | PKMYT1 | 55-kDa protein | — | — | 3 | 6 |
| IPI00383291 | — | PRO3078 | 5 | 69 | 4 | 54 |
| IPI00015927 | PTK6 | Tyrosine-protein kinase 6 | — | — | 3 | 6 |
| IPI00021917 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | 5 | 12 | 12 | 23 |
| IPI00739386 | SGK223 | Tyrosine-protein kinase SGK223 | 4 | 0.04 | — | — |
| IPI00465291 | SNF1LK2 | Serine/threonine-protein kinase SNF1-like kinase 2 (QIK) | 5 | 4 | 5 | 6 |
| IPI00641230 | SRC | Proto-oncogene tyrosine-protein kinase SRC | 19 | 41 | 21 | 42 |
| IPI00000878 | TEC | Tyrosine-protein kinase TEC | 6 | 11 | 19 | 32 |
| IPI00102677 | TESK2 | Dual-specificity testis-specific protein kinase 2 | 11 | 18 | 19 | 36 |
| IPI00218905 | TNIK | TRAF2 and NCK-interacting protein kinase | — | — | 7 | 4 |
| IPI00022353 | TYK2 | Non–receptor tyrosine-protein kinase TYK2 | — | — | 34 | 28 |
| IPI00013981 | YES1 | Proto-oncogene tyrosine-protein kinase YES | 12 | 22 | 13 | 21 |
| IPI00029643 | ZAK | MAPK kinase kinase MLT, isoform 2 | 9 | 21 | 12 | 26 |
| IPI00329638 | ZAK | MAPK kinase kinase MLT, isoform 1 | 7 | 1 | 17 | 22 |
HMC-1 kinases purified on dasatinib matrix and identified in both duplicate experiments. Proteins are sorted by the number of their unique peptides identified. Sequence coverages (“SC” in %) are based on these unique peptides. IPI-IDs are the protein identifiers from the IPI protein database.
MW indicates molecular weight in kilodaltons defined by the particular IPI-database entry; AC, acquisition number; PC, peptide count; and —, not applicable.
Effects of dasatinib on Lyn and Btk activation in neoplastic MCs. (A) Total cell lysates from HMC-1.1 cells (TCL) and proteins that were bound by immobilized dasatinib were immunoblotted for KIT and Btk. Ampicillin served as negative control. (B) HMC-1.1 cells and HMC-1.2 cells were cultured in control medium or in the presence of dasatinib (0.01-1μM for 4 hours). Thereafter, cells were subjected to Western blot analysis using Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and KIT. The β-actin loading control is also shown. To detect KIT activation, an immunoprecipitation was conducted using an anti-KIT Ab followed by immunoblotting using the anti-phosphotyrosine Ab 4G10. (C) HMC-1.2 cells were incubated in control medium (Co) or in medium containing dasatinib (1μM) for 4 hours or 24 hours at 37°C. Then, cells were spun on cytospin slides and stained with Abs specific for Lyn, p-LynTyr507, p-LynTyr396, Btk, and p-BtkTyr551 by indirect immunocytochemistry. Cells were analyzed using an Olympus AX-1 microscope equipped with 60×/1.35 UPlan-Apo objective lens. Image acquisition was performed using an Olympus DP11 camera and Adobe Photoshop CS2 software Version 9.0 (Adobe Systems). Magnification, ×600. (D) Primary neoplastic mast cells from one patient with aggressive systemic mastocytosis (ASM) and one with mast cell leukemia, MCL (no. 31 and no. 36 in Table 1) were cultured in control medium or in the presence of midostaurin (1μM) or dasatinib (1μM) for 12 hours. Thereafter, cells were subjected to Western blot analysis using Abs against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and β-actin. (E) Immunocytochemistry performed with primary mast cells (ASM patient 30) exposed to control medium or medium containing dasatinib (1μM) for 12 hours. Abs used and the microscopy technique applied were the same as that described for panel C. Magnification, ×600.
Effects of dasatinib on Lyn and Btk activation in neoplastic MCs. (A) Total cell lysates from HMC-1.1 cells (TCL) and proteins that were bound by immobilized dasatinib were immunoblotted for KIT and Btk. Ampicillin served as negative control. (B) HMC-1.1 cells and HMC-1.2 cells were cultured in control medium or in the presence of dasatinib (0.01-1μM for 4 hours). Thereafter, cells were subjected to Western blot analysis using Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and KIT. The β-actin loading control is also shown. To detect KIT activation, an immunoprecipitation was conducted using an anti-KIT Ab followed by immunoblotting using the anti-phosphotyrosine Ab 4G10. (C) HMC-1.2 cells were incubated in control medium (Co) or in medium containing dasatinib (1μM) for 4 hours or 24 hours at 37°C. Then, cells were spun on cytospin slides and stained with Abs specific for Lyn, p-LynTyr507, p-LynTyr396, Btk, and p-BtkTyr551 by indirect immunocytochemistry. Cells were analyzed using an Olympus AX-1 microscope equipped with 60×/1.35 UPlan-Apo objective lens. Image acquisition was performed using an Olympus DP11 camera and Adobe Photoshop CS2 software Version 9.0 (Adobe Systems). Magnification, ×600. (D) Primary neoplastic mast cells from one patient with aggressive systemic mastocytosis (ASM) and one with mast cell leukemia, MCL (no. 31 and no. 36 in Table 1) were cultured in control medium or in the presence of midostaurin (1μM) or dasatinib (1μM) for 12 hours. Thereafter, cells were subjected to Western blot analysis using Abs against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, and β-actin. (E) Immunocytochemistry performed with primary mast cells (ASM patient 30) exposed to control medium or medium containing dasatinib (1μM) for 12 hours. Abs used and the microscopy technique applied were the same as that described for panel C. Magnification, ×600.
Low concentrations of dasatinib block phosphorylation of Lyn and Btk, but not phosphorylation of KIT in neoplastic MCs: possible implications for synergistic effects seen with dasatinib and midostaurin
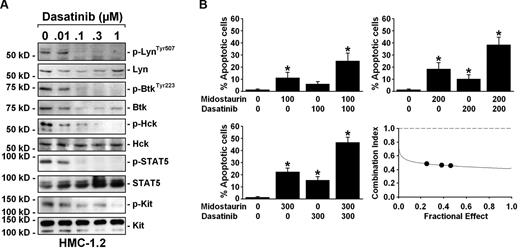
We have recently shown that midostaurin and dasatinib synergize in producing growth-inhibitory effects on KIT D816V+ neoplastic MCs.24 To explore whether this synergism is mediated by the specific effects of dasatinib on p-Btk and p-Lyn, we examined the phosphorylation status of various downstream signaling molecules including Hck and STAT5, both of which play a role in leukemogenesis,36-38 after drug exposure. We found that dasatinib at suboptimal concentrations (that work in synergism experiments: up to 300nM) completely inhibits the phosphorylation of multiple kinases including Lyn, Btk, and Hck, as well as phosphorylation of STAT5 in HMC-1.2 cells, but did not completely block phosphorylation of KIT D816V (Figure 5A). Such low concentrations of dasatinib slightly increased the number of apoptotic HMC-1.2 cells, whereas a significant induction of apoptosis was obtained when dasatinib was combined with suboptimal concentrations of midostaurin (Figure 5B). To exclude a cross-reactivity of anti–p-Lyn and anti–p-Hck Abs, immunoprecipitation was performed. These experiments confirmed that both p-Lyn and p-Hck are expressed in a constitutive manner in HMC-1.1 cells and HMC-1.2 cells (not shown).
Synergistic drug interactions between midostaurin and dasatinib may be mediated by the Lyn/Btk pathway and downstream STAT5. (A) HMC-1.2 cells were kept in control medium or in the presence of various concentrations of dasatinib (0.01-1μM) for 4 hours. Thereafter, Western blot analysis was performed using Abs directed against p-LynTyr507, Lyn, p-BtkTyr223, Btk, Hck, p-STAT5, STAT5, and KIT. To detect Hck and KIT activation, immunoprecipitation was performed using an anti–Hck-Ab or anti-KIT Ab, followed by immunoblotting with anti-phospho tyr-Ab 4G10. (B) HMC-1.2 cells were incubated in control medium (no drugs: 0) or in medium containing midostaurin and dasatinib (100-300nM each as indicated) either as single drugs or in combination for 48 hours. Thereafter, the number of apoptotic cells was counted by light microscopy. Results represent the mean ± SD of 3 independent experiments. *P < .05 compared with control (0). The bottom right panel shows combination index (CI) values obtained in these experiments using Calcusyn Software. As visible, the CI values are all below 1.0 indicating synergistic drug interactions.
Synergistic drug interactions between midostaurin and dasatinib may be mediated by the Lyn/Btk pathway and downstream STAT5. (A) HMC-1.2 cells were kept in control medium or in the presence of various concentrations of dasatinib (0.01-1μM) for 4 hours. Thereafter, Western blot analysis was performed using Abs directed against p-LynTyr507, Lyn, p-BtkTyr223, Btk, Hck, p-STAT5, STAT5, and KIT. To detect Hck and KIT activation, immunoprecipitation was performed using an anti–Hck-Ab or anti-KIT Ab, followed by immunoblotting with anti-phospho tyr-Ab 4G10. (B) HMC-1.2 cells were incubated in control medium (no drugs: 0) or in medium containing midostaurin and dasatinib (100-300nM each as indicated) either as single drugs or in combination for 48 hours. Thereafter, the number of apoptotic cells was counted by light microscopy. Results represent the mean ± SD of 3 independent experiments. *P < .05 compared with control (0). The bottom right panel shows combination index (CI) values obtained in these experiments using Calcusyn Software. As visible, the CI values are all below 1.0 indicating synergistic drug interactions.
Synergistic effects of midostaurin (PKC412) and bosutinib, another Lyn/Btk inhibitor, on growth of neoplastic MCs
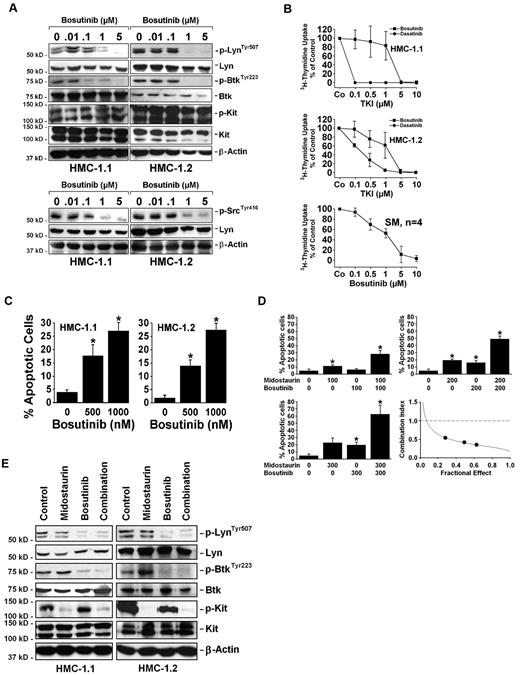
To demonstrate that drug-induced deactivation of Lyn and Btk is sufficient for cooperative drug effects obtained with midostaurin, we also applied bosutinib (SKI-606), a drug that (unlike dasatinib) deactivates Lyn and Btk but does not block KIT activation.39,40 In Western blot experiments, we were able to show that bosutinib (0.01-5μM) down-regulates expression of p-Lyn and p-Btk in HMC-1 cells, but does not deactivate KIT D816V (Figure 6A). Moreover, we were able to show that bosutinib inhibits the proliferation of primary neoplastic BM MCs (Figure 6B) and HMC-1 cells (Figure 6B), with IC50 values of 1-5μM. In addition, bosutinib induced apoptosis in HMC-1 cells (Figure 6C). Finally, similar to dasatinib, bosutinib was found to synergize with midostaurin in inducing apoptosis in HMC-1 cells (Figure 6D). As expected, the combination of midostaurin and bosutinib was found to block the phosphorylation of KIT, Lyn, and Btk (Figure 6E). These data suggest that Lyn and Btk represent drug targets that may be responsible for the synergistic effects of the TKI combinations “bosutinib + midostaurin” and “dasatinib + midostaurin.”
Midostaurin synergizes with bosutinib in producing apoptosis in HMC-1 cells. (A) HMC-1.1 and HMC-1.2 cells were kept in control medium or in the presence of bosutinib (0.01-5μM). After 4 hours, Western blot analysis was performed using Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, KIT, and β-actin. To detect expression of p-KIT, an immunoprecipitation was conducted using an anti-KIT Ab followed by immunoblotting using the anti-phosphotyrosine Ab 4G10. (B) HMC-1.1 cells and HMC-1.2 cells were kept in control medium (Co) or in various concentrations of bosutinib or dasatinib (as indicated) for 48 hours. BM cells of 4 patients with SM (nos. 13, 17, 19, and 25 in Table 1; all KIT D816V+) were incubated in control medium or bosutinib (0.1-10μM) for 48 hours. After incubation, proliferation was determined by 3H uptake. Results are expressed as percent of control and represent the mean ± SD of 3 independent experiments (cell lines), or the mean ± SD of 4 experiments performed with primary neoplastic mast cells (4 patients). (C) In HMC-1 cells, the percentage of apoptotic cells was determined by light microscopy after 48 hours of incubation with 500-1000nM bosutinib. Results represent the mean ± SD of 3 independent experiments. *P < .05. (D,E) HMC-1.1 cells and HMC-1.2 cells were incubated with midostaurin and bosutinib as single drugs or in combination. (D) HMC-1.2 cells were incubated in control medium (0/0) or in medium containing midostaurin and bosutinib alone or in combination (in fixed ratio: 1:1 concentration) as indicated. Three different drug concentrations are shown (100, 200, and 300nM). After 48 hours of incubation, the percentages of apoptotic cells were determined by light microscopy. Results represent the mean ± SD of 3 independent experiments. *P < .05 compared with control (0). The bottom right panel shows combination index (CI) values obtained in these experiments using Calcusyn Software. As visible, the CI values are all below 1.0 indicating synergistic drug interactions. (E) After 4 hours of incubation in control medium, midostaurin (1μM), bosutinib (1μM), or a combination of both drugs (1μM each), Western blot analysis was performed using Abs directed against p-LynTyr507, Lyn, p-BtkTyr223, Btk, KIT, and β-actin. To detect expression of p-KIT, an immunoprecipitation was conducted using an anti-KIT Ab followed by immunoblotting with anti-phosphotyrosine Ab 4G10.
Midostaurin synergizes with bosutinib in producing apoptosis in HMC-1 cells. (A) HMC-1.1 and HMC-1.2 cells were kept in control medium or in the presence of bosutinib (0.01-5μM). After 4 hours, Western blot analysis was performed using Abs directed against p-LynTyr507, p-SrcTyr416, Lyn, p-BtkTyr223, Btk, KIT, and β-actin. To detect expression of p-KIT, an immunoprecipitation was conducted using an anti-KIT Ab followed by immunoblotting using the anti-phosphotyrosine Ab 4G10. (B) HMC-1.1 cells and HMC-1.2 cells were kept in control medium (Co) or in various concentrations of bosutinib or dasatinib (as indicated) for 48 hours. BM cells of 4 patients with SM (nos. 13, 17, 19, and 25 in Table 1; all KIT D816V+) were incubated in control medium or bosutinib (0.1-10μM) for 48 hours. After incubation, proliferation was determined by 3H uptake. Results are expressed as percent of control and represent the mean ± SD of 3 independent experiments (cell lines), or the mean ± SD of 4 experiments performed with primary neoplastic mast cells (4 patients). (C) In HMC-1 cells, the percentage of apoptotic cells was determined by light microscopy after 48 hours of incubation with 500-1000nM bosutinib. Results represent the mean ± SD of 3 independent experiments. *P < .05. (D,E) HMC-1.1 cells and HMC-1.2 cells were incubated with midostaurin and bosutinib as single drugs or in combination. (D) HMC-1.2 cells were incubated in control medium (0/0) or in medium containing midostaurin and bosutinib alone or in combination (in fixed ratio: 1:1 concentration) as indicated. Three different drug concentrations are shown (100, 200, and 300nM). After 48 hours of incubation, the percentages of apoptotic cells were determined by light microscopy. Results represent the mean ± SD of 3 independent experiments. *P < .05 compared with control (0). The bottom right panel shows combination index (CI) values obtained in these experiments using Calcusyn Software. As visible, the CI values are all below 1.0 indicating synergistic drug interactions. (E) After 4 hours of incubation in control medium, midostaurin (1μM), bosutinib (1μM), or a combination of both drugs (1μM each), Western blot analysis was performed using Abs directed against p-LynTyr507, Lyn, p-BtkTyr223, Btk, KIT, and β-actin. To detect expression of p-KIT, an immunoprecipitation was conducted using an anti-KIT Ab followed by immunoblotting with anti-phosphotyrosine Ab 4G10.
Neoplastic MCs express wt Lyn and wt Btk: failure to detect a molecular defect in neoplastic mast cells that could explain Btk/Lyn activation
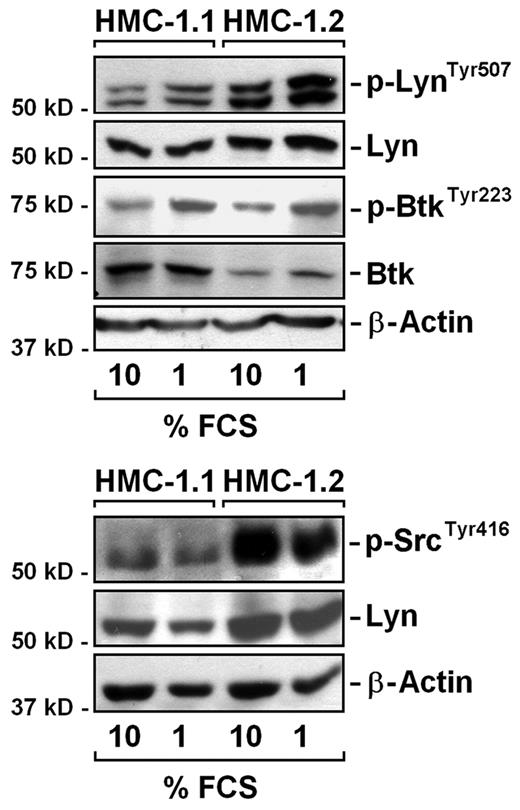
We next attempted to define the molecular mechanism leading to activation of Lyn and Btk in neoplastic MCs. In a first step, we excluded that Lyn and Btk are activated in HMC-1 cells by a serum-derived factor present in FCS. As visible in Figure 7, Lyn and Btk were found to be phosphorylated independent of the FCS concentration applied. Next, we screened for activating mutations. However, we were unable to detect a point mutation, single nucleotide polymorphism (SNP), or other gene defect in Lyn and Btk in HMC-1 cells. These data suggest that the primary defect may be located upstream of Lyn and/or Btk. Because the IgE receptor (R) reportedly triggers Lyn/Btk activation and growth in murine MCs,41,42 we examined HMC-1 cells for the presence of activating SNPs in the IgER β- and γ-chains. However, by sequence analysis, we were unable to detect mutated forms of IgERß or IgERγ. We next asked whether JAK2 may play a role in Btk/Lyn activation. However, the shRNA-induced knockdown of JAK2 did not lead to a decrease in expression of p-Lyn or p-Btk in HMC-1.2 cells (supplemental Figure 4). Finally, we asked whether HMC-1 cells would display leukemia-related molecular defects that would explain KIT-independent signaling and Lyn/Btk activation. However, no activating mutations in ABL, JAK2, RUNX1, PDGFR, and FGFR were found.
Activation of Lyn and Btk is not dependent on FCS in medium. HMC-1.1 and HMC-1.2 cells were kept in 1% or 10% FCS for 4 hours. Thereafter, the expression of Lyn, p-LynTyr507, p-SrcTyr416, Btk, and p-BtkTyr223 were analyzed by Western blotting. β-actin served as loading control.
Activation of Lyn and Btk is not dependent on FCS in medium. HMC-1.1 and HMC-1.2 cells were kept in 1% or 10% FCS for 4 hours. Thereafter, the expression of Lyn, p-LynTyr507, p-SrcTyr416, Btk, and p-BtkTyr223 were analyzed by Western blotting. β-actin served as loading control.
Discussion
Recent data suggest that KIT D816V exhibits only weak transforming potential in MC progenitors. In particular, KIT D816V alone is unable to induce proliferation in Ba/F3 cells,25 and transgenic mice usually develop slowly expanding nonaggressive MC accumulations, but rarely develop an aggressive MC disease.26 Correspondingly, the KIT D816V mutant is expressed not only in aggressive SM but also in the more frequent, indolent form of SM.1,4,6,7 Rarely, ISM may progress and then the prognosis is grave.1-4,6-9 So far however, little is known about factors underlying disease progression. We here show that Btk and Lyn are activated in a KIT-independent manner and contribute to malignant growth in neoplastic MCs. Both molecules are recognized by the multikinase inhibitors dasatinib and bosutinib. These 2 TKI apparently counteract neoplastic MC growth and synergize with midostaurin in producing growth inhibition. Collectively, these data suggest that p-Lyn and p-Btk are novel KIT-independent signaling molecules in neoplastic MCs that may contribute to disease progression and may serve as drug targets in advanced SM.
Expression of Lyn and Btk in neoplastic MCs was demonstrable by Western blotting as well as by immunostaining. An interesting observation was that p-Lyn and p-Btk are not only expressed in KIT D816V+ HMC-1.2 cells but also in KIT D816V-negative HMC-1.1 cells. These data suggest that Lyn and Btk activation in neoplastic MCs occurs independent of KIT D816V or may be triggered by other KIT variants. Notably, HMC-1.1 cells also display KIT G560V, another “transforming” KIT variant. However, the multikinase blocker midostaurin that deactivates wt KIT, KIT D816V, and KIT G560V, did not suppress expression of p-Lyn or p-Btk in HMC-1.1 or HMC-1.2 cells. Moreover, KIT D816V did not cause activation of Lyn or Btk in Ba/F3 cells, which was an unexpected result when considering previous data.43-45 However, in previous studies, Mo7e cells and murine BM MCs were used.43-45
We next were interested to learn whether Lyn and Btk contribute to abnormal growth and survival of neoplastic MCs. To address this issue, we applied siRNAs directed against Lyn or Btk on neoplastic MCs (HMC-1). The siRNA-induced knockdown of Lyn and Btk was followed by growth inhibition in HMC-1.2 cells suggesting that Lyn and Btk are indeed survival-related molecules and may serve as potential targets in neoplastic MC. To examine the mechanism underlying KIT-independent activation of Lyn and Btk, we screened for activating upstream- and downstream signaling molecules in neoplastic MCs. In these experiments, we found that neoplastic MCs display not only p-Lyn and p-Btk, but also p-Hck and p-STAT5, both of which are considered important signaling molecules. As expected, only p-STAT5 was dephosphorylated by midostaurin (PKC412) in HMC-1 cells, whereas expression of p-Hck, which is not an established KIT-downstream molecule, remained almost unchanged. Based on these data it is tempting to speculate that also p-Hck may play a role in KIT-independent growth and survival of neoplastic MC, an assumption that was confirmed by our siRNA studies. Notably, a Hck-specific siRNA was found to counteract growth of HMC-1 cells in the same way as siRNAs against Lyn or Btk.
In a next step, we asked whether Lyn and/or Btk display-activating mutations. However, no point mutations in Btk or Lyn were detected. We also screened for activating mutations in cytokine receptors and in the receptor for IgE. However, no mutations in the IgER β- or γ-chains or in other candidate genes (JAK2, PDGFR, FLT3) could be identified. We also excluded that an activating factor/cytokine was present in medium/FCS. Finally, we excluded that Lyn and Btk activation is a general phenomenon common to all leukemias. In particular, when screening various leukemias and cell lines, we found that only neoplastic MCs and HMC-1 cells display p-Btk and p-Lyn by Western blotting, whereas the other cell lines expressed p-Lyn without expressing substantial amounts of p-Btk.
Recent data suggest that dasatinib directly binds to Lyn and Btk in neoplastic cells in CML, and that the drug deactivates both signaling molecules.34,35,46 In the present study, we were able to show that dasatinib binds to and blocks Lyn and Btk in neoplastic MCs. In particular, chemical proteomic profiling and MS identified Lyn and Btk as major binders of dasatinib in HMC-1 cells. In addition, in contrast to midostaurin, dasatinib was found to block p-Btk, p-Lyn, and p-Hck in HMC-1.2 cells. These data are of interest as dasatinib has been described to inhibit growth of neoplastic MCs in vitro21,24 and is used in clinical trials.47
We have recently shown that dasatinib synergizes with midostaurin in producing growth inhibition in neoplastic MCs bearing KIT D816V.24 We were therefore interested to know whether the intriguing effects of dasatinib on Lyn and Btk activation may explain synergistic drug-combination effects. To address this question, we compared target responses to suboptimal concentrations of dasatinib in HMC-1 cells. We found that suboptimal concentrations of dasatinib (effective in synergism experiments) block Btk and Lyn phosphorylation, but do not block expression of p-KIT. These data suggest that the effect of dasatinib on Btk and Lyn activation may contribute to synergistic effects when combining with suboptimal concentrations of midostaurin, known to block p-KIT, but neither p-Btk nor p-Lyn. To further demonstrate that p-Lyn and p-Btk are targets responsible for mediating synergistic effects, we applied a second p-Lyn/p-Btk inhibitor that does not block KIT activation, namely bosutinib.39,40 This drug was found to block p-Lyn and p-Btk expression without suppressing p-KIT in neoplastic MCs. Furthermore, we found that similar to dasatinib, bosutinib synergizes with midostaurin in producing apoptosis in neoplastic MCs. Because bosutinib does not block KIT activation, these data strongly suggest that p-Lyn and p-Btk are important drug targets that can mediate cooperative effects of various kinase inhibitors on growth and survival of neoplastic MCs.
The observation that Lyn and Btk are important KIT-independent signaling molecules and targets in neoplastic MCs in advanced SM, may have clinical implications. First, the demonstration of p-Lyn and p-Btk in neoplastic cells may predict responses to dasatinib or to certain drug combinations. Moreover, our observations may explain the basis of synergistic effects of KIT inhibitors, and thus support preclinical and clinical concepts in this regard. In fact, based on our data, it seems reasonable to consider clinical trials using combinations of midostaurin/PKC412 with another drug that can suppress Lyn/Btk activation in neoplastic MCs in ASM and MCL. However, such combinations may also cause unforeseeable side effects, which has to be taken into account when planning such trials. In addition, such studies have to take the different pharmacologic behavior and half-life of the TKI partners into account.
Several previous data have shown that Lyn and Btk are involved in IgE receptor-dependent activation.48,49 Therefore, it is tempting to speculate that IgE-dependent mediator release and anaphylaxis in SM, a clinically relevant problem,1-5 may be triggered by activated signaling molecules like Lyn or Btk. Indeed, severe mediator-related problems often occur in SM.1-5 It is also noteworthy in this regard that dasatinib has been described to block IgE-dependent histamine release from basophils at high concentrations but even augments histamine release at low drug concentrations.46 This is of particular interest as Lyn activation may have positive and negative effects on IgER-dependent mediator release in MCs.50
Together, our data show that Btk and Lyn are activated in a KIT-independent manner in neoplastic MCs in advanced SM, and that these molecules are novel potential targets of therapy. Moreover, our data show that dasatinib and bosutinib bind to and deactivate Lyn and Btk in neoplastic MCs. The unique target spectrum of dasatinib and bosutinib may explain synergistic growth-inhibitory effects of these drugs when combined with midostaurin (PKC412), and may thus provide a basis for the development of new combination strategies in advanced SM.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Harald Herrmann for skilful technical assistance and Wolfgang R. Sperr for assistance in statistical analysis.
This work was supported by Fonds zur Förderung der Wissenschaftlichen Forschung in Österreich (FWF) grant P-21 173-B13; the Austrian Federal Ministry for Science and Research (grants GZ 200.136/1-VI/1/2005 and GZ 200.142/1-IV/1/2006); grant 20030 from the Center of Molecular Medicine (CeMM) of AustrianAcademy of Sciences; and a research grant of The Mastocytosis Society (TMS).
Authorship
Contribution: K.V.G performed Western blotting, proliferation assays, PCR, drug combination assays, and siRNA technique, designed the study, and established the research plan; M.M., G.H., and G.M.-H. contributed by performing PCR and sequencing experiments; S.C.-R. performed immunocytochemistry and immunohistochemistry; U.R., K.L.B., and G.S.-F. performed drug-affinity purification and mass spectrometry; E.H. performed TUNEL assay experiments; R.A.M. performed flow cytometry experiments; W.F.P. contributed by performing key laboratory experiments on cell growth and proliferation and by analyzing respective data; J.G. and A.R. contributed patients' samples; H.-P.H. contributed patients' samples and evaluation of immunohistochemistry results; and P.V. contributed by providing logistic and budget support, and by approving the data and the final version of the manuscript.
Conflict-of-interest disclosure: P.V. is a consultant in a Novartis trial and received research grants from Novartis and BMS. J.G. is consultant in a Novartis trial. The remaining authors declare no competing financial interests.
Correspondence: Peter Valent, MD, Department of Internal Medicine I, Division of Hematology & Hemostaseology, Medical University of Vienna & Ludwig Boltzmann Cluster Oncology, Waehringer Guertel 18-20, A-1090 Vienna, Austria; e-mail: peter.valent@meduniwien.ac.at.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal