Abstract
An important problem in translational cancer research is our limited ability to functionally characterize behaviors of primary patient cancer cells and associated stromal cell types, and relate mechanistic understanding to therapy selection. Functional analyses of primary samples face at least 3 major challenges: limited availability of primary samples for testing, paucity of functional information extracted from samples, and lack of functional methods accessible to many researchers. We developed a microscale cell culture platform that overcomes these limitations, especially for hematologic cancers. A key feature of the platform is the ability to compartmentalize small populations of adherent and nonadherent cells in controlled microenvironments that can better reflect physiological conditions and enable cell-cell interaction studies. Custom image analysis was developed to measure cell viability and protein subcellular localizations in single cells to provide insights into heterogeneity of cellular responses. We validated our platform by assessing viability and nuclear translocations of NF-κB and STAT3 in multiple myeloma cells exposed to different conditions, including cocultured bone marrow stromal cells. We further assessed its utility by analyzing NF-κB activation in a primary chronic lymphocytic leukemia patient sample. Our platform can be applied to myriad biological questions, enabling high-content functional cytomics of primary hematologic malignancies.
Introduction
One challenging area of translational cancer research is the difficulty of performing functional analyses of primary patient samples to increase our understanding of human cancer biology. Highly sensitive genomic and proteomic methods, approaches that primarily evaluate the state of cancer samples, have significantly added to, and continue to increase, our understanding of the biology and the stratification of human malignancies.1-4 In contrast, functional analyses, assessing biological responses to various experimental conditions, with primary patient samples are challenging for two reasons: (1) standard in vitro models and cell culture platforms (eg, Petri dishes, well plates, Transwell inserts) do not accurately reflect the complex spatiotemporal dynamics of the physiological microenvironment5,6 ; and (2) current laboratory techniques often require more biological starting material than can be adequately obtained from patients. For example, electrophoretic mobility shift assays (EMSAs) to detect transcription factor-DNA interactions typically require a minimum of 105 to 106 cultured cells per condition (ie, per lane).7,8 This quantity, in some cases, may not be obtainable from patient samples with particularly low cell counts. EMSAs also belong to a broad class of population-average cellular assays that provide only a single readout for the entire cultured cell population. Such population-average approaches not only restrict experimentation to samples with abundant populations and limit the number of replicates and experiments that can be performed, but more importantly, mask potentially valuable single-cell data that could be crucial to understanding cell-cell heterogeneity.
Recently, microscale technologies have emerged as advanced tools for examining and measuring cell behavior and function in controlled microenvironments, with high-throughput potential and the capability for single-cell analysis.6,9-11 These advances have been useful for demonstrating the broad capabilities of microscale technologies for cell biology applications and have importantly moved us closer to high-content functional cytomics12-14 that provide more elaborate datasets at higher resolution and in a more physiological context such that valuable biological information contained within samples is preserved. In several instances, microfluidics have led to commercialized products that offer new functionality, increased control, and ease of use (Fluidigm, ibidi, and Bellbrook Labs). Despite this progress, however, many other microfluidic systems under development continue to face significant challenges when attempting to gain acceptance and adoption by the biology research community, partly because of operational complexity. For microscale systems to make a significant impact in cell biology research, these systems must consider not only how to get more data out of less starting material but also provide improved accessibility so that less effort from the end user is needed for them to transition from established methods to newly developed technologies.
To overcome these challenges, we pursued an interdisciplinary approach to develop, test, and apply a microscale cell culture and analysis platform capable of handling low sample volume, examining multiple cell types (both adherent and suspension cells) in complex microenvironments, and providing readouts at single-cell resolution, all while using only a micropipette and an epifluorescence microscope for operation. This accessibility was the result of a concerted effort to integrate passive pumping and indirect immunostaining techniques into the platform, thus obviating the need for external pumps, valves, and tubing common in other microfluidic systems.
Here we describe the design and validation of the platform within the context of hematologic malignancies, including multiple myeloma (MM) and chronic lymphocytic leukemia (CLL). MM served as an attractive cancer model for testing this system because it represents a hematologic malignancy often studied in the research laboratory by isolating relevant cell types from limited volumes of patient-derived bone marrow aspirates to understand complex direct contact and soluble factor signaling between both adherent and suspension cells in the local bone marrow microenvironment.7,15,16 CLL cells are also useful because of a relatively ready availability of patient blood samples. To demonstrate the effectiveness of the platform in providing functional endpoints, we designed two cell-based assays that measure cell viability and nuclear translocation events in suspension cells under various cytokine and drug treatments. We further demonstrated the robustness of the platform by using the geometry to study effects of neighboring bone marrow stromal cell (BMSC) line on cocultured myeloma cells and applying the system to primary samples from CLL patients.
Methods
Microchamber fabrication
Established soft lithography techniques17,18 were used to fabricate single-use devices consisting of 12 independent microscale cell culture chambers (microchambers) arranged in a 3 × 4 array (Figure 1). Two separate master molds were made: one for the channel layer containing the central well, side chambers, diffusion ports, and inlet and outlet channels, and one for the access port layer. Polydimethylsiloxane (PDMS) was mixed at a 10:1 base-to-curing agent ratio and cured at 80°C for 4 hours. The 2 PDMS layers were then sequentially bonded via plasma treatment to a glass slide to produce the final device (supplemental Methods; see the Supplemental Materials link at the top of the article)
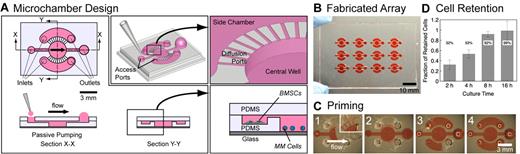
Microchamber design, fabrication, and characterization. (A) A single microchamber consisted of a central well connected to 2 side chambers via diffusion channels, and access ports to each of the compartments. Fluid replacement is performed via passive pumping using a micropipette (Section X-X). The central well was designed to have low wall shear stress at the bottom to retain settled nonadherent MM cells. Side chambers were used for experiments involving BMSCs in coculture with MM cells (Section Y-Y, inset). Scale bar represents 3 mm. (B) A single device consisted of a 3 × 4 array of microchambers on a 75 × 50-mm slide. Scale bar represents 10 mm. (C) Priming of the microchamber with 70% ethanol containing red food coloring dye. Scale bar represents 3 mm. (D) Characterization of the retention of suspension cells in microchambers. Fraction of retained cells, after 25 VRs of 8 μL per VR into the central well, increased with increasing culture time, up to 99% retention for 16 hours of culture before VRs.
