Abstract
The introduction of imatinib in the treatment of chronic myeloid leukemia (CML) represents the most successful example of targeted therapy in human cancer. However, leukemic stem cells are insensitive to tyrosine kinase inhibitors (TKIs) and contribute to the persistence of disease by representing a reservoir of selfrenewing cells that replenish the disease after drug discontinuation. This finding has refocused the interest of scientists toward drug combinations, ie, treating with TKIs and simultaneously targeting alternative survival mechanisms. One candidate target mechanism is autophagy, a cellular recycling process that acts as a cytoprotective shield in CML cells in response to TKI-induced stress and in other cancer cells surviving in an inhospitable microenvironment. On that basis, inhibition of autophagy has now become an exciting option for combination treatment in cancer, and clinical trials have been initiated in solid and hemopoietic tumors such as CML, chronic lymphocytic leukemia, and multiple myeloma. This review describes the biology of CML and elucidates how the molecular driver BCR-ABL led to the development of TKIs. We then discuss the molecular regulation of autophagy and the potential for autophagy inhibition as the next step in our attempt to tackle the problem of CML persistence to offer a curative option.
Introduction
Fifty years ago a genetic link to cancer was provided by Peter C. Nowell and David Hungerford, who for the first time described an unusual small chromosome present in leukocytes from patients with chronic myeloid leukemia (CML). This abnormality, designated as the Philadelphia (Ph) chromosome after the city in which it was discovered, was found in all malignant cells of CML patients and is now considered the hallmark of CML.1 In 1973 Janet Rowley demonstrated that the Ph chromosome resulted from a reciprocal translocation between the long arms of chromosomes 9 and 22, t(9:22)(q34;q11).2 Later, it was shown that this alteration generates a fusion between c-ABL (human homologue of the Abelson murine leukemia virus), a tyrosine kinase encoding oncogene, and BCR (breakpoint cluster region), the function of which is still not clear.3 The chimeric BCR-ABL protein possesses cellular-transforming ability that is ascribed to the elevated tyrosine kinase (TK) activity of the molecule compared with the native c-ABL.4,5 Further studies have established BCR-ABL as a leukemogenic oncogene, and both mouse models and in vitro assays have shown that BCR-ABL, as the sole oncogenic event, is sufficient to induce leukemia.6
Molecular mechanisms of BCR-ABL
There are 3 major forms of the BCR-ABL oncoprotein, depending on the break point occurring within the BCR gene. The most common is the p210-kDa protein (e13a2 or e14a2 transcript), which is found in most CML cases and in approximately one-third of adults with Ph-positive acute lymphoblastic leukemia.7 A p190-kDa protein (e1a2 transcript) is expressed in the remaining Ph-positive patients with acute lymphoblastic leukemia and rarely in patients with CML. The third fusion protein (p230 kDa; e19a2 transcript) has been identified in the rare neutrophilic CML.8 All 3 major BCR-ABL fusion proteins induce a similar CML-like syndrome in mice but differ in their ability to induce lymphoid leukemia.9
Key BCR-ABL–activated signaling pathways
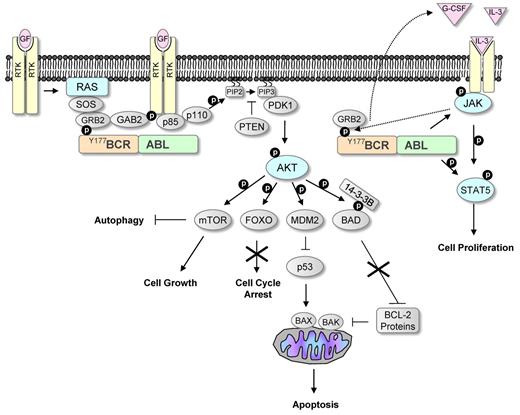
Once BCR-ABL was established as the oncogene causing CML, significant effort was invested in understanding its molecular mechanism of action. To date several signaling pathways affected by the constitutively active BCR-ABL have been identified, as well as numerous binding partners and substrates that provide a link between these pathways and the defects that characterize CML (Figure 1).
BCR-ABL signaling mimics growth factor activation. Growth factors are known to activate receptor tyrosine kinases (RTKs), leading to the activation of the RAS and PI3K/AKT pathways. These pathways are commonly activated in human cancers. Phosphorylation of tyrosine 177 within the BCR part of BCR-ABL leads similarly to activation of these pathways by interacting with the SH2 domain in GRB2. GRB2 activates RAS by interacting with Son of Sevenless (SOS), and it also activates PI3K by mediating phosphorylation of GAB2. Constitutive activation of PI3K leads to conversion of phosphatidylinositol bisphosphate (PIP2) to phosphatidylinositol triphosphate (PIP3). This can be inhibited by phosphatase and tensin homolog (PTEN), a phosphatase that is down-regulated in many cancers. PIP3 provides a platform for the recruitment of the serine/threonine kinases AKT and 3-phosphoinositide-dependent protein kinase-1 (PDK1), leading to phosphorylation and activation of AKT. Activated AKT phosphorylates many downstream targets that affect proliferation and survival, including the FOXO transcription factors and the proapoptotic protein BAD. AKT-mediated phosphorylation of FOXO inhibits its nuclear entry and therefore suppresses its activity, leading to increased cell proliferation. The proapoptotic activity of BAD is also suppressed after AKT-mediated phosphorylation. Phosphorylation of BAD prevents it binding to and inhibiting the function of antiapoptotic BCL-2 proteins that inhibit the proapoptotic proteins BAK and BAX. AKT activation leads to down-regulation of the tumor suppressor p53 by increasing the levels of the negative regulator of p53, MDM2. AKT also phosphorylates the serine/threonine kinase mTOR. Activated mTOR promotes protein translation and inhibits autophagy. BCR-ABL also leads to the activation of the JAK/STAT pathway, a pathway that is frequently activated in myeloproliferative diseases. JAK is a nonreceptor tyrosine kinase and is normally activated by growth factors. BCR-ABL has been shown to induce IL-3 and G-CSF production that could lead to activation of this pathway. Active JAK phosphorylates the transcription factor STAT5 and BCR-ABL has also been shown to directly phosphorylate STAT5, leading to increased proliferation.
BCR-ABL signaling mimics growth factor activation. Growth factors are known to activate receptor tyrosine kinases (RTKs), leading to the activation of the RAS and PI3K/AKT pathways. These pathways are commonly activated in human cancers. Phosphorylation of tyrosine 177 within the BCR part of BCR-ABL leads similarly to activation of these pathways by interacting with the SH2 domain in GRB2. GRB2 activates RAS by interacting with Son of Sevenless (SOS), and it also activates PI3K by mediating phosphorylation of GAB2. Constitutive activation of PI3K leads to conversion of phosphatidylinositol bisphosphate (PIP2) to phosphatidylinositol triphosphate (PIP3). This can be inhibited by phosphatase and tensin homolog (PTEN), a phosphatase that is down-regulated in many cancers. PIP3 provides a platform for the recruitment of the serine/threonine kinases AKT and 3-phosphoinositide-dependent protein kinase-1 (PDK1), leading to phosphorylation and activation of AKT. Activated AKT phosphorylates many downstream targets that affect proliferation and survival, including the FOXO transcription factors and the proapoptotic protein BAD. AKT-mediated phosphorylation of FOXO inhibits its nuclear entry and therefore suppresses its activity, leading to increased cell proliferation. The proapoptotic activity of BAD is also suppressed after AKT-mediated phosphorylation. Phosphorylation of BAD prevents it binding to and inhibiting the function of antiapoptotic BCL-2 proteins that inhibit the proapoptotic proteins BAK and BAX. AKT activation leads to down-regulation of the tumor suppressor p53 by increasing the levels of the negative regulator of p53, MDM2. AKT also phosphorylates the serine/threonine kinase mTOR. Activated mTOR promotes protein translation and inhibits autophagy. BCR-ABL also leads to the activation of the JAK/STAT pathway, a pathway that is frequently activated in myeloproliferative diseases. JAK is a nonreceptor tyrosine kinase and is normally activated by growth factors. BCR-ABL has been shown to induce IL-3 and G-CSF production that could lead to activation of this pathway. Active JAK phosphorylates the transcription factor STAT5 and BCR-ABL has also been shown to directly phosphorylate STAT5, leading to increased proliferation.
BCR-ABL mimics growth factor stimulation in many ways; activation of these signaling pathways leads to (1) increased proliferation and decreased apoptosis of hemopoietic stem and progenitor cells, giving rise to a massive increase in myeloid cell numbers; (2) reduced growth factor-dependence; and (3) abnormal interaction with the extracellular matrix and stroma, leading to premature release of immature myeloid cell into the circulation. These events, together with the fact that BCR-ABL promotes genomic instability, ultimately drive disease progression.7
RAS/MAP kinase pathway
The RAS pathway is well studied, both in normal and cancer cells, and activating mutations in RAS are found in many human cancers. Phosphorylation of tyrosine 177 within the BCR region of BCR-ABL allows for interaction with the adapter molecule GRB2, an important factor in human hemopoietic progenitor transformation.10 Although BCR-ABL does not affect the level of GRB2 expression, this binding enables the formation of complexes between GRB2 and SOS (ie, Son of Sevenless; a guanine-nucleotide exchange factor of RAS), leading to activation of RAS signaling.11
PI3K/AKT pathway
Another pathway commonly activated in various cancers and a crucial event in tumorigenesis is the PI3K/AKT pathway.12 Activation of receptor TKs by growth factors or BCR-ABL expression induces the activation of this pathway.13 Many molecules downstream of AKT have been shown to be important players in mediating the leukemogenic effects of BCR-ABL, either through activation or inactivation, including BAD, MDM2, mammalian target of rapamycin (mTOR), and the FOXO subclass of forkhead factors (FOXO1, FOXO3a, and FOXO4).12 (1) BAD mediates its proapoptotic effect by binding to antiapoptotic BCL-2 proteins and preventing their functions.14 (2) MDM2 is a negative regulator of p53 and has been shown to be up-regulated by BCR-ABL.15 (3) mTOR functions mainly in the translational control of proteins involved in cellular growth, size, and cycle regulation.16 In BCR-ABL–expressing cells, mTOR is active and rapamycin, an inhibitor of mTOR, suppresses the growth of BCR-ABL–transformed cells.17 mTOR also negatively regulates autophagy (discussed in “Molecular mechanism of autophagy”). Finally, (4) the FOXO transcription factors are involved in cell-cycle arrest and/or apoptosis.18 BCR-ABL transformation has been shown to inhibit FOXO3a activity by maintaining its constitutive phosphorylation and cytoplasmic retention.19
JAK-STAT pathway
Another antiapoptotic pathway activated by BCR-ABL is the JAK-STAT pathway. Recent in vivo experiments in mouse models have portrayed Stat5 as an indispensible factor for induction and maintenance of Bcr-Abl–positive leukemia, underlining the critical role of STAT5 in CML.20,21 Furthermore, a recent study by Warsch et al22 explored the role of STAT5 in the development of TK inhibitor (TKI) resistance, demonstrating that enhanced STAT5 levels reduced sensitivity to TKIs in a STAT5-dependent but JAK2-independent manner. The role of the RAS and the JAK-STAT pathways in the cellular response to various growth factors could explain why many growth factor–dependent cell lines become independent after BCR-ABL expression.6 Evidence has emerged illustrating an autocrine loop dependent on BCR-ABL–induced secretion of growth factors. BCR-ABL induces an IL-3 and G-CSF–positive feedback mechanism in early progenitor cells that could alter critical cell-cycle regulators.23 Autocrine production of IL-3 and G-CSF may therefore play a role in promoting cell-cycle entry of primitive leukemic stem cells and early progenitors during the chronic phase (CP) of disease, where the G-CSF receptor is expressed.24
CML treatment
Disease course and progression
The natural history of CML is triphasic, involving an initial CP, lasting 3-5 years, and followed by progression either to accelerated phase and then blast crisis or directly to blast crisis. These advanced phases of the disease carry a much poorer prognosis for patients, with their median survival measured in months. CML progression is generally a gradual and complex process promoted by the accumulation of secondary genetic and epigenetic changes rather than isolated alterations. As stated by Perrotti et al25 in their review, BCR-ABL expression and activity may be significantly elevated at advanced stage via gene amplification, increased promoter activity, or other less-direct mechanisms, such as decreased miR-203 levels, inhibition of SHP-1 phosphatase, and inactivation of PP2A. BCR-ABL–driven manipulation of the transcriptome plays a key role in disease progression by activating mitogenic, antiapoptotic, and antidifferentiation mediators (eg, MYC, JAK2, hnRNP-E2, MDM2, STAT5, BMI-1, and BCL-2), inhibiting tumor suppressors (eg, p53 or CEBPa), or through aberrant splicing of modulators such as GSK3β and PYK2.25
TKIs in the management of CML
During the 1980s increased understanding of the mechanism of BCR-ABL as the main driver of the disease led to the development of an Abl-specific TKI, imatinib (Gleevec, Norvartis Pharmaceuticals Corp; and Glivec, Pharmalink Ltd).26 Preclinical studies followed by clinical trials verified the success of imatinib in specifically targeting BCR-ABL–positive cells, and subsequently, imatinib has been established as the standard therapy for CML.
The problem of resistance
The mechanisms of imatinib resistance have been extensively investigated, and it is now known that approximately 50% of patients who relapse on imatinib have BCR-ABL point mutations in more than 40 different amino acids within the ABL kinase domain.27 These findings led to the rapid development of second-generation TKIs to circumvent this problem. Nilotinib and dasatinib both demonstrate increased potency over imatinib and inhibit most imatinib-resistant mutants except T315I.28,29 Both inhibitors have now been approved by Food and Drug Administration and European Medicines Agency and are used routinely and very effectively as second-line therapy for patients with resistance or intolerance to imatinib.30,31 Interestingly, as reported by Saglio et al32 and Kantarjian et al,33 the administration of nilotinib or dasatinib to newly diagnosed CML patients in CP resulted in greater frequencies and faster achievement of cytogenetic and molecular responses compared with standard-dose imatinib (400 mg daily). By 12 months of treatment, there was a trend toward reduced progression rates that achieved significance for nilotinib.32,33 On the basis of these remarkable results, both drugs are now undergoing approval for first-line use in many countries. For the T315I mutant inhibitors including ponatinib (AP24534), a pan–BCR-ABL inhibitor (phase 2), and DCC-2036, a “switch-control” inhibitor (phase 1), have advanced to clinical trials.34-37
Combination studies
In an effort to make patients' responses more robust, clinical trials have been launched involving the use of TKIs in combination with other drugs that could potentially augment TKI-induced cell death. In one such study, researchers have combined imatinib with peginterferon alfa-2a or cytarabine and currently demonstrate a significant advantage for the interferon combination in terms of molecular responses (overall survival and progression-free survival unchanged).38 Further smaller trials are being conducted in which researchers have combined imatinib with zileuton (phase 1; NCT01130688), panobinostat (NCT00686218), sodium valproate (phase 1; NCT01011998), arsenic trioxide (phase 2; NCT00250042), or hydroxychloroquine (phase 2; NCT01227135), and there is a phase 1 evaluation of dasatinib with a SMO inhibitor underway (NCT01218477).39
The problem of persistence
The persistence of disease refers to the presence of primitive BCR-ABL–positive CD34+ cells that are inherently insensitive to TKIs in patients who are otherwise responding well. Although most of these patients are in major molecular response with BCR-ABL transcripts detectable by quantitative RT-PCR, recent data also confirm the presence of BCR-ABL–expressing cells by genomic PCR in patients in complete molecular remission on TKIs.40,41 We and others have shown that 4 of the currently available TKIs (imatinib, nilotinib, dasatinib, and bosutinib) have strong antiproliferative effects but fail to induce cell death in primitive stem/progenitor cells. This finding may explain why primitive leukemic cells are present in the BM of patients with established complete cytogenetic response during the course of 5 years on imatinib.42-46 It is also in keeping with the rapid recurrence of the ability to detect BCR-ABL transcripts in most patients with apparent complete molecular remission after the discontinuation of imatinib.40,47 In recent studies by the French and Australian groups, disease recurrence rates were 61% (of 100 patients) and 56% (of 18 patients), respectively, in selected patients who had undetectable disease maintained for > 2 years before TKI discontinuation. These clinical observations have prompted the development of stem cell–directed therapy and have raised the debate as to whether CML stem cells are oncogene addicted, that is, whether CML stem cells are dependent on BCR-ABL for survival. Recent results from Corbin et al48 and our laboratory (Hamilton et al, unpublished data, submitted December 22, 2010) suggest that CML stem cells can survive after complete BCR-ABL inhibition and are therefore not oncogene addicted.
