Abstract
Deletions of chromosome 5q are associated with poor outcomes in acute myeloid leukemia (AML) suggesting the presence of tumor suppressor(s) at the locus. However, definitive identification of putative tumor suppressor genes remains controversial. Here we show that a 106-nucleotide noncoding RNA vault RNA2-1 (vtRNA2-1), previously misannotated as miR886, could potentially play a role in the biology and prognosis of AML. vtRNA2-1 is transcribed by polymerase III and is monoallelically methylated in 75% of healthy individuals whereas the remaining 25% of the population have biallelic hypomethylation. AML patients without methylation of VTRNA2-1 have a considerably better outcome than those with monoallelic or biallelic methylation (n = 101, P = .001). We show that methylation is inversely correlated with vtRNA2-1 expression, and that 5-azanucleosides induce vtRNA2-1 and down-regulate the phosphorylated RNA-dependent protein kinase (pPKR), whose activity has been shown to be modulated by vtRNA2-1. Because pPKR promotes cell survival in AML, the data are consistent with vtRNA2-1 being a tumor suppressor in AML. This is the first study to show that vtRNA2-1 might play a significant role in AML, that it is either mono- or biallelically expressed in the blood cells of healthy individuals, and that its methylation state predicts outcome in AML.
Introduction
Interstitial deletions or loss of the long arm of chromosome 5 are frequent events in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS), suggesting the presence of tumor suppressor gene(s) (TSG) at this locus. Two critical deleted regions (CDRs) have been identified on 5q. One region is linked to de novo AML and high-risk MDS (CDR1 at 5q31),1,2 and a second region to the low-risk MDS 5q− syndrome (CDR2 at 5q32-33).3 In AML, deletion of 5q is a poor prognostic factor, while the prognostic significance of 5q loss in MDS is variable depending on whether or not 5q− is the sole genetic abnormality.4
Several candidate tumor suppressors have been suggested to be located in the CDRs on 5q, but targeted disruption of single genes by somatic mutations has not been identified despite intense efforts. Instead, several studies suggest that a gene dosage effect may be critical and that haploinsufficiency of certain genes on 5q can recapitulate BM failure syndromes.5-8 Interestingly, disruption of noncoding RNAs (ncRNAs) and ribonucleoproteins has been implicated in a variety of BM failure diseases. These include, among others, genes involved in ribosomal biogenesis in the 5q− syndrome (RPS14),5 Diamond-Blackfan anemia (RPS19, RPS24, RPS17, RPS7, RPL35A, RPL5, and RPL11) and Shwachman-Diamond syndrome (SBDS), and the telomerase and telomere-associated proteins in dyskeratosis congenita/aplastic anemia (TERT, TERC, and DKC1).9-11
The CTNNA1 and the RIL genes have been suggested as tumor suppressors in the CDR1 region because their promoters are hypermethylated in primary MDS and AML cases with and without 5q loss.2,12,13 Although association to advanced-stage MDS was shown for both genes, no prognostic influence has so far been demonstrated in AML. Hence, we searched for other candidate genes in the CDR1 region and identified the putative microRNA (miR) MIR886, which became induced by azanucleoside treatment in an AML cell line, suggesting that it was regulated by promoter methylation.14 However, 3 previous studies and our own confirmatory analyses showed that this is not a bona fide miR but a different type of longer ncRNA with homology to vault RNAs (vtRNAs).15-17 Accordingly, “pre-miR-886” has subsequently been annotated as VTRNA2-1.15,16 vtRNAs are small, ∼100 nt long, polymerase III (pol III) transcripts, which all locate to chromosome 5q31. The vtRNAs and the associated ribonucleotide (vault) complex has been linked with a multitude of cellular functions among which is multidrug resistance and innate immunity,18 but many of these observations are still controversial and may be cell-type dependent.17 Indeed, a recent study implied that vtRNA2-1 is distinct from the genuine vtRNAs and fails to associate to the vault complex.17 Instead, it was suggested to be involved in the regulation of the double-stranded RNA-dependent kinase, PKR. Down-regulation of vtRNA2-1 lead to activation of PKR and its downstream targets including NFκB, leading the authors to suggest that vtRNA2-1 might be a novel tumor suppressor.17 Because constitutive PKR and NFκB activity are well-documented features in AML,19,20 we speculated whether this may at least in part be mediated via loss of VTRNA2-1 at the CDR at 5q31.1.
Here, we show that VTRNA2-1 may be directly implicated in AML, that expression of vtRNA2-1 is regulated by promoter methylation, and that a majority of the healthy Danish population (∼ 75%) carry a monoallelically silenced VTRNA2-1 in normal hematopoietic cells. Our data suggest that the gene dosage of this particular type of ncRNA may play an important role in tumor progression or response to therapy because patients with hypomethylation of both alleles of the VTRNA2-1 promoter have a significantly better prognosis, while those with hypermethylation or loss of the second VTRNA2-1 copy have a poorer outcome.
Methods
Cell lines and drug treatment
The HL60 cell line was originally derived from a female patient with AML-FAB M2.21 Its origin was confirmed by M-FISH.14 All hematopoietic cancer cell lines were cultured in RPMI 1640 medium with Glutamax-1 plus 10% FBS. T24, LD419, UROtsa, and UMUC3 cells were cultured in McCoy 5A medium supplemented with 10% FBS. To all cultures, 100 U/mL penicillin and 100 μg/mL streptomycin was added. As previously described, HL60 cells were treated with azanucleosides14 and cytosine arabinoside (AraC; Sigma-Aldrich) 20nM or 100nM and harvested on day 2 and 8.22
Patients and healthy donors
BM cells (BMCs) were obtained from 101 patients with AML at the time of diagnosis. All patients were diagnosed and treated heterogeneously according to standard regimens or in the AML15 MRC protocol23 at the Department of Hematology, Rigshospitalet from 2003-2009. Patient characteristics at diagnosis are summarized in Table 1. The distribution according to the French-American-British classification was M0: 5%, M1:13%, M2:26%, M3: 5%, M4:23%, M5:9%, M6: 0%, M7:0%.