Microchamber design, fabrication, and characterization. (A) A single microchamber consisted of a central well connected to 2 side chambers via diffusion channels, and access ports to each of the compartments. Fluid replacement is performed via passive pumping using a micropipette (Section X-X). The central well was designed to have low wall shear stress at the bottom to retain settled nonadherent MM cells. Side chambers were used for experiments involving BMSCs in coculture with MM cells (Section Y-Y, inset). Scale bar represents 3 mm. (B) A single device consisted of a 3 × 4 array of microchambers on a 75 × 50-mm slide. Scale bar represents 10 mm. (C) Priming of the microchamber with 70% ethanol containing red food coloring dye. Scale bar represents 3 mm. (D) Characterization of the retention of suspension cells in microchambers. Fraction of retained cells, after 25 VRs of 8 μL per VR into the central well, increased with increasing culture time, up to 99% retention for 16 hours of culture before VRs.
Cell preparation
RPMI 8226 (human-derived MM cell line of B-cell origin) and HS-5 (human-derived BMSC line) were obtained from ATCC. RPMI 8226 cells were routinely cultured at 37°C with 5% CO2 in high-glucose DMEM containing 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin (1% P/S), and 10mM HEPES buffer at 1.0 to 1.5 × 105 cells/mL in tissue culture-treated flasks. Cells were passaged every 2 to 3 days. For experiments conducted in microchambers, RPMI 8226 cells were collected and resuspended at 1.0 to 1.2 × 106 cells/mL. A total of 5 μL of the concentrated cell suspension was dispensed by passive pumping into each microchamber. HS-5 cells were routinely cultured in high-glucose DMEM containing 10% FBS and 1% P/S (without HEPES). Cells were fed every other day and passaged every 4 days at confluence. For coculture experiments in microchambers, HS-5 cells were collected and resuspended at 1.0 × 106 cells/mL in regular HS-5 media supplemented with 10mM HEPES (same as RPMI 8226 media), and 2 μL of the cell suspension was dispensed per side chamber. Blood samples were obtained with informed consent from patients diagnosed with CLL at the University of Wisconsin-Madison Hospital in accordance with UW-Madison institutional review board requirements (H-2009-1070).
Cell retention assay
Cells were incubated for 2, 4, 8, and 16 hours at 37°C with 5% CO2 before applying volume replacements (VRs). For each time point, a total of 25 VRs of 8 μL each to the central well were applied. Phase-contrast images, in a 2 × 2 grid, were captured at room temperature using an Olympus IX-71 microscope (Olympus America) with an UPlanFl 4× objective (NA = 0.13, through air; Olympus) equipped with a Hamamatsu ORCA-ER CCD camera (model C4742-80-12AG, Hamamatsu). Images were acquired using Metamorph Version 7.5.0.0 software (Molecular Devices) and stitched to reconstruct the whole 3-mm-diameter central well. A custom ImageJ macro was used to count all cells in the chamber (supplemental Methods). Images were taken before the first VR to establish the initial cell count and were then taken successively after different numbers of VRs. As a guide to the reader, average flow rates generated by passive pumping in these tests were approximately 1 to 2 μL/s, estimated by dividing the dispensed volume (8 μL) with the estimated total time needed for the volume to transfer from inlet to outlet (∼ 4-5 seconds).
Reagents
TNF-α was from Calbiochem, reconstituted at 10 μg/mL in PBS containing 0.1% BSA, and stored at −20°C in separate aliquots. Bortezomib (PS-341 or Velcade) was obtained as a 2.6mM clinical saline solution from Millenium Pharmaceuticals and stored at −20°C in separate aliquots. Both reagents were thawed 5 minutes before each treatment was applied. Reagents were serially diluted to the desired concentrations in media warmed to 37°C, and dispensed into microchambers in 3 sequential VRs followed by aspiration of the outlet port. LIVE/DEAD Viability/Cytotoxicity Assay Kit from Invitrogen was used to detect live and dead cells in microchambers. Both calcein AM (green) and ethidium homodimer (red) were used at a working concentration of 4μM.
Immunocytochemical staining
Antibody sources were as follows: rabbit polyclonal IgG antibodies for RelA from Santa Cruz Biotechnology, rabbit polyclonal IgG antibodies for prostate specific antigen (negative control for RelA nonspecific binding) from Abcam, mouse monoclonal IgG antibodies for STAT3 from Cell Signaling Technologies, mouse monoclonal IgG antibodies for cadherin-5 (VE-cadherin; negative control for STAT3 nonspecific binding) from BD Transduction Laboratories, and AlexaFluor-488 goat anti–rabbit and AlexaFluor-488 goat anti–mouse secondary antibodies, Hoechst 33342 nuclear dye, and Texas Red-X phalloidin from Invitrogen. Cells cultured and treated in microchambers were immunostained following a standard procedure of fixation, permeabilization, blocking, and antibody binding steps. The procedure involved sequential VRs of reagents by passive pumping (supplemental Methods).
Fluorescence image analysis
All fluorescent images for nuclear translocation assays were taken at room temperature using a Nikon Eclipse Ti inverted fluorescent microscope (Nikon Instruments) with a PlanFluor 10× objective (NA = 0.30, through air, additional 1.5× intermediate magnification; Nikon Instruments) coupled to a Nikon DS-Qi1Mc CCD camera (Nikon Instruments). Images were acquired with NIS-Elements Version D 3.10 software (Nikon Instruments). Image analysis was performed in ImageJ with custom algorithms supported by an in-house database management system (Je'Xperiment) to manage 2280 raw images acquired during this study. The algorithm accepted raw stained images as inputs and generated binary cytoplasmic masks that isolated cells in the view field, and separated the RelA stain into nuclear and cytoplasmic regions based on the location of the nucleus (see supplemental Methods for details).
Results
Microchamber design, fabrication, and characterization
We designed and fabricated a microchamber using soft lithography consisting of a 3-mm-diameter × 500-μm-deep central chamber (the central well) surrounded by 2 1.5-mm-wide × 200-μm-deep crescent-shaped lobes (the side chambers; Figure 1A). A set of 100-μm-wide × 20-μm-deep radial diffusion ports connected the well to the side chambers, allowing soluble molecules to pass freely via diffusion while restricting bulk flow between compartments. Independent inlet and outlet ports provided separate access to the well and side chambers and were sized to facilitate passive pumping and efficient replacement of fluidic volumes for feeding, treatment, and immunostaining operations. Because of the small microchamber footprint, arrays of these microchambers could be fabricated on single devices (Figure 1B), thus streamlining experimentation and increasing throughput. Passive pumping relies on surface tension of small dispensed droplets to move fluids and is governed by the Laplace law where the pressure within a small droplet is inversely proportional to the droplet radius.19,20 Small droplets dispensed at the inlet of the microchamber were therefore pumped toward larger droplets at the outlet port. Importantly, passive pumping is scalable to the needs of the researcher and is equally amenable to manual pipetting operations at the bench with small or modestly sized microchamber arrays, as well as automated pipetting operations with large arrays that can be interfaced with robotic liquid handlers.21,22
The size and geometry of the central well were chosen to accommodate on the order of 103 cells in uniform spatial distribution and to facilitate settling of suspension cells onto a surface below the direct path of fluid flow from inlet to outlet, thereby minimizing shear flow on the cells once they had settled to the bottom surface. We estimated by numerical simulation a minimum wall shear stress of τw approximately 8 × 10−3 Pa at the widest cross section perpendicular to the direction of flow (ie, the 3-mm-diameter cross section), and a maximum wall shear stress of τw approximately 23 × 10−3 Pa near the inlet and outlet recirculation zones, which was sufficiently low to permit efficient and repeated VRs without significant cell depletion. Shear stress can be further reduced by simply increasing the depth of the well or by adjusting other dimensions.
To capture and retain suspension cells in microfluidic systems, other researchers successfully incorporated arrays of cell traps in their microscale designs for different applications, including for cell fusion23 and for capture and treatment of leukemia cells.24 Because of the large number of microfeatures, these systems have high fluidic resistances that are not a major concern when integrated with active pumps capable of producing sufficient pressures but are a significant issue when passive pumping pressures are used. We were able to use relatively simple geometries without cell traps to retain and culture suspension cells with minimal cell loss, thereby allowing us to incorporate passive pumping in lieu of externally applied pumping methods. A design without cell traps was advantageous because spatial arrangement of cells was less restrictive, permitting long-term cultures where cells could freely proliferate and migrate. To test whether cell retention in these microchambers was dependent on culturing time, RPMI 8226 cells were cultured in wells for 2, 4, 8, and 16 hours, after which repeated VRs (8 μL each to central well) were applied. For 2 hours culturing time, only 8 VRs were needed to deplete 50% of the cell population; after 25 VRs, only 32% remained in the well. By increasing culturing time, cell retention improved, and after 16 hours, 99% of cells were retained (Figure 1D). Based on these results, we performed all subsequent experiments after first incubating cells overnight for at least 12 hours before any VRs. Because RPMI 8226 cells are known to form loose adhesions via integrins,25 we suspected that the longer culturing times facilitated these cell-substrate interactions enough to resist the shear applied in our microchambers.
Cell viability assay
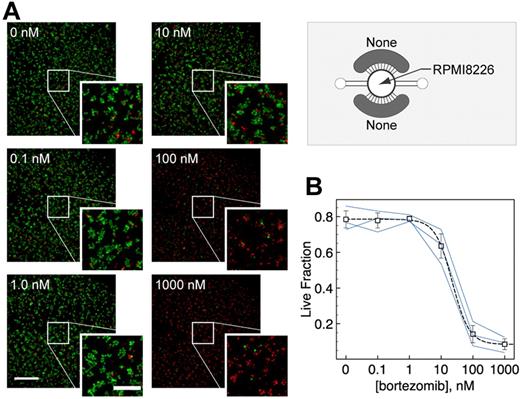
We first used microchamber arrays to develop a simple and fast fluorescence-based cell viability assay for suspension cells. Our main objectives were to demonstrate that the suspension RPMI 8226 cells could be cultured in the arrays over days, that drug treatments had measurable effects on the RPMI 8226 cells, and that the entire assay from cell seeding to readout could be performed with minimal operations. RPMI 8226 cells were allowed to settle and acclimate to the microchamber environment overnight before drug treatment. Bortezomib (Velcade, PS-341), a proteasome inhibitor that can block NF-κB activation and is known to be clinically effective as a therapeutic agent in MM,26 was added directly into each microchamber via a micropipette at the desired concentrations. After 24 hours of bortezomib treatment, RPMI 8226 cells were stained with both calcein AM and ethidium homodimer to indicate live (green) and dead (red) cells, respectively (Figure 2A). Nontreated RPMI 8226 cells showed approximately 80% viability after 2 days in the microchambers (compared with cell viability of 74% ± 5% in Petri dishes). RPMI 8226 cells treated with bortezomib for 24 hours showed lower viability at higher concentrations, with a half-maximal inhibitory concentration IC50 of approximately 22nM (Figure 2B). Thus, RPMI 8226 cells were viable in the microchambers over a minimum of 2 days and were responsive to bortezomib treatments in a dose-dependent manner.
Microchamber cell viability assay for suspension cells. (A) Calcein AM (green) and ethidium homodimer (red) were used to label live and dead RPMI 8226 cells, respectively, which were cultured and treated with bortezomib for 24 hours. (B) Cell viability after 24-hour treatment with bortezomib was dose-dependent with average IC50 ∼ 22nM. Blue lines indicate dose-responses predicted by each of 3 independent experiments (n = 3; independent averages of duplicates). Error bars represent SE.
Microchamber cell viability assay for suspension cells. (A) Calcein AM (green) and ethidium homodimer (red) were used to label live and dead RPMI 8226 cells, respectively, which were cultured and treated with bortezomib for 24 hours. (B) Cell viability after 24-hour treatment with bortezomib was dose-dependent with average IC50 ∼ 22nM. Blue lines indicate dose-responses predicted by each of 3 independent experiments (n = 3; independent averages of duplicates). Error bars represent SE.
μSCeNT assay
We next extended the capabilities of the platform by developing a microfluidic single-cell nuclear translocation (μSCeNT) assay, which has broad applicability in cell signaling studies involving transcription factor activation.27 Transcription factors such as NF-κB and STAT3 translocate from cytoplasm to nucleus before initiating expression of genes that regulate important cellular processes in cancer and other diseases. NF-κB in particular has been implicated as a crucial regulator of MM cell survival and acquired drug resistance,7,28,29 with STAT3 being similarly involved.30 Activation of transcription factors can be measured by several biochemical and functional assays, including EMSAs, luciferase reporter assays or nuclear translocation assays. We used our platform, in conjunction with custom image analysis, to develop a nuclear translocation assay with single-cell resolution for minute cell samples. Custom image analysis was critical to our assay development and is further described in supplemental Methods under “Fluorescence image analysis.”
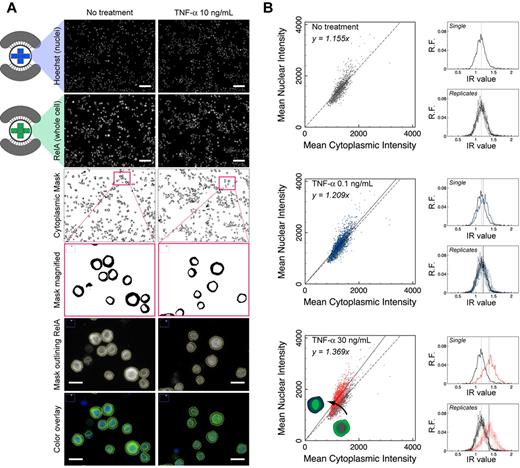
RPMI 8226 cells cultured and treated with drugs or cytokines in microchambers were fixed and immunostained following a procedure that involved sequential VRs of reagents by passive pumping. After staining both the nucleus and the transcription factor of interest, raw images of the stains for each condition were collected, processed, and used to produce a binary mask that separated the cytoplasmic and nuclear regions of single cells (Figure 3A). Single cells within a given population were analyzed, and mean nuclear and cytoplasmic intensities for each cell were calculated and compared graphically in scatter plots (Figure 3B). For each microchamber, 5 images were acquired covering 34% of the well area; approximately 1000 to 2000 cells were analyzed of 5000 to 6000 seeded cells. The choice to acquire 5 images was for convenience. In scatter plots of mean nuclear intensity versus mean cytoplasmic intensity, cells clustered around a line with slope determined via linear regression. For conditions that revealed nuclear translocation (Figure 3B red vs gray), the slope of the fitted line increased compared with control, indicating an overall increase in mean nuclear intensity of the entire population. In contrast, when the slope of the fitted line was similar to the slope of the control, nuclear translocation was absent (Figure 3B blue vs gray). To summarize the data, the ratio of mean nuclear and cytoplasmic intensity, or intensity ratio (IR), for each cell was presented in histograms to reveal features of the population distribution. In this format, nuclear translocation was recognized as a rightward shift in the population distribution of IR values.
Fluorescence image analysis for μSCeNT assay. (A) Representative raw and filtered images from TNF-α dose-response experiments comparing NF-κB activation via TNF-α treatment at 10 ng/mL for 20 minutes to no treatment. (Top to bottom) Raw image of Hoechst stain (nucleus only), and raw image of RelA (NF-κB subunit) stain (whole cell). Scale bar represents 100 μm. Binary cytoplasmic mask generated by subtracting nuclear region from whole cell; inset of cytoplasmic mask; magnified RelA stain with yellow outline of cytoplasmic mask to indicate boundaries separating nucleus and cytoplasm, and color overlay of Hoechst (blue) and RelA (green) stains. Scale bar represents 20 μm. (B) Scatter plots of mean nuclear intensity versus mean cytoplasmic intensity per cell analyzed in a single microchamber. Intensity ratio IR per cell can be calculated, and a histogram can be displayed to show features of the population distribution of IR values (x-axis), where y-axis is relative frequency (R.F.). Gray represents no treatment; blue, TNF-α at 0.1 ng/mL for 20 minutes; and red, TNF-α at 30 ng/mL for 20 minutes. “Replicates” indicates distributions from 9 independent microchambers overlapped.
Fluorescence image analysis for μSCeNT assay. (A) Representative raw and filtered images from TNF-α dose-response experiments comparing NF-κB activation via TNF-α treatment at 10 ng/mL for 20 minutes to no treatment. (Top to bottom) Raw image of Hoechst stain (nucleus only), and raw image of RelA (NF-κB subunit) stain (whole cell). Scale bar represents 100 μm. Binary cytoplasmic mask generated by subtracting nuclear region from whole cell; inset of cytoplasmic mask; magnified RelA stain with yellow outline of cytoplasmic mask to indicate boundaries separating nucleus and cytoplasm, and color overlay of Hoechst (blue) and RelA (green) stains. Scale bar represents 20 μm. (B) Scatter plots of mean nuclear intensity versus mean cytoplasmic intensity per cell analyzed in a single microchamber. Intensity ratio IR per cell can be calculated, and a histogram can be displayed to show features of the population distribution of IR values (x-axis), where y-axis is relative frequency (R.F.). Gray represents no treatment; blue, TNF-α at 0.1 ng/mL for 20 minutes; and red, TNF-α at 30 ng/mL for 20 minutes. “Replicates” indicates distributions from 9 independent microchambers overlapped.
Our assay is similar in concept to other nuclear translocation methods developed to study NF-κB activation,31,32 but with several important distinctions. First, whereas these previous studies used conventional culture platforms and focused on adherent cell types, our microscale platform was designed to accommodate adherent and suspension cells in complex microenvironments. To ensure that our assay could be immediately applied to primary patient samples, we focused on integrating indirect immunostaining into our platform (similar to Ding et al31 ) instead of relying on, for example, GFP-labeled cell lines, as in the work of Bartfeld et al.32 Although GFP-labeled cell lines can provide real-time live-cell monitoring of intracellular dynamics, immunostaining has the advantage of being more accessible and adaptable because it is a ubiquitous technique in research laboratories, any protein can be studied given appropriate antibodies are available, and it can be directly translated to primary patient cell samples, enabling clinical studies that would be practically challenging or impossible via transfection methods.
Our ability to perform efficient immunostaining of suspension cells is important because it has both streamlined traditional suspension immunostaining procedures and extended microfluidic immunostaining techniques beyond adherent cells.22 The current standard procedure for immunostaining suspension cells requires the transfer of cells from culture plate to staining substrate via cytocentrifugation or chemical linkers so that cells can resist repeated rinsing.33 Cell loss during transfer and cell distortion during cytocentrifugation both lead to potential biases. These issues can be avoided with the current platform, which facilitates culture, treatment, fixation, staining, and analysis of suspension cells all in the same system. Whereas immunostaining of adherent cells in microfluidic channels is relatively straightforward, immunostaining of suspension cells has been challenging because of the inability of suspension cells to resist high shear stresses typically found in microscale channels. Recently, Lecault et al achieved this in a high-throughput microfluidic array for studying hematopoietic stem cell proliferation.11 Our system achieved this with simple geometries, avoiding high resistance cell traps and permitting the use of passive pumping, thus obviating the need for tubing and complex interfacing.
Finally, our analysis retains the entire distribution of IR values and obviates the need for gating procedures, as was used by Bartfeld et al.32 Reporting whole distributions is particularly important in unique cases where subpopulations in samples lead to multimodal distributions, which may ultimately reflect distinct behaviors of activation between subgroups of cells that would otherwise be masked by averaging or by gating. Our current method has some limitations, namely: (1) the fluorescence intensity depends on nonlinear antibody binding efficiencies inherent in indirect immunostaining; and (2) captured signals are integrated over a finite depth of focus that spans a significant section of the cell. These issues contribute to limited dynamic range and affect our ability to accurately quantitate nuclear and cytoplasmic fractions of stained proteins. Use of confocal or deconvolution microscopy, if available, would help minimize signals from outside the focal plane, whereas direct immunostaining with fluorescently labeled primary antibodies would further eliminate nonlinearities in antibody binding. Given that our current readout is based on IR, which only reports ratio of intensities and not absolute values, our method was not designed to provide accurate quantitation of molecular events but instead offer a semiquantitative endpoint for capturing protein trafficking tendencies of single cells or subpopulations within a larger population.
NF-κB activation via TNF-α treatment
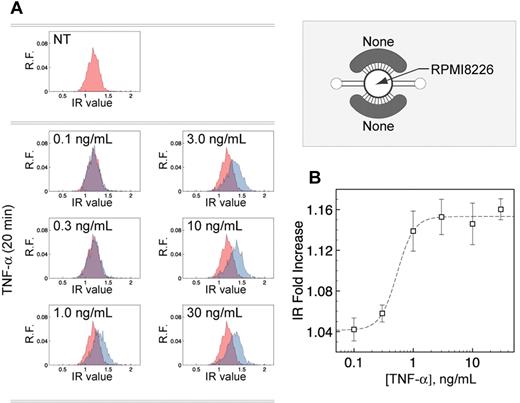
The μSCeNT assay was used to examine a series of cytokine and drug treatments known to activate the NF-κB and STAT3 pathways in MM cells. These tests served as validation for both the microscale platform and for the nuclear translocation assay. Importantly, to our knowledge, this was the first report of NF-κB and STAT3 activation of MM cells in microscale environments, and it was thus necessary to establish a baseline of cellular responses at this scale. We treated RPMI 8226 cells with 10 ng/mL TNF-α for 20 minutes, 1 hour, 2 hours, and 4 hours to establish a time course for NF-κB activation within microchambers, and observed that the IR values of the cell population shifted to the right for all time points (supplemental Figure 4), indicating nuclear translocation of RelA (an NF-κB subunit) began as early as 20 minutes after treatment, consistent with typical activation of the canonical NF-κB pathway.34,35 Based on time-course results, we treated RPMI 8226 cells with different TNF-α concentrations for 20 minutes. RelA nuclear translocation was detected for all concentrations between 1.0 and 30 ng/mL but was not detected for 0.1 or 0.3 ng/mL (Figure 4A). The shapes of the IR-cell population distribution at different doses were similar with untreated cells, except for the variable right shifts. This indicates that all cells were similarly responsive to TNF-α treatment under the microchamber condition, similar to what was observed using a κB-GFP reporter assay under macroscale conditions.36 The fold change in population mean IR between the detected and nondetected groups was statistically significant (P < .05). Sigmoidal curve fitting of the dose-response revealed a half-maximal effective concentration (EC50) of approximately 0.53 ng/mL TNF-α (Figure 4B). In total, measurements were made for 9 different microchambers (3 independent experiments, each in triplicate) at each concentration, with low SD in population mean IR between the 9 samples (Figure 3B, “Replicates”), thus demonstrating the reproducibility of cellular responses in individual channels in repeated experiments.
NF-κB activation via TNF-α treatment as measured by nuclear translocation of major NF-κB subunit RelA. (A) Histograms of IR values (x-axis) for no treatment control (red) versus TNF-α treatment for 20 minutes at varying concentrations (blue), where y-axis is relative frequency (R.F.). (B) Dose-response curve of population mean IR, normalized to the population mean IR of the no treatment control (“IR Fold Increase” in graph). EC50 ∼ 0.53 ng/mL TNF-α (n = 3; independent averages of triplicates). Error bars represent SE.
NF-κB activation via TNF-α treatment as measured by nuclear translocation of major NF-κB subunit RelA. (A) Histograms of IR values (x-axis) for no treatment control (red) versus TNF-α treatment for 20 minutes at varying concentrations (blue), where y-axis is relative frequency (R.F.). (B) Dose-response curve of population mean IR, normalized to the population mean IR of the no treatment control (“IR Fold Increase” in graph). EC50 ∼ 0.53 ng/mL TNF-α (n = 3; independent averages of triplicates). Error bars represent SE.
Effect of bortezomib treatment on NF-κB activation
We further tested the effects of bortezomib treatment on basal and TNF-α–mediated NF-κB activation. Because bortezomib blocks TNF-α–induced NF-κB nuclear translocation by inhibiting IκB degradation and sequestering NF-κB in the cytoplasm, treatment with bortezomib was expected to block activation of this canonical NF-κB pathway.7 RPMI 8226 cells were pretreated with various concentrations of bortezomib for 4 hours in the microchambers. Treatment was then replaced with either TNF-α at 10 ng/mL for 20 minutes or with control media for 20 minutes. Bortezomib treatment successfully abrogated TNF-α–mediated NF-κB activation in a dose-dependent manner (Figure 5). Sigmoidal curve fitting estimated the half-maximal inhibitory concentration (IC50) approximately 7.2nM bortezomib, which is in the similar range observed for this cell type in the macroscale.7 Examination of the distributions revealed an interesting observation for populations with both TNF-α–untreated and –treated conditions. At bortezomib concentrations of 3nM and higher without TNF-α treatment, the distribution was slightly shifted to the right and the distribution was more spread as indicated by the blunted peak compared with the no treatment control. The lack of bortezomib-induced inhibition of basal NF-κB activity using EMSA was previously observed in RPMI 8226 cells under macroscale.7 In addition, paradoxical NF-κB activation by bortezomib treatment alone has also been reported in both RPMI 8226 and primary MM cells7,37 ; however, details of the behavior of the entire MM cell population under bortezomib-treated conditions were not described at the single-cell level. The blunting of the peak and increased spreading of the IR values suggested that the bortezomib-induced NF-κB activation probably occurred in a subpopulation of cells, as opposed to an equal contribution of nuclear translocation from all cells (as in the case with TNF-α–induced activation, Figure 4).
Effect of bortezomib treatment on NF-κB activation via TNF-α induction. (A) Histograms of IR values (x-axis) for 4-hour bortezomib treatment only (left column) versus 4-hour bortezomib treatment followed by 20 minutes of 10 ng/mL TNF-α treatment, over a range of bortezomib concentrations (y-axis is relative frequency [R.F.]). Red represents no treatment; and blue, treatments. (B) Representative images of RelA stains comparing bortezomib treatment only (left column) versus 4-hour bortezomib treatment followed by 20 minutes of 10 ng/mL TNF-α treatment. Scale bar represents 20 μm. (C) Dose-response curve of population mean IR, normalized to the population mean IR of the no treatment control (“IR Fold Increase” in graph). IC50 ∼ 7.2nM bortezomib (n ≥ 3; independent averages of duplicates). Error bars represent SE.
Effect of bortezomib treatment on NF-κB activation via TNF-α induction. (A) Histograms of IR values (x-axis) for 4-hour bortezomib treatment only (left column) versus 4-hour bortezomib treatment followed by 20 minutes of 10 ng/mL TNF-α treatment, over a range of bortezomib concentrations (y-axis is relative frequency [R.F.]). Red represents no treatment; and blue, treatments. (B) Representative images of RelA stains comparing bortezomib treatment only (left column) versus 4-hour bortezomib treatment followed by 20 minutes of 10 ng/mL TNF-α treatment. Scale bar represents 20 μm. (C) Dose-response curve of population mean IR, normalized to the population mean IR of the no treatment control (“IR Fold Increase” in graph). IC50 ∼ 7.2nM bortezomib (n ≥ 3; independent averages of duplicates). Error bars represent SE.
Similarly, for cells pretreated with bortezomib and then treated with TNF-α, population distributions were not only shifted back to the left to indicate a dose-dependent decrease in nuclear translocation but were also more negatively skewed (ie, distribution concentrated on the right side), particularly at concentrations of 3nM and lower. This suggests that a subpopulation of highly NF-κB–activated cells was more sensitive to inhibition by bortezomib at higher drug doses. However, the distributions were not found to be significantly bimodal in nature based on the Hartigan dip test (P > .05). Altogether, the μSCeNT assay revealed the presence of subpopulations of MM cells that weakly activated NF-κB in response to bortezomib treatment and that were highly susceptible to bortezomib inhibition of TNF-α–induced NF-κB activation. Thus, our system offers valuable information not attainable by population-averaged methods while using a very limited number of cells.
STAT3 activation in MM-BMSC coculture
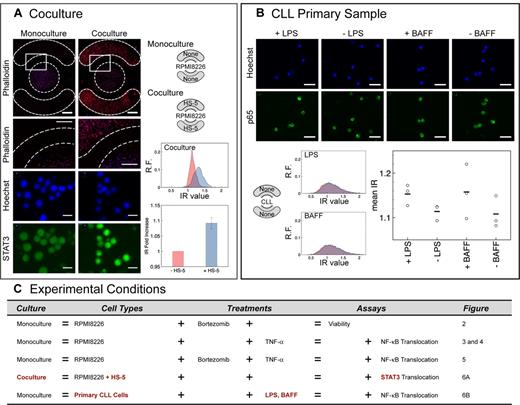
To demonstrate coculture capabilities of the platform, we studied the effect of neighboring adherent BMSCs (HS-5 cell line) cocultured with suspension RPMI 8226 cells in the microchambers. BMSCs are known to activate the STAT3 pathway in MM cells via paracrine signaling.38,39 We cultured HS-5 cells for 24 hours in the 2 side chambers to allow adhesion, cell spreading, and conditioning of media with factors secreted by HS-5 cells. RPMI 8226 cells were then seeded and cultured for 24 hours either in monoculture or in coculture with HS-5 cells. RPMI 8226 cells in monoculture displayed low levels of constitutive STAT3 activity in the nucleus, whereas RPMI 8226 cells in coculture displayed marked increases in nuclear STAT3 levels, with the entire distribution of cultured RPMI 8226 cells significantly shifted to higher IR values (Figure 6A). This result demonstrated that all of the MM cells in microchambers were responsive to stromal influence. The data also showed that the microchambers were capable of properly compartmentalizing cell types and allowing paracrine signaling between cocultured cells via soluble factors. Additional levels of complexity can be achieved using the side chambers for different cell types (eg, bone marrow endothelial cells, osteoblasts) other than BMSCs and by simple design modifications to accommodate more side chambers or different geometries. Moreover, this system offers the possibility of analyzing all combinations of these cell-cell interaction studies using cells derived entirely from individual patient samples. Such “intra-patient” cell signaling studies using MM patient samples, which have not been described in the literature, has the potential to significantly advance our understanding of patient-to-patient heterogeneity within a cancer type and provide valuable information of individualized therapy.
Robustness of the microchamber array was demonstrated by coculture and primary cell experiments. (A) Effect of HS-5s on STAT3 activation. Phalloidin (red) and Hoechst (blue) stains showed compartmentalization of cell types in their respective chambers. Scale bar represents 1 mm (inset scale bar represents 500 μm). Hoechst (blue) and STAT3 (green) stains showed marked increase in nuclear STAT3 for cocultured RPMI 8226 cells compared with monoculture. Histograms of IR values (x-axis) for monoculture (red) versus coculture (blue), where y-axis is relative frequency (R.F.). Population mean IR for coculture was normalized to the population mean IR for monoculture (IR fold increase in bar graph, n = 3; independent averages of quadruplicates). Error bars represent SE. (B) CLL primary patient sample showed detectable NF-κB activation by BAFF and lipopolysaccharide. Shape of distribution differed from cocultured RPMI 8226 distribution in panel A. Scale bar represents 20 μm; graph, n = 1. ○ represents triplicates; and bar, triplicate average. (C) List of experimental conditions tested.
Robustness of the microchamber array was demonstrated by coculture and primary cell experiments. (A) Effect of HS-5s on STAT3 activation. Phalloidin (red) and Hoechst (blue) stains showed compartmentalization of cell types in their respective chambers. Scale bar represents 1 mm (inset scale bar represents 500 μm). Hoechst (blue) and STAT3 (green) stains showed marked increase in nuclear STAT3 for cocultured RPMI 8226 cells compared with monoculture. Histograms of IR values (x-axis) for monoculture (red) versus coculture (blue), where y-axis is relative frequency (R.F.). Population mean IR for coculture was normalized to the population mean IR for monoculture (IR fold increase in bar graph, n = 3; independent averages of quadruplicates). Error bars represent SE. (B) CLL primary patient sample showed detectable NF-κB activation by BAFF and lipopolysaccharide. Shape of distribution differed from cocultured RPMI 8226 distribution in panel A. Scale bar represents 20 μm; graph, n = 1. ○ represents triplicates; and bar, triplicate average. (C) List of experimental conditions tested.
NF-κB activation in a CLL patient sample
Finally, the microchamber array was used to investigate NF-κB activation in a primary CLL patient sample. NF-κB in CLL can be activated by B-cell activating factor (BAFF) and a proliferation-inducing ligand.40 CLL cells, which were initially isolated from the blood of a patient and cryopreserved, were thawed, cultured for several days in Petri dishes, and then seeded into microchambers in preparation for a μSCeNT assay. For this patient, we found that CLL cells showed a minor but detectable activation of NF-κB from both BAFF (P = .13) and lipopolysaccharide (P < .05) treatments. Of interest, the shape of the NF-κB IR distributions for the CLL samples (both basal and stimulated) differed significantly from the shape of the distributions for RPMI 8226 cells (both basal and with HS-5 coculture; compare Figure 6A vs Figure 6B). For the same histogram binning width, this particular CLL sample appeared to have a much larger spread in distribution. Determination of whether this is a general phenomenon of primary CLL cells compared with MM cells requires further investigation.
Discussion
By combining microscale technology, high-content image processing, and accessible design, we have developed a microscale cell culture and analysis platform that has expanded both the quantity and quality of experiments in an accessible manner for cell biologists. Our platform is capable of handling minute samples of suspension (and adherent) cells and extracting high-content functional information from complex microenvironments, all using only a micropipette and fluorescence microscopy for operation. The ability of the nuclear translocation assay to analyze samples at single-cell resolution offers the potential for new biological insight regarding the heterogeneity of patient-derived cell populations. Furthermore, accessibility was designed directly into the platform by integrating passive pumping to handle fluids and by employing convenient suspension cell immunostaining to allow detection of different proteins, aspects that will facilitate direct translation to patient samples and more immediate adoption by other research laboratories.
Although devices in the current work were fabricated in PDMS and glass, our platform can also be fabricated in polystyrene,41 a thermoplastic widely used in common cell culture labware and useful for studies involving small hydrophobic molecules (eg, estrogen, fluoxetine) that are known to absorb into bulk PDMS.42-44 We validated the platform in the context of MM and showed that other suspension cell types (CLL) can also be studied. We showed the platform can detect cell viability, dose-dependent activation, and inhibition of NF-κB after cytokine and drug treatments, and activation of STAT3 in MM cells in the presence of cocultured BMSCs. Our platform can therefore be applied to a myriad of biological problems and be used to reveal new and previously unattainable biological insights on cell behavior and function in human diseases.
Based on its robustness of handling various cell types, culture arrangements, treatment conditions, and offering different assays (Figure 6C), we envision the platform being used in various ways. First, the platform will be useful in basic research as a flexible platform for elucidating complex cell signaling mechanisms between all the different cell types in the microenvironment, thereby enabling a deeper understanding of how complex cell-cell interactions affect disease etiology, development, and treatment. Second, it has potential to find application in translational research as a functional microculture array screening tool in personalized therapy where different cell types isolated from individual patient-derived bone marrow (BMSCs, MM cells, endothelial cells, and osteoblasts) are cultured in appropriate spatial arrangements in the platform, and drug panels are tested on the cells to determine the appropriate drug cocktail or regimen most effective for the individual. Ultimately, our platform allows us to overcome major current challenges to progress in translational research, moving us toward high-content functional cytomics.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr E. Berthier and Dr J. Warrick for providing Je'Xperiment custom software for interfacing and running the nuclear translocation algorithm.
This work was supported by Molecular & Cellular Pharmacology Training Grant (C.P.), National Institutes of Health-National Cancer Institute (R33 CA137673, D.J.B.), Forward Lymphoma/UWCCC (S.M. and D.J.B.), National Institutes of Health (R01 CA081065 and R56 CA081065), the Multiple Myeloma Research Foundation (Senior Award), and the Lymphoma Research Foundation (S.M.).
National Institutes of Health
Authorship
Contribution: E.W.K.Y., C.P., S.M., and D.J.B. designed research; E.W.K.Y. and C.P. performed research; B.S.K., D.T.Y., and N.S.C. contributed new reagents and primary samples; E.W.K.Y. analyzed data and made the figures; and E.W.K.Y., C.P., S.M., and D.J.B. wrote the paper.
Conflict-of-interest disclosure: D.J.B. has ownership interest in Bellbrook Laboratories LLC, which has licensed technology reported in this publication. The remaining authors declare no competing financial interests.
Correspondence: David J. Beebe, Wisconsin Institutes for Medical Research, Department of Biomedical Engineering, University of Wisconsin-Madison, 1111 Highland Ave, Rm 6009, Madison, WI 53705; e-mail: djbeebe@wisc.edu.

![Figure 5. Effect of bortezomib treatment on NF-κB activation via TNF-α induction. (A) Histograms of IR values (x-axis) for 4-hour bortezomib treatment only (left column) versus 4-hour bortezomib treatment followed by 20 minutes of 10 ng/mL TNF-α treatment, over a range of bortezomib concentrations (y-axis is relative frequency [R.F.]). Red represents no treatment; and blue, treatments. (B) Representative images of RelA stains comparing bortezomib treatment only (left column) versus 4-hour bortezomib treatment followed by 20 minutes of 10 ng/mL TNF-α treatment. Scale bar represents 20 μm. (C) Dose-response curve of population mean IR, normalized to the population mean IR of the no treatment control (“IR Fold Increase” in graph). IC50 ∼ 7.2nM bortezomib (n ≥ 3; independent averages of duplicates). Error bars represent SE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/10/10.1182_blood-2011-10-384347/4/m_zh89991287280005.jpeg?Expires=1765984200&Signature=EFeZ8R50f9OMJTlzpNG5erZUbKPKRYUO95ntGFUswrjO1D8h3Zu9hvR9H8IKVzolrR7fkZmm58spgwbaQv6uaXxfZAHuKGlqtEt5400fiC0MpxvFduyB-ejpYah~a9SN4SQGrYV2s1Z7BJE0APq~E22SiBwl1z2VO7lLwmjVopDmyyn1qFFDcYHw-Ha2JJBrzwrCheIC68Y97z7KbB8fFbPWjX2TVLhyLVcK~pDKkycxt0UuR-0MsTliQuJ5NvVKWlhVsypjzR1g8dDk2Oyk-2SxEEcxoA12vNkk2H7WfZXtl76TRVIgOpKUR~8NGXxLzxOSeRuByfgEy8wn1gHA-Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)