Given that BCR-ABL–expressing stem cells can survive without BCR-ABL signaling, combination therapy is likely to be one way to circumvent the problem of disease persistence. However, improving existing or combining different BCR-ABL inhibitors is unlikely to tackle the problem of persisting CML stem cells that are able to survive despite full BCR-ABL inhibition.48 Therefore, alternative survival pathways responsible for leukemic stem cell survival after oncogene inactivation must be examined to apply successful stem cell–directed therapy, potentially leading to patient cure. One example of such a survival pathway is macroautophagy (autophagy).49,50
Molecular mechanism of autophagy
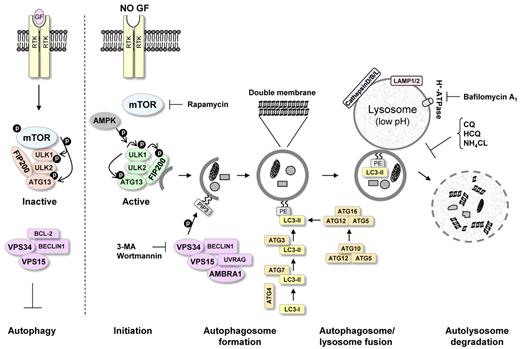
Autophagy (self [auto]-eating [phagy]) is a recycling process that leads to sequestration and degradation of damaged proteins and other intracellular material within lysosomes (Figure 2).51 Recycling of these intracellular constituents can serve as an alternative source of energy during periods of metabolic stress or starvation to maintain cellular homeostasis and survival.52
Molecular mechanism of autophagy. Autophagy initiation starts with activation of the ULK1/2 kinase complex, which also includes ATG13 and FIP200. mTOR suppresses the activity of this complex by phosphorylating ULK1 and ATG13 on “negative” sites. This complex then becomes active after AMPK activation and mTOR inhibition, for example, during starvation or treatment with rapamycin. Phosphorylated and now-active ULK1 promotes phosphorylation of ATG13 and FIP200 and dissociates from mTOR. The PI3K-III VPS34 is critical for further autophagosome formation. VPS34 forms a complex with VPS15, UVRAG, AMBRA1, and BECLIN1. This complex can be inhibited by the antiapoptotic protein BCL-2, which can interact with BECLIN1 through the BH3 domain in BECLIN1. Autophagosome completion is mediated by the ATG8 (LC3)-PE and ATG12/ATG5 conjugation systems. These systems perform the lipid modification of LC3-I, leading to LC3-II-PE binding to the autophagosomal membrane (see “Autophagosome formation”). The conversion of LC3-I to LC3-II is commonly used to monitor autophagy by various assays. Completed autophagosomes contain materials such as proteins and organelles that can be digested after autophagosome fusion with lysosomes. Lysosomes have low pH and an abundance of pH-sensitive enzymes that can break down waste materials and cellular debris. The autolysosome degradation can therefore remove unwanted materials such as damaged proteins in addition to providing the cell with new building blocks for cellular maintenance. The mTOR inhibitor rapamycin can be used to promote autophagy induction. VPS34 inhibitors, such as 3-MA and wortmannin, can be used to inhibit the early stages of autophagy. Bafilomycin A1, CQ, HCQ, and NH4Cl, which inhibit the fusion of autophagosomes and lysosomes, can be used to inhibit autophagy at later stages.
Molecular mechanism of autophagy. Autophagy initiation starts with activation of the ULK1/2 kinase complex, which also includes ATG13 and FIP200. mTOR suppresses the activity of this complex by phosphorylating ULK1 and ATG13 on “negative” sites. This complex then becomes active after AMPK activation and mTOR inhibition, for example, during starvation or treatment with rapamycin. Phosphorylated and now-active ULK1 promotes phosphorylation of ATG13 and FIP200 and dissociates from mTOR. The PI3K-III VPS34 is critical for further autophagosome formation. VPS34 forms a complex with VPS15, UVRAG, AMBRA1, and BECLIN1. This complex can be inhibited by the antiapoptotic protein BCL-2, which can interact with BECLIN1 through the BH3 domain in BECLIN1. Autophagosome completion is mediated by the ATG8 (LC3)-PE and ATG12/ATG5 conjugation systems. These systems perform the lipid modification of LC3-I, leading to LC3-II-PE binding to the autophagosomal membrane (see “Autophagosome formation”). The conversion of LC3-I to LC3-II is commonly used to monitor autophagy by various assays. Completed autophagosomes contain materials such as proteins and organelles that can be digested after autophagosome fusion with lysosomes. Lysosomes have low pH and an abundance of pH-sensitive enzymes that can break down waste materials and cellular debris. The autolysosome degradation can therefore remove unwanted materials such as damaged proteins in addition to providing the cell with new building blocks for cellular maintenance. The mTOR inhibitor rapamycin can be used to promote autophagy induction. VPS34 inhibitors, such as 3-MA and wortmannin, can be used to inhibit the early stages of autophagy. Bafilomycin A1, CQ, HCQ, and NH4Cl, which inhibit the fusion of autophagosomes and lysosomes, can be used to inhibit autophagy at later stages.
An understanding of the molecular regulation of autophagy has been gained only in the past 10 years. Autophagy has been thoroughly examined in yeast, where many autophagy-related genes (ATG) have been identified, and several of these genes have now been shown to have mammalian orthologs. The process can be divided into sequential steps that ultimately result in breakdown of cytoplasmic material within the lysosomes (Figure 2).
Initiation
It has recently been elucidated that the initiation of autophagy starts with activation of a serine/threonine kinase complex containing ULK1 and 2 (yeast Atg1), ATG13, and FIP200.53 During glucose starvation, AMP-activated protein kinase phosphorylates ULK1, promoting activation of autophagy. Alternatively, when nutrient sufficiency is established, mTOR blocks this interaction.54,55 Hence, mTOR acts as the main regulator of autophagy by directly inhibiting the ULK1/2/ATG13/FIP200 complex and other ATG proteins.56 Consequently, on mTOR inhibition (ie, cell starvation or rapamycin treatment), the UKL1 complex dissociates from mTOR, allowing it to accumulate at an isolation cup-shaped membrane, called the phagophore.53
Autophagosome formation
The class III PI3K (PI3K-III) VPS34 is critical for further expansion of phagophores to autophagosomes, double-membraned vesicles that are the main mediators of autophagy.57 VPS34 forms a complex with VPS15 and autophagy protein BECLIN1 (yeast Atg6).51 The antiapoptotic protein BCL-2 can interact with the BH3 domain of BECLIN1 and inhibit autophagy.58 This interaction is reduced on starvation, freeing BECLIN1 to activate autophagy.59 It has been suggested that this antiautophagy function of BCL-2 might maintain autophagy at levels that are optimal for cell survival. BCL-2 has also been shown to suppress autophagy by inhibiting cytosolic calcium elevation, which has been shown to induce autophagy.60 UVRAG (UV irradiation resistance-associated gene) and AMBRA1 are other proteins that have been shown to interact with BECLIN1 and positively regulate autophagy, indicating that BECLIN1-dependent autophagy might be tightly regulated by BECLIN1 binding partners.61
Autophagosome completion is mediated by 2 ubiquitin-like conjugation systems: the ATG12/ATG5 and ATG8-PE (ATG8 is also known as microtubule-associated protein 1 light chain 3, LC3; PE, phosphatidylethanolamine) conjugation systems, which are both essential for autophagy.62 The ATG8-PE process is mediated by ATG4, ATG7 (E1-ubiquitin enzyme), and ATG3 (E2-ubiquitin enzyme), which perform the lipid modification of ATG8. ATG4 also cleaves the amide bond between ATG8 and PE to release the protein from membranes.63 The ATG12/ATG5 process starts with conjugation of ATG12 to ATG5, with ATG10 acting as an E2 enzyme. The ATG12/ATG5 conjugate then interacts with ATG16, and this complex (ATG12/ATG5/ATG16) exerts an E3 enzyme-like function and regulates the ATG8-PE binding to the autophagosomal membrane.64 LC3 (ATG8) has been investigated most thoroughly and acts now as the main marker for autophagosomes. LC3 maturation completes with the reversible conjugation of LC3-I to PE (by ATG7 and ATG3) to form LC3-II on the surface of autophagosomes.65 This alteration is used to monitor autophagy by various assays. However, correct interpretation of LC3-I/II conversion is very important because accumulation of autophagosomes could, for example, reflect either increased autophagosome formation because of an increase in autophagic activity or inhibition of autophagosome turnover. Leading scientists in the field have published detailed guidelines and a standard set of criteria for monitoring autophagy that should help new investigators that join this growing field.66
Maturation and degradation
The maturation process includes the fusion of the outer membrane of autophagosomes with lysosomes to form autolysosomes. The mechanism of this process is not very well understood, but it has been shown that LAMP1/2 (lysosomal proteins), RAB7 (small GTPase), and cathepsin D/B/L are all involved in the autophagosome maturation, with RAB7 and LAMP1/2 potentially facilitating autophagosome and lysosome fusion.67 UVRAG has also been shown to be involved in the maturation step, where it stimulates RAB7 GTPase activity and autophagosome fusion with lysosomes.57 At the final autolysosomes, the inner single membrane of the autophagosomes and its cargo is lysed by lysosomal hydrolases, especially cathepsins and the content degraded.68
However, autophagy should not be viewed only as recycling machinery. This process is constantly taking place at basal levels in the majority of mammalian cells, as early as the stage of oocyte fertilization, and is involved in a plethora of mechanisms and pathways, including cell development, differentiation, homeostasis, quality control, aging, infection and immunity, neurodegeneration, and cancer.69
The paradoxical role of autophagy in cancer
Autophagy has been suggested to work as a double-edged sword in cancer development and progression by inducing both tumor cell survival and death. These diverse effects depend to a large degree on the type of tumor, stage of disease, and nature of treatment.70 Basal autophagy within normal cells is pivotal because it (1) promotes genetic stability by removing damaged mitochondria that would otherwise produce reactive oxygen species and damage the DNA and (2) functions as a “guardian” of the proteome and promotes adaption under changing conditions and/or stress.69 The first strong link between autophagy and cancer was published in 1999 by Levine's laboratory, in which they demonstrated that BECLIN1 is a haploinsufficient tumor suppressor.71 Subsequent studies revealed that BECLIN1 is lost at high frequencies in human breast, ovarian, and prostate cancers and that BECLIN1-heterozygous mice are prone to tumor development.72
However, autophagy inhibition leads to enhanced apoptotic effect of irradiation, alkylating agents, and arsenic trioxide in tumor cells.70 Hence, autophagic activity may suppress tumorigenesis in healthy cells by removing damaged organelles and proteins, reducing reactive oxygen species, and suppressing necrosis-induced inflammation that can lead to the development of cancer when chronic.73 However, it seems to have a cytoprotective role in established tumor cells and thus, its inhibition could enhance apoptosis. Greater understanding of the functional role of autophagy with regard to cancer in the future will be critical to enable specific autophagy inhibition/promotion in a range of tumors for therapeutic gain.
Pharmacologic inhibition of autophagy
Several inhibitors that disrupt the autophagy process are available and frequently used to study the role of autophagy in tumorigenesis and in response to cancer therapy. 3-Methyladenine (3-MA) is a PI3K-III inhibitor and therefore inhibits autophagosome formation. Wortmannin (pan-PI3K inhibitor) has also been used to inhibit autophagy at its early stage. In contrast to the PI3K-III inhibitors, bafilomycin A1, chloroquine (CQ), hydroxychloroquine (HCQ), and NH4Cl inhibit autophagy at a later stage by preventing the fusion of autophagosomes and lysosomes.74 Bafilomycin A1 is a specific inhibitor of vacuolar-type H+-ATPase, whereas CQ, HCQ, and NH4Cl are lysosomotropic drugs that increase intracellular pH and therefore impair autophagosome and lysosome fusion and autophagic protein degradation. Although these drugs are not specific autophagy inhibitors and will have an effect on other pathways, they have proven very useful in studying and inhibiting autophagy in the laboratory. Autophagy inhibition in a specific manner is currently performed in vitro via siRNA-mediated knockdown of essential autophagy genes, such as BECLIN1, ATG5, and ATG7. The challenge for the future is the development of autophagy-specific drugs. With our ever-increasing understanding of the main regulators of this key process, this step should not be far away. There are already some critical players that appeal as attractive drug targets, such as the kinases in the ULK1 protein complex, VPS34, and essential ATG autophagy proteins (eg, ATG4 or ATG7), for which activity could be repressed with small inhibitory compounds.70
Autophagy in CML
BCR-ABL signaling, like growth factor activation, leads to activation of the PI3K/AKT pathway and mTOR. In line with this, it has been demonstrated that BCR-ABL–expressing mouse hemopoietic precursor cells have low basal levels of autophagy but are highly dependent on this process.75 Inhibition of BCR-ABL by TKIs has now been shown to not only induce apoptosis but also autophagy, a similar effect to that seen after growth factor withdrawal.49 However, the questions of (1) what level of autophagy is taking place within HSCs and (2) what is its role remain unanswered. However, the authors of 2 recent studies have elucidated the field. First, Liu et al76 reported that FIP200 is indispensible for the function and maintenance of fetal murine HSCs in a cell-autonomous manner. The hypothesis that autophagy is essential for the maintenance of HSCs was further supported by Mortensen et al,77 who showed in mice that adult Atg7-deficient HSCs fail to engraft lethally irradiated recipients, clearly depicting autophagy as an essential modulator of HSCs maintenance.
Many recent publications have shown that chemotherapeutic agents used in the treatment of hemopoietic malignancies, including CML, induce both apoptosis and autophagy. The ultimate fate of a cell resembles a hub that integrates all the signals from the apoptosis, autophagy, necrosis, and senescence responses. Hence, the choice between death and survival depends critically on the balance between these mechanisms. However, the ultimate decision between apoptosis and survival may also differ depending on the maturational status of the cell. As hypothesized in a recent review by Calabretta and Salomoni,78 TKI treatment in CML could potentially trigger different signaling pathways in primitive CD34+CD38− cells versus their progenitors, ie, inhibition of BCR-ABL could trigger apoptosis within the progenitors but protective autophagy at the primitive stem cell level.
In many situations targeting autophagy has increased the effect of the anticancer drug, although it is not yet clear whether autophagy inhibition is enhancing programmed or necrotic cell death. Nonetheless, with the use of mouse models it has been convincingly shown that autophagy serves as a survival pathway, and its inhibition enhances p53 or drug-induced cell death in lymphoma cells.79 This finding indicates that autophagy inhibitors, such as CQ, in combination with therapies designed to induce cell death, could be beneficial in the treatment of hematologic neoplasms. Recently several publications have linked CML with autophagy (Table 1).49,50,80-87 In 2006 it was shown that the neurotoxin crotoxin induced autophagy in K562 cells and blockade of autophagy with 3-MA and NH4Cl potentiated the neurotoxin's cytotoxicity.80 One year later, Carew et al81 showed that targeting autophagy by using CQ enhanced the anticancer activity of the histone deacetylase inhibitor suberoylanilide hydroxamic acid in imatinib-resistant primary CML cells. These reports represented the first evidence that CML cells induce autophagy as a cell-survival response after treatment with anticancer drugs, suggesting autophagy inhibition as a novel strategy with therapeutic implications for imatinib resistance.82 The first evidence that inhibition of BCR-ABL activity induced autophagy came 1 year later, when bafetinib (INNO-406), a potent c-ABL and LYN kinase inhibitor, was shown to induce autophagy in K562 cells.83 Again, this autophagy response had a protective role as treatment with CQ-enhanced cell death induced by bafetinib. A few months later, imatinib was also shown to induce autophagy in K562 cells, and cotreatment with imatinib and CQ markedly enhanced imatinib-induced death.84
Autophagy induction in CML cells after various drug treatments
| Drug . | Main drug target . | Cell type . | Drug affect . | Autophagy inhibitor . | Affect of drug and autophagy inhibitor . | Reference . |
|---|---|---|---|---|---|---|
| Crotoxin | Not known | Cell line (K562) | Apoptosis and autophagy induction | 3-MA NH4Cl | Increased cell death | 80 |
| SAHA | Histone deacetylases | Cell lines, primary CML MNC | Apoptosis and autophagy induction | 3-MA CQ | Increased cell death | 81,82 |
| Bafetinib | BCR-ABL | Cell line (K562) | Apoptosis and autophagy induction | CQ 3-MA | Increased cell death | 83 |
| Imatinib | BCR-ABL | Cell line (K562) | Apoptosis and autophagy induction | CQ | Increased cell death | 84 |
| Imatinib, dasatinib | BCR-ABL | Cell lines, primary CD34+ cells, including CD34+38− | Apoptosis and autophagy induction | CQ Bafilomycin A1 | Increased cell death | 49,50 |
| Imatinib | BCR-ABL | K562 | Apoptosis and autophagy induction | 3-MA | Increased cell death | 85 |
| OSI-027 | mTOR | Cell line (K562) | Apoptosis and autophagy induction | CQ | Increased cell death | 88 |
| Resveratrol | Not known AMPK? | Cell lines, primary CD34+ cells | Apoptosis and autophagy induction | Bafilomycin A1 | Decreased cell death | 89 |
| Drug . | Main drug target . | Cell type . | Drug affect . | Autophagy inhibitor . | Affect of drug and autophagy inhibitor . | Reference . |
|---|---|---|---|---|---|---|
| Crotoxin | Not known | Cell line (K562) | Apoptosis and autophagy induction | 3-MA NH4Cl | Increased cell death | 80 |
| SAHA | Histone deacetylases | Cell lines, primary CML MNC | Apoptosis and autophagy induction | 3-MA CQ | Increased cell death | 81,82 |
| Bafetinib | BCR-ABL | Cell line (K562) | Apoptosis and autophagy induction | CQ 3-MA | Increased cell death | 83 |
| Imatinib | BCR-ABL | Cell line (K562) | Apoptosis and autophagy induction | CQ | Increased cell death | 84 |
| Imatinib, dasatinib | BCR-ABL | Cell lines, primary CD34+ cells, including CD34+38− | Apoptosis and autophagy induction | CQ Bafilomycin A1 | Increased cell death | 49,50 |
| Imatinib | BCR-ABL | K562 | Apoptosis and autophagy induction | 3-MA | Increased cell death | 85 |
| OSI-027 | mTOR | Cell line (K562) | Apoptosis and autophagy induction | CQ | Increased cell death | 88 |
| Resveratrol | Not known AMPK? | Cell lines, primary CD34+ cells | Apoptosis and autophagy induction | Bafilomycin A1 | Decreased cell death | 89 |
AMPK indicates AMP-activated protein kinase; BCR-ABL, breakpoint cluster region-Abelson murine leukemia; CML, chronic myeloid leukemia; CQ, chloroquine; 3-MA, 3-methyladenine; MNC, mononuclear cell; mTOR, mammalian target of rapamycin; and SAHA, suberoylanilide hydroxamic acid.
The strongest evidence that autophagy inhibition might enhance the therapeutic effects of imatinib in the treatment of CML was shown in 2009.49,50 Then, we demonstrated that autophagy inhibition combined with TKIs (imatinib, dasatinib, or nilotinib) resulted in near-complete elimination of phenotypically and functionally defined CML stem cells. Our data were further supported by a study from Crowley et al, 85 who also showed that autophagy works as a cellular protection against imatinib-induced stress. Despite a recently published role for ABL kinases in the regulation of late-stage autophagy, the effect in our study was specific to BCR-ABL inhibition.88 Autophagy was not induced on imatinib treatment in a cell line carrying the T315I mutation, and ectopic expression of resistant c-ABL (T315I) mutant did not block the induction after imatinib exposure. It is likely that imatinib induces autophagy by inhibiting mTOR, the negative regulator of autophagy, and it was shown to potentially be associated with endoplasmic reticulum stress, but the exact molecular mechanism is still not clear and is being investigated in our laboratory. Importantly, our data were supported by in vivo experiments in a mouse model. As published by Altman et al,89 transplantation of Bcr-Abl–expressing hemopoietic cells that were depleted of Atg3 by conditional deletion to lethally irradiated mice revealed that Bcr-Abl failed to induce leukemia in an autophagy ablated background.
All the aforementioned promising results within the CML stem cell population have led to the initiation of the phase 2 CHOICES (CHlOroquine and Imatinib Combination to Eliminate Stem cells) clinical trial. The CHOICES trial is a randomized trial of imatinib versus HCQ and imatinib for CML patients in major cytogenetic response with residual disease detectable by quantitative PCR and is the first clinical trial using autophagy inhibition in CML.
To support the role of active mTOR in blocking autophagy in BCR-ABL–expressing cells, inhibition of mTOR with the use of OSI-027, which inhibits both mTORC1 and mTORC2 (the 2 separate mTOR complexes), has recently been shown to induce autophagy in K562 cells.86 Furthermore, the combination of OSI-027 with CQ resulted in increased apoptosis, indicating that autophagy was acting as a survival mechanism after OSI-027 treatment.
Paradoxically, drug treatment has also been shown to induce autophagic cell death (type II apoptosis) in imatinib-sensitive and imatinib-resistant K562 cells.87 Puissant et al87 demonstrated that resveratrol (phytoalexin produced naturally by several plants) treatment induced apoptosis and autophagy through AMP-activated protein kinase–dependent inhibition of the mTOR pathway. After autophagy inhibition with either bafilomycin A1 or knockdown of ATG5 or LC3, resveratrol-induced apoptosis was partially rescued. It was also shown that bafilomycin A1 reduced resveratrol-induced cell death in primary CD34+ CML cells, with the opposite effect found on imatinib-induced death, where bafilomycin A1 enhanced the imatinib effect (consistent with our previous results).49,87
These results underline the importance of understanding the type of autophagy induction after specific types of stress and anticancer treatment so the most appropriate manipulation of autophagy can be applied for therapeutic intervention. Ongoing and future investigation regarding the use of autophagy regulators in vivo will shed light on this enigma because autophagy response differs not only among different patients and different organs within the same patient but also among cells within the same tumor.69 Thus, how CHOICES and other clinical trials' patients respond to combination therapy with an autophagy inhibitor will provide us with essential information on whether modulation of autophagy in human tumors is a future therapeutic option of merit.
Autophagy inhibition in the clinic
None of the available autophagy inhibitors is specific for autophagy, and hence, affect other pathways. Nevertheless, CQ and HCQ are used as antimalarial drugs and have been approved for treatment of other diseases, eg, rheumatoid arthritis. Because HCQ is less toxic than CQ to the retina, several clinical trials have now been initiated in patients with hemopoietic or solid tumors with the use of HCQ as an autophagy inhibitor.90,91 Investigators in these trials, like the CHOICES trial, use the combination of HCQ with a targeted therapy, or in combination with cytotoxic chemotherapy or metabolic stress inducers, for example, in multiple myeloma, where HCQ is used in combination with the proteasome inhibitor bortezomib (velcade).90 However, the overall hypothesis in all these trials is that autophagy is a survival mechanism of therapeutic resistance and that blocking autophagy might increase the effect of the anticancer treatment. Although CQ has convincingly been shown to block autophagy in vitro, it still remains to be shown that drug/radiation-induced autophagy can be blocked with the use of a 400- to 800-mg daily dosing of HCQ in human tumors in vivo. Therefore, developing rigorous biomarkers to monitor autophagy in vivo will be critical.
Conclusion
In many ways CML represents a model disease and a paradigm for cancer stem cells, oncogene addiction and targeted therapies. With the introduction of TKI for the treatment of CML, we have embarked on a journey aiming to cure the disease with orally administered, nontoxic agents. The major current impediment to cure for patients in CP resides in the cancer stem cell population that is neither oncogene addicted nor sensitive to TKI. The recent discovery that TKI induce the process of autophagy, which in turns protects CML stem cells, has presented the field with an exciting opportunity to kill one bird, CML, with two stones, TKI and autophagy inhibitors, with potential for cure.
Acknowledgments
G.V.H. is currently funded by the Kay Kendall Leukaemia Fund (KKL404). M.K. is funded by a Glasgow University PhD scholarship. Our earlier work on autophagy was conducted with MRC and LLR funding.
Authorship
Contribution: G.V.H., M.K., and T.L.H wrote the paper and contributed to revisions; and T.L.H. checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tessa L. Holyoake, Paul O'Gorman Leukaemia Research Centre, Institute of Cancer Sciences, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, G12 OYN, United Kingdom; e-mail: Tessa.Holyoake@glasgow.ac.uk.