Patient characteristics
| . | All . | Methylated . | Unmethylated . | P . |
|---|---|---|---|---|
| 101 | 63 | 38 | ||
| Cytogenetic risk | ||||
| Favorable | 17 | 6 | 11 | |
| Intermediate | 68 | 49 | 19 | |
| Poor | 12 | 7 | 5 | .017 |
| Missing | 4 | 1 | 3 | |
| Leukocytes, ×109/L | ||||
| < 10 | 29 | 18 | 11 | |
| 10-19.9 | 22 | 12 | 10 | |
| 20-49.9 | 16 | 7 | 9 | |
| 50-99.9 | 18 | 14 | 4 | |
| > 100 | 12 | 10 | 2 | .134 |
| Missing | 4 | 2 | 2 | |
| Sex | ||||
| Female | 45 | 30 | 15 | |
| Male | 56 | 33 | 23 | .536 |
| Age, y | 56.87 (SD 16.3) | 57.8 (SD 15.1) | 55.33 (SD 18.3) | |
| < 60 | 50 | 30 | 20 | |
| ≥ 60 | 51 | 33 | 18 | .684 |
| . | All . | Methylated . | Unmethylated . | P . |
|---|---|---|---|---|
| 101 | 63 | 38 | ||
| Cytogenetic risk | ||||
| Favorable | 17 | 6 | 11 | |
| Intermediate | 68 | 49 | 19 | |
| Poor | 12 | 7 | 5 | .017 |
| Missing | 4 | 1 | 3 | |
| Leukocytes, ×109/L | ||||
| < 10 | 29 | 18 | 11 | |
| 10-19.9 | 22 | 12 | 10 | |
| 20-49.9 | 16 | 7 | 9 | |
| 50-99.9 | 18 | 14 | 4 | |
| > 100 | 12 | 10 | 2 | .134 |
| Missing | 4 | 2 | 2 | |
| Sex | ||||
| Female | 45 | 30 | 15 | |
| Male | 56 | 33 | 23 | .536 |
| Age, y | 56.87 (SD 16.3) | 57.8 (SD 15.1) | 55.33 (SD 18.3) | |
| < 60 | 50 | 30 | 20 | |
| ≥ 60 | 51 | 33 | 18 | .684 |
In a separate patient cohort from the Nordic MDS collaboration, BM mononuclear cells (BM MNCs) and sorted CD34+ cells were obtained from heavily pretreated MDS (n = 12) and AML (n = 9) patients with deletion of 5q before entry to a clinical lenalidomide trail.24
In addition, BM MNCs and sorted CD34+ cells were obtained from 5 healthy donors. Peripheral blood MNCs were obtained from 20 healthy donors after informed consent in accordance with the Declaration of Helsinki. The ethical committees of all participating institutions approved the study.
Cell sorting
BMCs were isolated by red cell depletion and the median blast count was 62% (range 21%-93%) as measured by flow cytometry. BM MNCs were isolated from heparinized BM aspirates on Ficoll Paque PLUS (GE Healthcare) gradient. CD34+ cells were sorted from BM MNCs either by the StemCell CD34+ positive selection kit on a RoboSep device (StemCell Technologies) or by the MACS CD34+ isolation kit (Miltenyi Biotec)24 ; the purity of the sorted cells was above 95%. Normal granulocytes, CD19+ B cells, and CD4+ T cells were isolated using the relevant kits on a RoboSep Device. The CD34+ cell population from a normal donor was sorted by FACS into stem and progenitor cells. The following marks were used for sorting: HSCs (Lin−, CD34+, CD38−, CD90+, 45RA−), multipotent progenitors (MPPs; Lin−, CD34+, CD38−, CD90−, 45RA−), common myeloid progenitors (CMPs; Lin−, CD34+, CD38+, CD45RA−, CD123+), megakaryocyte-erythroid progenitors (MEPs; Lin−, CD34+, CD38+, CD45RA−, CD123−) and granulocyte-macrophage progenitors (GMPs; Lin−, CD34+, CD38+, CD45RA+, CD123+).25,26 Lineage cocktail contained Abs against: CD3, CD7, CD14, CD19, CD56, CD61, CD235a, CD15, CD11b.
Probes, primers, and conditions
For all oligonucleotides and conditions in the following experiments, see supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
5′- and 3′-rapid amplification of cDNA ends
The 5′ and 3′ ends of vtRNA2-1 were determined using the FirstChoice RLM-RACE kit (Ambion) according to the manufacturer's instruction.
Methods for methylation detection
Bisulfite treatment.
One microgram of each DNA sample was converted as previously described with a slight modification.27 DNA from hematopoietic stem and progenitor cells was converted with the EZ DNA Methylation kit (Zymo Research).
Melting curve analysis.
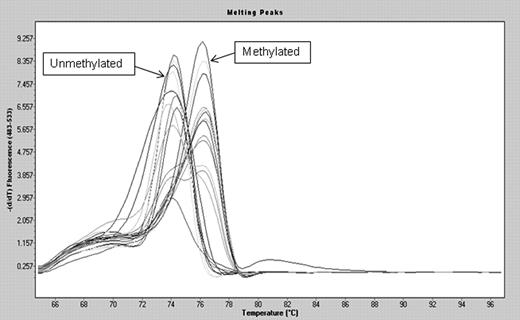
Methylation screening of was done by Ms-MCA as described.28 The melting peaks were calculated using the LightCycler 480 Software Release 1.5.0SP3. SssI-treated DNA (Millipore) was bisulfite converted and used as positive control. DNA from peripheral blood was whole genome amplified twice29 and used as unmethylated control after bisulfite conversion.
Bisulfite sequencing.
To analyze the methylation status of individual alleles, bisulfite-treated genomic DNA was amplified and cloned into the pCR2.1 vector using the TOPO-TA cloning kit (Invitrogen). Colonies were screened for the respective inserts. Plasmid DNA was extracted using the PureLink Quick Miniprep Kit (Invitrogen). Plasmid DNA from individual clones was automatically sequenced using M13 primers.
Pyrosequencing.
For pyrosequencing, VTRNA2-1 and LINE1 were amplified using the PyroMark PCR kit (QIAGEN). The number of cycles was raised to 50 when running the VTRNA2-1 PCR for stem and progenitor cells because of the low amount of material. The PCR product was purified, denatured, pyrosequenced, and analyzed according to the manufacturer's protocols.
Mutation analysis
The VTRNA2-1 gene was analyzed for point mutations by PCR and denaturing gradient gel electrophoresis (DGGE). PCR products were analyzed in a 10% denaturant/6% polyacrylamide-70% denaturant/12% polyacrylamide double-gradient gel (100% denaturant = 7M urea and 40% formamide). The gel was run at 180V for 5 hours in 1× TAE buffer kept at a constant temperature of 58°C.
RT-qPCR
RNA was RT using SuperScript III Reverse Transcriptase (Invitrogen) and random hexamers (Promega). RT-quantitative PCR for miR886-3p and -5p was performed using TaqMan MicroRNA assays (Applied Biosystems), vtRNA2-1 expression was analyzed using TaqMan probes (Applied Biosystems). For normalization, U6 RNA was used in the miR886-3p and -5p analysis, while vtRNA2-1 was normalized to GAPDH.
Northern blot analysis
Total RNA (20 μg) was loaded onto a 15% polyacrylamide denaturing gel and transferred to a nylon membrane. The StarFire radiolabeled probes (Integrated DNA Technologies) were prepared by incorporation of [α-32P] dATP 6000 Ci/mmol following the manufacturer's instruction. Prehybridization and hybridization were carried out using ExpressHyb Hybridization Solution (Clontech). Hybridization was carried out at 42°C overnight. U6 was used as a loading control.
RNA interference
siRNA duplexes (ON-TARGETplus SMARTpool L-016996-00-0005, Human RNASEN) were transfected into T24 cells using DharmaFECT 4 siRNA Transfection Reagent (Thermo Fisher Scientific). Cells were split again 1 day after the first transfection, transfected once more on the third day, and incubated for 3 more days. The cells were incubated for 5 days in total from the time of the first transfection.
α-amanitin (RNA polymerase II inhibitor) treatment
T24 cells were plated at 1.5 × 105 cells/well in a 6-well dish and treated the next day with 50 μg/mL α-amanitin (Sigma-Aldrich) as described.30 Cells were collected 24 hours and 48 hours after drug treatment. Two independent experiments were performed.
Western blot analysis
HL60 cells were treated with 0.1μM 5-aza-CdR and harvested at day 8 after treatment. Cells were lysed in RIPA buffer and 50 μg of protein of each sample were subjected to SDS/PAGE gel. Western blots were probed with the following Abs: anti-PKR (phosphor T446; ab32036) and anti-PKR (ab32052; Abcam).
Statistics
Differences in clinical characteristics of patients were assessed using the Pearson χ2 or Fisher exact tests. Overall survival was estimated using the Kaplan-Meier method. Multivariate Cox proportional hazards models31 were used to assess whether methylation of VTRNA2-1 or any known risk factor, such as age, cytogenetic category,32 or leukocyte counts at diagnosis, were independently associated with overall survival. Statistical analyses were performed in SPSS 17.0 for Windows (SPSS Inc). Any differences were considered to be statistically significant when P < .05.
Results
vtRNA2-1 is up-regulated by 5-azanucleosides and is independent of Drosha processing
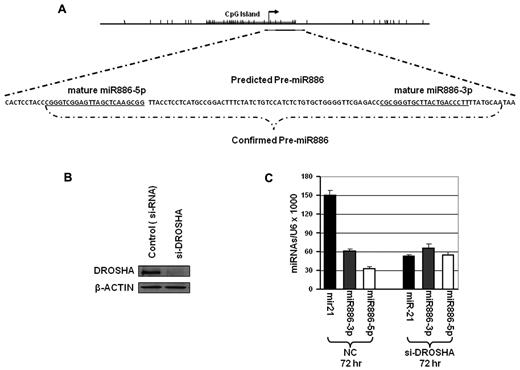
We began our investigations by examining the spectrum of miRNAs up-regulated in the HL60 AML cell line following treatment with 5-azacytidine (5-aza-CR) or 5-aza-2′-deoxycytidine (5-aza-CdR). These experiments showed up-regulation of a group of miRs by the demethylating agents.14 Among the most differentially expressed miRs were the so-called “miR886-3p and -5p” which have subsequently been recognized as small vault-RNAs (svRNAs)33 or degradation products of vtRNA2-1. VTRNA2-1 locates to chromosome 5q31.1 in the CDR1 of high-risk MDS/AML,1,2 and is embedded in a CpG island (Figure 1A). We determined its transcription start site (TSS) by 5′ rapid amplification of cDNA ends (RACE; data not shown). These results showed that the TSS was located at the start site of the pre-MIR88 indicating that the expected longer “pri-miR” transcript was not present. The results summarized in the DNA sequence shown in Figure 1A suggest that this vtRNA might be formed independently of Drosha processing. The formation of the svRNAs was also independent of the presence of Drosha because its down-regulation by siRNA knockdowns (Figure 1B) did not alter the levels of the processed svRNAs (Figure 1C).
Pre-MIR886/VTRNA2-1 lacks a pri-miR transcript and is independent of Drosha. (A) Map and genomic sequence showing pre-MIR886/VTRNA2-1, mature “MIR886-5p and -3p” and the transcription start site (TSS) as determined by 5′ RLM-RACE in T24 cells. Pre-MIR886/VTRNA2-1 is depicted as the black line in the map and the gray lines represent the mature MIR886. The tick marks represent individual CpG sites. The CpG island is labeled on the map by the gray bar. The TSS is shown as the arrow. (B-C) Processing of vtRNA2-1 is independent of Drosha. T24 cells were transfected twice with Drosha siRNAs, and collected 72 hours after the second transfection. (B) Drosha protein levels were detected by Western blots. (C) Stem-loop RT-PCR show unchanged levels of mature “miR886-5p and -3p,” while miR-21 expression is inhibited by Drosha knockdown.
Pre-MIR886/VTRNA2-1 lacks a pri-miR transcript and is independent of Drosha. (A) Map and genomic sequence showing pre-MIR886/VTRNA2-1, mature “MIR886-5p and -3p” and the transcription start site (TSS) as determined by 5′ RLM-RACE in T24 cells. Pre-MIR886/VTRNA2-1 is depicted as the black line in the map and the gray lines represent the mature MIR886. The tick marks represent individual CpG sites. The CpG island is labeled on the map by the gray bar. The TSS is shown as the arrow. (B-C) Processing of vtRNA2-1 is independent of Drosha. T24 cells were transfected twice with Drosha siRNAs, and collected 72 hours after the second transfection. (B) Drosha protein levels were detected by Western blots. (C) Stem-loop RT-PCR show unchanged levels of mature “miR886-5p and -3p,” while miR-21 expression is inhibited by Drosha knockdown.
vtRNA2-1 is a pol III transcript
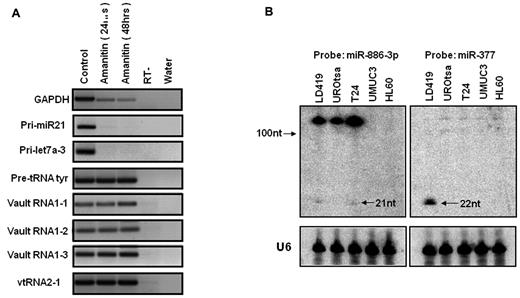
We next confirmed the findings by others34 that vtRNA2-1 is indeed a pol III transcript (Figure 2A). Unlike the transcription of the bona fide pri-miR21 or pri-Let7a-3 and GAPDH, its transcription was not inhibited by α-amanitin treatment, which is a relatively specific inhibitor of pol II. Likewise the transcription of pre-tRNAs and vtRNAs was not affected by α-amanitin treatment.
VTRNA2-1 is transcribed by RNA pol III and the transcript is ∼ 100 nt long. (A) VTRNA2-1 is transcribed by RNA pol III. T24 cells were treated with α-amanitin (RNA pol II inhibitor) and harvested at 24 and 48 hours after treatment. vtRNA2-1 is insensitive to α-amanitin treatment as are the known pol III transcripts (pre-tRNAtyr, vtRNA1-1, vtRNA1-2, and vtRNA1-3), while the formation of pol II transcripts is inhibited (GAPDH, pri-miR-21, pri-let-7a-3). (B) Expression of vtRNA2-1 in different cell lines by Northern blot analysis showing that very little is processed to the mature miR (21 nt). U6 indicates loading control; and miR-377, small RNA control.
VTRNA2-1 is transcribed by RNA pol III and the transcript is ∼ 100 nt long. (A) VTRNA2-1 is transcribed by RNA pol III. T24 cells were treated with α-amanitin (RNA pol II inhibitor) and harvested at 24 and 48 hours after treatment. vtRNA2-1 is insensitive to α-amanitin treatment as are the known pol III transcripts (pre-tRNAtyr, vtRNA1-1, vtRNA1-2, and vtRNA1-3), while the formation of pol II transcripts is inhibited (GAPDH, pri-miR-21, pri-let-7a-3). (B) Expression of vtRNA2-1 in different cell lines by Northern blot analysis showing that very little is processed to the mature miR (21 nt). U6 indicates loading control; and miR-377, small RNA control.
The longer ncRNA transcript (vtRNA2-1) predominates
Northern blot analyses of RNA extracted from human fibroblasts, immortalized urothelial cells or the indicated cancer cell lines showed that the predominant transcript was ∼ 100 nucleotides in length with only a minor fraction being processed to the 21 nucleotide product expected for a mature miR (Figure 2B).
Taken together, these data confirm the findings of 3 other groups who recently stated that “miR886” has been misannotated and is either a vtRNA or another longer type of ncRNA.15-17
vtRNA2-1 expression is regulated by promoter methylation
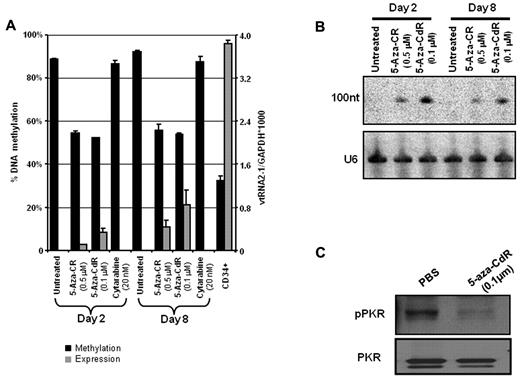
Since we originally focused on this ncRNA because of its up-regulation following 5-aza-CR treatment, we measured the methylation status of the CpG island at the TSS by melting curve analysis and pyrosequencing assays before and after treatment with 5-azanucleoside drugs. The CpG island was extensively methylated in untreated HL60 cells and several other AML cell lines with and without deletion of 5q (supplemental Figure 1). In HL60, the methylation level was strongly reduced both 2 and 8 days after treatment with 5-aza-CR or 5-aza-CdR, but not by cytosine arabinoside (Figure 3A). Interestingly, the TSS was methylated to ∼ 35% in sorted CD34+ cells where the vtRNA2-1 was also strongly expressed. Northern analysis probed with vtRNA2-1 probes showed a strong up-regulation of vtRNA2-1 2 and 8 days after treatment start with the demethylating drugs (Figure 3B). However, the svRNAs/miR886-5p or -3p were not detected on these Northern blots (supplemental Figure 2). These data show that the CpG island at the TSS was partially methylated in normal CD34+ cells and became extensively methylated in the HL60 cell line, and that the vtRNA2-1 expression could be reactivated by demethylating agents.
Expression of vtRNA2-1 is derepressed by 5-azanucleosides. Derepression of vtRNA2-1 by both 5-azacytidine (5-aza-CR) and 5-aza-2′-deoxycytidine (5-aza-CdR) but not by cytosine arabinoside treatment (cytarabine). The level of methylation correlates to expression. RT-qPCR (A) and Northern hybridization (B). On day 8 after treatment with 5-aza-CdR, vtRNA2-1 is up-regulated (A-B) and pPKR is down-regulated (C). Reactions were at least done in biologic duplicates.
Expression of vtRNA2-1 is derepressed by 5-azanucleosides. Derepression of vtRNA2-1 by both 5-azacytidine (5-aza-CR) and 5-aza-2′-deoxycytidine (5-aza-CdR) but not by cytosine arabinoside treatment (cytarabine). The level of methylation correlates to expression. RT-qPCR (A) and Northern hybridization (B). On day 8 after treatment with 5-aza-CdR, vtRNA2-1 is up-regulated (A-B) and pPKR is down-regulated (C). Reactions were at least done in biologic duplicates.
Functional consequences of VTRNA2-1 deregulation
Since a previous study has shown that vtRNA2-1 may associate with and modulate the activity of the RNA-activated protein kinase PKR,17 we analyzed whether treatment of the HL60 cell line by 5-aza-CdR (and thus up-regulation of vtRNA2-1) would alter the level of phosphorylated PKR (pPKR). Indeed, significant down-regulation of its phosphorylation without a change in the amount of PKR protein was observed in HL60 cells after 5-aza-CdR treatment (Figure 3C).
VTRNA2-1 regulation in healthy individuals
All CD34+ progenitors and mature blood cells of a healthy individual carry similar levels of methylation.
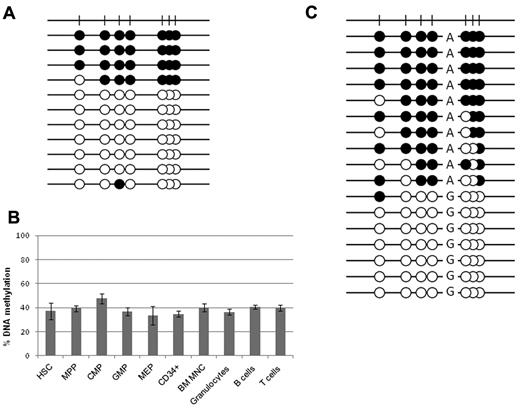
Our initial melting curve analysis of DNA from random donor peripheral blood MNCs and isolated normal CD34+ cells (which express vtRNA2-1), showed a methylation level of ∼ 35%. Bisulfite genomic sequencing revealed that the individual alleles were either almost fully methylated or essentially fully unmethylated (Figure 4A), strongly suggesting allele-specific methylation rather than partial methylation of both alleles. This made us question whether a specific subgroup of CD34+ cells was biallelically unmethylated.
Methylation level of VTRNA2-1 in hematopoietic cells. Methylation of the VTRNA2-1 promoter in HSCs, myeloid progenitors, and mature hematopoietic cells from normal donors. (A) Bisulfite sequencing of pooled CD34+ cells from random donors showing fully methylated and fully unmethylated alleles. (B) Results from pyrosequencing of sorted hematopoietic cells and myeloid progenitors from a donor with a biphasic methylation pattern. All cell types carry a methylation level ∼ 35%. MPP indicates multipotent progenitors; CMP, common myeloid progenitors; MEP, megakaryocyte-erythroid progenitors; GMP, granulocyte-macrophage progenitors; and BM MNC, BM mononuclear cells. (C) Bisulfite sequencing of a region −250 bp from the TSS reveals allele-specific methylation, in an individual carrying an A/G SNP.
Methylation level of VTRNA2-1 in hematopoietic cells. Methylation of the VTRNA2-1 promoter in HSCs, myeloid progenitors, and mature hematopoietic cells from normal donors. (A) Bisulfite sequencing of pooled CD34+ cells from random donors showing fully methylated and fully unmethylated alleles. (B) Results from pyrosequencing of sorted hematopoietic cells and myeloid progenitors from a donor with a biphasic methylation pattern. All cell types carry a methylation level ∼ 35%. MPP indicates multipotent progenitors; CMP, common myeloid progenitors; MEP, megakaryocyte-erythroid progenitors; GMP, granulocyte-macrophage progenitors; and BM MNC, BM mononuclear cells. (C) Bisulfite sequencing of a region −250 bp from the TSS reveals allele-specific methylation, in an individual carrying an A/G SNP.
CD34+ cells are a heterogeneous population consisting of stem and progenitor cells. Because some degree of methylation was detected in the normal CD34+ population, different subtypes of CD34+ cells from a healthy donor were isolated by FACS to evaluate if methylation was linked to a particular CD34+ subtype. The CD34+ cell population was sorted into HSCs, MPPs, CMPs, MEPs, and GMPs. All 5 subtypes display a median methylation level of 37% (range 33%-47%) by pyrosequencing (Figure 4B). Normal mature B, T cells, and granulocytes display a similar level of methylation of VTRNA2-1. Accordingly, a similar methylation level is present in all subtypes of the normal CD34+ population and mature blood cells, thus excluding that the methylation was cell-type specific.
VTRNA2-1 is monoallelically methylated in the majority of healthy individuals.
We next wondered whether interindividual differences in VTRNA2-1 methylation exist among healthy individuals. Fifteen of a panel of 20 healthy individuals carried ∼ 40% methylation of the VTRNA2-1 promoter; however, 5 carried completely unmethylated VTRNA2-1 promoters, while none showed increased methylation. These data were confirmed by bisulfite sequencing in selected cases (data not shown).
Given that 75% of healthy individuals showed a level of methylation close to 50% in the normal tissues because of the presence of either almost fully methylated or mostly completely unmethylated alleles, we then considered whether this monoallelic methylation leads to monoallelic expression. Unfortunately, the expressed vtRNA2-1 sequence does not carry a SNP that could confirm this. However, a SNP (A/G; rs9327740) was present −250 bp from the TSS in some individuals, and bisulfite genomic sequencing showed methylation of only the A allele, indicating that VTRNA2-1 is allele-specifically methylated and likely monoallelically expressed because methylation correlates with expression (Figure 4C, supplemental Figure 3). The sequencing was repeated twice for each individual and the same allele was always methylated in a given individual, thus excluding cloning bias.
VTRNA2-1 regulation in AML patients
Pretreated patients with loss of 5q display either a methylated or an unmethylated allele.
In a sample of BM MNC from 21 advanced cases of heavily pretreated AML/MDS patients with loss of chromosome 5q (EGR1 FISH probe) obtained from a clinical relapse trial,24 we observed that in the majority of cases, the VTRNA2-1 promoter was either essentially methylated (n = 10) or unmethylated (n = 6; Figure 5). This is consistent with the possibility that the malignant clones are derived from cells in which either the methylated or unmethylated allele was lost. Alternatively, the originally unmethylated allele might have become methylated in the tumor cells. However, in this small sample of end-stage AML/MDS patients, selection toward loss of the unmethylated allele was only borderline significant (P = .12). Analysis of RNA from the CD34+ cells showed that none of the patients with a fully methylated promoter had any expression of vtRNA2-1. RNA was available before and after lenalidomide treatment in 4 cases, in 2 of these with a fully methylated promoter lenalidomide did not influence vtRNA2-1 expression. However, in the 2 patients with an unmethylated promoter and no expression, vtRNA2-1 was up-regulated during lenalidomide treatment (supplemental Figure 4). Thus, hypermethylation of the VTRNA2-1 promoter silences expression in vivo; however, in hypomethylated cases with no basal expression, exposure to agents such as lenalidomide leads to vtRNA2-1 up-regulation. This data show that although basal expression was absent in some cases without methylation vtRNA2-1 was inducible by suitable stimulus.
Allelic methylation levels in del 5q AML/MDS patients. BM MNCs from del(5q) patients preferentially carry either a methylated or an unmethylated allele, suggesting their origin from normal cells with either a methylated or an unmethylated VTRNA2-1. The bimodal melting profile seen in a few cases is likely caused by normal MNCs in the sample.
Allelic methylation levels in del 5q AML/MDS patients. BM MNCs from del(5q) patients preferentially carry either a methylated or an unmethylated allele, suggesting their origin from normal cells with either a methylated or an unmethylated VTRNA2-1. The bimodal melting profile seen in a few cases is likely caused by normal MNCs in the sample.
VTRNA2-1 methylation levels predicts prognosis in diagnostic AML samples
The methylation level of the VTRNA2-1 promoter was examined in BMC samples from 101 AML patients obtained at the time of diagnosis. Thirty-eight (37.6%) cases showed hypomethylation of the VTRNA2-1 promoter (< 10% methylation by pyrosequencing), while 63 cases (62.4%) showed a methylation level above 10% (16%-75%). The patient characteristics at diagnosis are listed in Table 1. The methylated and unmethylated cases only differed by cytogenetic risk group, with relatively more patients with intermediate cytogenetic risk in the methylated group.
A Kaplan-Meier analysis of overall survival revealed that the AML patients with VTRNA2-1 hypomethylation (< 10% methylation) had significantly better overall survival compared with those with methylation (> 10% methylation; P = .019; supplemental Figure 5). The relative impact of VTRNA2-1 methylation on overall survival was evaluated by Cox proportional hazard analysis.31 Factors included in the model were: age, cytogenetic risk classification,32 leukocyte count at diagnosis, and methylation status. The model revealed that the methylation status of VTRNA2-1 is an independent prognostic factor for overall survival (P = .022; Table 2). To unravel whether the positive effect of VTRNA2-1 hypomethylation was specific to this gene or if it is an indicator of a general positive effect of DNA hypomethylation, the level of LINE1 methylation was compared with VTRNA2-1 hypomethylation or intermediate/hypermethylation, respectively. No significant difference in the level of LINE1 methylation was observed between the 2 groups indicating that the positive effect of hypomethylation is specific for VTRNA2-1 (supplemental Figure 6).
Multivariate analysis of cases
| Multivariate analysis . | P . | Hazard ratio . | 95% CI for HR . | |
|---|---|---|---|---|
| Lower . | Upper . | |||
| Hypomethylated vs methylated cases | ||||
| Age | .000 | 4.666 | 2.684 | 8.113 |
| Cytogenetic risk group | .004 | 2.007 | 1.241 | 3.246 |
| VTRNA2-1 | .022 | 1.947 | 1.101 | 3.443 |
| WBC count at diagnosis | .000 | 1.007 | 1.003 | 1.011 |
| Hypomethylated vs intermediate methylated vs hypermethylated cases | ||||
| Age | .000 | 4.156 | 2.396 | 7.209 |
| Cytogenetic risk group | .003 | 2.029 | 1.267 | 3.248 |
| VTRNA2-1 | .027 | 1.537 | 1.049 | 2.253 |
| WBC count at diagnosis | .000 | 1.007 | 1.003 | 1.011 |
| Multivariate analysis . | P . | Hazard ratio . | 95% CI for HR . | |
|---|---|---|---|---|
| Lower . | Upper . | |||
| Hypomethylated vs methylated cases | ||||
| Age | .000 | 4.666 | 2.684 | 8.113 |
| Cytogenetic risk group | .004 | 2.007 | 1.241 | 3.246 |
| VTRNA2-1 | .022 | 1.947 | 1.101 | 3.443 |
| WBC count at diagnosis | .000 | 1.007 | 1.003 | 1.011 |
| Hypomethylated vs intermediate methylated vs hypermethylated cases | ||||
| Age | .000 | 4.156 | 2.396 | 7.209 |
| Cytogenetic risk group | .003 | 2.029 | 1.267 | 3.248 |
| VTRNA2-1 | .027 | 1.537 | 1.049 | 2.253 |
| WBC count at diagnosis | .000 | 1.007 | 1.003 | 1.011 |
WBC indicates white blood cells.
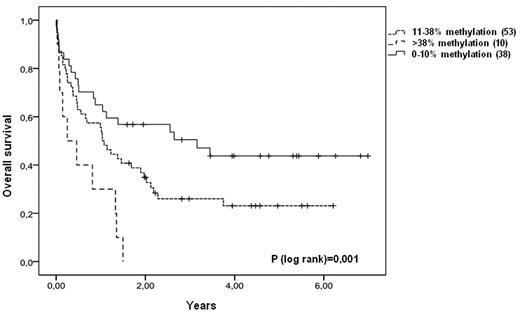
We next asked whether patients with methylation levels above that of normal hematopoietic cells were associated with a poorer prognosis. Because the available samples contain a mixture of normal- and blast cells, a cut off for hypermethylation was defined as 38% because the normal BM MNC has a methylation level of 35% ± 2.5% by pyrosequencing. Accordingly, patients with methylation of VTRNA2-1 were split in a group with an intermediate methylation level defined as 11-38% (mean: 32%, range 20%-37%) and a group with hypermethylation defined as > 38% methylation (mean: 44%, range 39%-75%). A Kaplan-Meier estimate shows that patients with hypermethylation of VTRNA2-1 (> 38%) have a significantly poorer survival (P = .001; Figure 6). Also in this model, multivariate analysis revealed that methylation of VTRNA2-1 is an independent prognostic factor for survival (P = .027; Table 2). Because none of the hypermethylated cases in these samples carried deletion of 5q as determined by conventional cytogenetics or FISH (EGR1 probe), methylation may occur at both alleles; however, a microdeletion of VTRNA2-1 could result in a similar methylation pattern. To unravel whether there is also hypermethylation of neighboring genes, we analyzed the prognostic impact of methylation of an adjacent TSG (CTNNA1) located in the CDR1 region. However, only 1 of 101 AML patients had a methylated CTNNA1 promoter in these diagnostic samples, indicating that the negative effect of hypermethylation at diagnosis is specific to VTRNA2-1 silencing.
Overall survival of AML patients corresponds to VTRNA2-1 methylation level. Patients are divided into 3 groups depending on VTRNA2-1 methylation level (< 10% methylation, 11%-38% methylation, > 38% methylation). Patients with < 10% methylation of VTRNA2-1 have a significant better survival, while patients with methylation > 38% have a much poorer survival.
Overall survival of AML patients corresponds to VTRNA2-1 methylation level. Patients are divided into 3 groups depending on VTRNA2-1 methylation level (< 10% methylation, 11%-38% methylation, > 38% methylation). Patients with < 10% methylation of VTRNA2-1 have a significant better survival, while patients with methylation > 38% have a much poorer survival.
To further investigate whether vtRNA2-1 function could be altered by mechanisms other than promoter methylation, all patient samples were analyzed for point mutations of the VTRNA2-1 gene by PCR/DGGE. No mutations or SNPs were identified.
Discussion
Tumor suppressors have long been sought for in the 5q31.1 region in MDS/AML. Although most deletions are large, studies of a few AML/high-risk MDS cases with minor deletions have narrowed down a critical region. Several TSGs in this region have been suggested, and in some cases knock down studies in mice and hematopoietic cells35 mirror an MDS/AML phenotype. For several of these putative TSGs, it has been suggested that haploinsufficiency by 5q loss in itself leads to tumorigenesis,35-37 and hypermethylation of the CTNNA12 and RIL12 genes has been associated with advanced stages of AML/MDS. However, to the best of our knowledge, no studies have previously identified genes in this region which can already be monoallelically methylated in a majority of normal individuals, and where both hypo- and hypermethylation is associated with outcome in AML patients.
Here, we show that VTRNA2-1 is monoallelically methylated in the majority of healthy individuals and AML patients. However, hypomethylation is seen in approximately one-third of de novo AMLs with better prognosis. Because hypomethylation is also seen in a similar fraction of healthy individuals this may represent a diversity in normal cells that may influence the outcome of treatment. Hypermethylation of the VTRNA2-1 promoter was never observed in any of the normal tissues analyzed, but was seen in BMCs in a fraction of AML patients with very poor prognosis. From our studies in cell lines we know that the alternate, unmethylated allele can be either methylated or lost, and we believe both mechanisms may act in patients because none of the 10 patients with increased levels of methylation were shown to have a 5q deletion as determined by conventional karyotyping or FISH. Of note, we show that in advanced, aggressive cases with deletion of 5q the methylated allele may be lost, indicating that in those cases vtRNA2-1 is not the target tumor suppressor that associates with poor prognosis. Other tumor suppressors present in this region2,6,12 may also contribute to poor prognosis in del 5q cases.
Multivariate analysis showed that in the diagnostic AML samples VTRNA2-1 methylation is independent of the well-known prognostic markers in AML such as cytogenetic risk group, leukocyte count and age at diagnosis, indicating that quantitative determination of VTRNA2-1 methylation may serve as a useful prognostic tool. While lowering of the gene dosage because of haploinsufficiency has been reported as a common mechanism of tumor suppressor loss in MDS/AML, the observation that a part of the population carries hypomethylated alleles is intriguing, because hypomethylation associates with an increase in the basal level of expression relative to individuals with monoallelic methylation. Our data indicate that AML patients with hypomethylation do better than patients with intermediate (monoallelic) methylation levels, and taken together this may probably imply that an increased gene dosage influences outcome. This may not only apply to this particular disease category but to cancer in general, and because this is most likely a normal variant, it could potentially be crucial for an individuals' cancer susceptibility or general reaction to chemotherapy.
The mechanisms that lead to allele specific methylation in some cases are currently unknown, however interindividual methylation differences exist independently of imprinting which may be related to genotype.38 It has been suggested that a nearby SNP (within 5 kb) can direct methylation to one allele or the other, and these SNPs can act both in cis and trans.39,40 However, the SNP we examine here does not determine which allele becomes methylated, because homozygous (AA) individuals carry both a methylated and an unmethylated A allele, while the heterozygote (AG) carries a methylated A allele and an unmethylated G allele. However, we cannot exclude the possibility that SNPs in the neighboring regions are involved in this monoallelic methylation which appears to present on the same allele in all cells, or at least all blood cells, of a single individual.
It is still controversial as to whether the vtRNA2-1 or the svRNA/miR886-3p and -5p are functionally dominant in vivo. We have shown that they are coordinately regulated with by far the largest fraction of the transcripts being ∼ 100 nt long (vtRNA2-1), in agreement with the observations of others.17,41 Lee et al did not detect the svRNA/mature miR886-3p or -5p by Northern hybridization17 ; however, it is unclear whether this is related to the specific cellular context or technical issues. Alternatively, the detected “mature miRs” are actually degradation products of vtRNA2-1. Another issue is the exact function of the longer transcript, which may or may not associate to the vault complex.15,17 Again it is likely that the exact function of this ncRNA may be cell type dependent.17
A recent study shows that vtRNA2-1 associates with PKR and modulates its phosphorylation activity, so that inhibition of vtRNA2-1 leads to activation of PKR (pPKR).17 Our finding that azanucleosides induce expression of vtRNA2-1 and that this occurs in concert with down-regulation of the pPKR is therefore particularly interesting. PKR was originally identified as a component of the host defense mechanism against viral infections. Later, it has been shown that PKR activation also can be induced by various stress stimuli, for example, cytotoxic cytokines, growth factor deprivation, and DNA damage.42 PKR has been implicated in a variety of signaling pathways including eiF2α, NF-κB, PI3K/AKT, and C/EBPα and was originally viewed as a tumor suppressor. This has, however, been questioned because PKR is reported to be constitutively active in many human tumors including AML.19,43 Indeed, it was demonstrated that PKR kinase activity is required for AML blast maintenance and growth, and inhibition of PKR activity blocks cell proliferation, increases chemosensitivity, and cause cell death.19,44 In addition, novel studies show that PKR may have specific functions in the nucleus of leukemic blasts.45
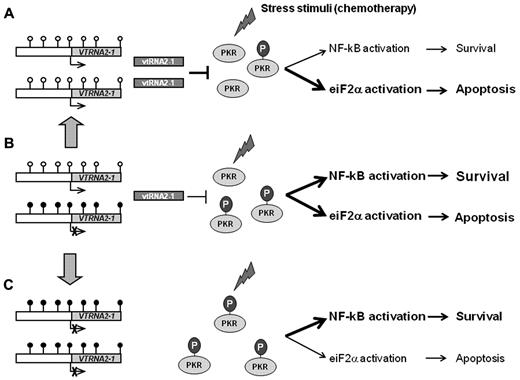
Although none of the reported functions of the mature miRs41,46 seem to apply directly to the current study, this proposed negative regulatory function of vtRNA2-1 on PKR kinase activity, the increased chemosensitivity by PKR inhibition, and the need for PKR signaling in AML blast maintenance are in support of the current data. Both PKR and NF-κB are constitutively active in AML, which could be a consequence of loss of vtRNA2-1–mediated PKR inhibition (Figure 7).
The role of vtRNA2-1 in PKR activation. A simplified theoretical model of PKR activation and its influence on downstream pathways with (A) hypomethylation of the promoter, (B) monoallelic methylation of the promoter, or (C) biallelic hypermethylation of the promoter. On stimulation by various stress stimuli, for example, DNA damage induced by chemotherapy, PKR becomes autophosphorylated to an active kinase (pPKR) that activates multiple signaling pathways. (A) In AML blasts with a hypomethylated VTRNA2-1 promoter, vtRNA2-1 inhibits pPKR and favors the apoptotic pathways. (B) In AML blast with monoallelic methylation, pPKR increases because of the decrease in vtRNA2-1 and balance the different downstream signaling pathways. (C) When vtRNA2-1 expression is lost, the inhibitory effect disappears, pPKR increases and favor the activation of survival pathways (using data from Lee et al,17 Blalock et al,19 Blalock et al,43 and Pataer et al44 ).
The role of vtRNA2-1 in PKR activation. A simplified theoretical model of PKR activation and its influence on downstream pathways with (A) hypomethylation of the promoter, (B) monoallelic methylation of the promoter, or (C) biallelic hypermethylation of the promoter. On stimulation by various stress stimuli, for example, DNA damage induced by chemotherapy, PKR becomes autophosphorylated to an active kinase (pPKR) that activates multiple signaling pathways. (A) In AML blasts with a hypomethylated VTRNA2-1 promoter, vtRNA2-1 inhibits pPKR and favors the apoptotic pathways. (B) In AML blast with monoallelic methylation, pPKR increases because of the decrease in vtRNA2-1 and balance the different downstream signaling pathways. (C) When vtRNA2-1 expression is lost, the inhibitory effect disappears, pPKR increases and favor the activation of survival pathways (using data from Lee et al,17 Blalock et al,19 Blalock et al,43 and Pataer et al44 ).
In line with the current study, Lee et al showed that PKR-mediated inhibition of proliferation seems to be dose dependent. Here, we observe that the level of methylation corresponds to the level of expression, that hypomethylation (and presumably up-regulation) is associated with a good prognosis, while hypermethylation or loss of the unmethylated allele (and presumably down-regulation) is associated with a bad prognosis in primary AML patients. While hypomethylation may be a normal variant that protects against cancer or, more likely, support the function of chemotherapy, our coordinated observations in myeloid cell lines and the 5q-deleted cases confirm that an increase in methylation can occur in the myeloid blasts. Accordingly, vtRNA2-1 most likely functions as a tumor suppressor. This is supported by the fact that the VTRNA2-1 promoter region binds numerous transcription factors (based on ENCODE data, http://genome.ucsc.edu), which was recently shown to be typical of so-called HOT genes of high importance to basic cellular functions.47
In summary, we have identified the noncoding VTRNA2-1 as a novel putative tumor suppressor on 5q31.1, which is monoallelically methylated and therefore probably monoallelically expressed in 75% of the Danish population. Allelic loss of the alternate 5q region in these individuals would be expected to lead to its inactivation. Our data indicate that vtRNA2-1 is an important target, because biallelic methylation is another plausible mechanism for its inactivation. We also describe a novel phenomenon in cancer; that hypomethylation of the same promoter is associated with a better prognosis, suggestive of a dose-effect, and this is potentially a normal variant that influences the patients' sensitivity to chemotherapy by modulating the level of pPKR.44 Our studies in cell lines show that azanucleosides up-regulate vtRNA2-1 and down-regulate pPKR, and taken together, this creates a rationale for a treatment schedule where 5-azanucleosides are given before the administration of combination chemotherapy.
In addition, the vtRNA2-1 promoter methylation status is a prognostic factor for overall survival, independently of well-known prognostic markers and with a predictive value comparable with the recently published complex “quantitative global methylation predictor.”48 Our findings are done in a rather heterogeneously treated patient group and obviously need confirmation in a uniformly treated cohort, with sorted blasts and normal cells isolated at the day of diagnosis. In addition, we did not include other known molecular markers in our model, for example, disrupted epigenetic regulators such as the recently discovered DNMT3A49 and IDH50 mutation status. At present the clinical use of these markers is still hampered by the requirement of relatively costly sequencing facilities. Accordingly, we suggest that a measure of the VTRNA2-1 methylation pattern may be a simple, inexpensive and fast alternative prognostic marker in newly diagnosed AML. Finally, the current data combined with the recent observation that loss of vtRNA2-1 may cause constitutive activation of PKR,17 which is necessary for the maintenance of the leukemic blasts,19 makes VTRNA2-1 an attractive putative novel tumor suppressor on chromosome 5q. It shall be interesting to determine whether the VTRNA2-1 methylation pattern is also crucial to the outcome of 5-azanucleoside treatment and whether it also applies to different types of cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Anja Pedersen and Yvonne Tsai for skillful technical assistance, and are grateful to Christian Geisler and all members of the Nordic MDS group for donation of patient material.
This study was supported by The Lundbeck Foundation, The Novo Nordisk Foundation, Rigshospitalets Research Foundation, The Danish Cancer Society (M.B.T. and K.G.), the National Institutes of Health (R37 CA82422, P.A.J.; R01 CA1378794, G.L.), and the China Scholarship Council (X.Q.).
B.T.P. and K.G. are members of the European Program for Cooperation in Science and Technology (Cost Action BM0801).
National Institutes of Health
Authorship
Contribution: M.B.T. and X.Q. designed and performed the research, analyzed the data and wrote the manuscript; A.S., X.Y., C.N.-B., C.H., and J.J. performed the research and analyzed the data; M.K.A. performed research, analyzed data, and contributed with material; L.K., L.M., and E.H.-L. contributed with material; BTP contributed with reagents and analytical tools; P.A.J., G.L., and K.G. designed the research, analyzed the data, contributed with reagents/analysis tools, and wrote the manuscript; and all authors critically read and commented on the manuscript.
Conflict-of-interest disclosure: P.A.J. serves as a consultant for Eli Lilly. E.H.-L. is on the Celgene advisory board and holds a Celgene research grant (clinical trial 07A). K.G. is supported by scientific research grants from the Novo Nordisk and the Lundbeck Foundations. The Novo Nordisk Foundation and the Lundbeck Foundation are scientific research foundations that are independent of any commercial interests by the Novo Nordisk and Lundbeck pharmaceutical companies. Accordingly, K.G. has no financial or nonfinancial, professional, or personal affiliation to these commercial companies. The remaining authors declare no competing financial interests.
Correspondence: Kirsten Grønbæk, MD, DMSc, Department of Hematology, L4042, Rigshospitalet, Blegdamsvej 9, DK-2100 Copenhagen Ø, Denmark; e-mail: kirsten.groenbaek@rh.regionh.dk; or Gangning Liang, PhD, Department of Urology, USC/Norris Comprehensive Cancer Center, Keck School of Medicine of the University of Southern California, Los Angeles, CA 90089; e-mail: gliang@usc.edu.
References
Author notes
M.B.T., X.Q., G.L., and K.G. contributed equally to this manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal