Abstract
This systematic review was designed to provide more precise effect estimates of inhibitor development for the various types of F8 gene mutations in patients with severe hemophilia A. The primary outcome was inhibitor development and the secondary outcome was high-titer-inhibitor development. A systematic literature search was performed to include cohort studies published in peer-reviewed journals with data on inhibitor incidences in the various F8 gene mutation types and a mutation detection rate of at least 80%. Pooled odds ratios (ORs) of inhibitor development for different types of F8 gene mutations were calculated with intron 22 inversion as the reference. Data were included from 30 studies on 5383 patients, including 1029 inhibitor patients. The inhibitor risk in large deletions and nonsense mutations was higher than in intron 22 inversions (pooled OR = 3.6, 95% confidence interval [95% CI], 2.3-5.7 and OR = 1.4, 95% CI, 1.1-1.8, respectively), the risk in intron 1 inversions and splice-site mutations was equal (pooled OR = 0.9; 95% CI, 0.6-1.5 and OR = 1.0; 95% CI, 0.6-1.5), and the risk in small deletions/insertions and missense mutations was lower (pooled OR = 0.5; 95% CI, 0.4-0.6 and OR = 0.3; 95% CI, 0.2-0.4, respectively). The relative risks for developing high titer inhibitors were similar.
Introduction
Patients with severe hemophilia A have a factor VIII (FVIII) activity level of less than 0.01 IU/mL. Over the past decades, severe hemophilia has changed from a debilitating disease to a condition with a good quality of life.1 Great advances in efficacy and safety of FVIII products and treatment strategies have made this possible. The current standard of care for children is primary prophylaxis—regular FVIII infusions aimed at preventing joint bleeds and joint damage. However, in 1 of 4-5 patients with severe hemophilia A, treatment with FVIII is complicated by the occurrence of neutralizing inhibitory Abs against FVIII at a young age.2 These inhibitors not only make adequate correction of the bleeding diathesis more difficult in case of bleeding and surgery, but also make regular prophylaxis with FVIII impossible. This situation may last until immune tolerance is achieved or may even persist for a person's lifetime if immune tolerance therapy fails.
One of the most important predictors of the risk of inhibitor development in severe hemophilia A is the F8 gene mutation type.3,4 Reported absolute and relative risks of inhibitor development according to the different F8 mutation types vary markedly between studies, because the estimates per study are based on relatively few patients. A meta-analysis can yield more precise estimates of the risk for different types of F8 mutations. These data may contribute to the knowledge on the risk factors of inhibitor development that allows clinicians to assess whether an individual patient is at high risk. In the future, when treatment strategies such as immune-modifying treatments become available to prevent the development of inhibitors, this information will be essential to weighing the potential harmful side effects of such treatments against the inhibitor risk.
We performed a systematic review to summarize the currently available evidence on the relationship between inhibitor development and the various types of F8 mutations and a meta-analysis to obtain more precise estimates of the relative risks of inhibitor development according to the F8 genotypes in patients with severe hemophilia A.
Methods
This systematic review is reported in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement (www.prisma-statement.org).5
Inclusion criteria and methodological quality criteria were specified and documented in a protocol in advance. Methods of the analysis were specified in advance, except for the type of random effects model, which was adjusted to an appropriate model during data analysis.
Study eligibility criteria
Types of studies.
Observational cohort studies studying the relationship between F8 genotype and inhibitor development published as an article or letter in a peer-reviewed journal in English, German, Dutch, French, Italian, or Spanish without publication date restrictions were eligible for inclusion.
Types of participants.
Cohorts of patients with severe congenital hemophilia A (baseline FVIII activity level of < 0.01 IU/mL) in whom inhibitor tests had been performed were considered. Study inclusion was not restricted to cohorts of unrelated patients.
Types of outcome measures.
The primary outcome was inhibitor development, defined according to the methods of the investigators of the original study. The secondary outcome was high-titer-inhibitor development, defined as a peak inhibitor titer of at least 5 Bethesda units/mL.
Type of determinants.
The determinant was the F8 mutation type as assessed by any strategy. The F8 genotype was classified as large deletions (single exon or multiple exons), nonsense mutations (light chain or non-light chain), intron 1 and 22 inversions, small deletions/insertions/combined deletions and insertions (in poly-A runs or outside poly-A runs), missense mutations (light chain or non-light chain), splice site (conserved or nonconserved nucleotide positions). A poly-A run was defined as at least 6 adenines in a row. Conserved splice-site mutations were defined as those affecting the +1, +2, −1, or −2 nucleotide position at a splice site. Nonconserved splice-site mutations were all other splice-site mutation types.
Only studies that had determined the whole spectrum of F8 mutations and reached a mutation detection rate of at least 80% were eligible for inclusion. Studies that reported only gross gene derangements such as partial gene deletions or intron 22 inversions detected by restriction enzyme cleavage with Southern blotting were excluded. Studies in which the study population only included patients who were negative for intron 22 inversions were also excluded, because the reference group was lacking.
Information sources
Studies were identified by searching electronic databases, by screening the bibliographic references of retrieved studies and reviews, and by contacting experts in the field (J.O. and J.A.) to identify any studies that were not retrieved by the literature search. The following bibliographic databases were searched: PubMed (1966-present), EMBASE (1980-present), Web of Science (1945-present), Cochrane database (1992-present), CINAHL (1982-present), Academic Search Premier (1975-present), and ScienceDirect (1995-present).
Search
We used the following search terms to search all databases: hemophilia A, FVIII deficiency, mutation,* gene,* inhibitor,* and antibody. The full search for each database is listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The aim of the search was a high sensitivity. The search was designed and supervised by an experienced librarian (J. W. Schoones, MA, Walleus Library of the Leiden University Medical Center, Leiden, The Netherlands). The search was run on February 22, 2010. After February 2010, additional studies were included by performing monthly searches in PubMed up to February 28, 2011.
Study selection
Titles and abstracts were examined to identify potentially relevant studies by one investigator (S.G.) because the eligibility criteria were apparent and the risk of rejecting relevant reports was low. All potentially relevant studies were retrieved as complete manuscripts. S.G. examined the studies for compliance with the inclusion criteria. In case of any doubt for eligibility of the study for inclusion, this was discussed with a methodological expert (J.G.v.d.B.).
Data collection process
Duplicate reports of studies were excluded by checking the authors' names, authors' affiliations, and catchment areas. Duplicate inclusion of individual patients who participated in more than one study was avoided by systematically evaluating patient recruitment periods and catchment areas. In case of duplicate patients, the investigators of the original studies were contacted to provide the data after exclusion of the duplicate patients. In several cases, reported anonymous patient identifiers enabled the investigator to exclude the individual patients who were reported in duplicate.
Data extraction and management
Data extraction from manuscripts was performed by one investigator (S.G.). A structured electronic data collection form was used. In 22 studies, the published reports did not provide all required information. Therefore, we contacted the corresponding authors of these studies by e-mail for further information. We requested to restrict the data for patients with severe hemophilia only, to categorize patients in subclassifications of genotypes, and to provide numbers of patients with high responder inhibitor development. Of the contacted investigators, 17 responded and 16 provided the required data.
Data items
The following data were extracted from the included studies: manuscript identifier, study identifier, year of publication, eligibility for inclusion, number of patients with severe hemophilia A, city/country, study period, definition of inhibitor development, frequency of inhibitor screening, inhibitor assay used, methods of mutation analysis, mutation detection rate, ethnicity, methodological quality items (see “Sensitivity analyses”), total number of patients with inhibitor development, total number of patients with high-titer-inhibitor development, total number of patients with inhibitor development, cumulative incidence of inhibitor development in the different F8 genotype categories, and the total study population.
Summary measures
Odds ratios (ORs) with 95% confidence intervals (95% CIs) for each category of F8 mutation in every study were calculated with the patient group with intron 22 inversions as the reference. Because approximate 95% CIs cannot be computed in studies with 0% or 100% events in the genotype category of interest and/or reference category, the exact mid-P 95% CIs were calculated.6 No continuity correction was used. No data were imputed.
Data exploration
To explore the within-study and between-study variability (heterogeneity), we visually assessed the extent of overlap in 95% CIs in forest plots. In addition, we estimated τ,2 which is an estimate of the between-study variance. We did not test for statistical significance of heterogeneity with the Cochran Q test because of its known limitations.7
Data synthesis
Because of limited clinical and methodological heterogeneity, we pooled the results of each study in a meta-analysis. We calculated the pooled ORs for patients in each F8 genotype category with the patient group with intron 22 inversions as the reference because this was the largest group. To account for variability between studies in frequency of inhibitor screening, we used a random-effects model. Because of the small sample sizes and low number of inhibitor patients in some F8 genotypes, conventional methods for meta-analysis can be severely biased.8 Therefore, we applied exact methods for meta-analysis using a mixed-effect hypergeometric-normal model.8 Furthermore, this model avoids the potential problems associated with the use of continuity corrections.9 In case the between-study variance (τ2 ) was zero, a fixed-effect hypergeometric normal model was used.
For each genotype and outcome (inhibitor development and high-titer-inhibitor development), the ORs and 95% CIs and the pooled estimate were plotted in forest plots together with the numbers of patients, numbers of inhibitor patients, and percentages of inhibitors.
Data evaluation
Small study data trends.
We evaluated whether small study data trends were present by arranging the studies in the forest plots by study sample size. To evaluate the impact of the small study bias, we visually assessed asymmetry of the forest plots.
Sensitivity analyses.
To verify whether the associations were influenced by the methodological quality of the studies, we repeated the calculations in a subgroup of studies that we considered to have a low chance of bias for this specific research question. Because hemophilia centers that publish their data are generally the larger and more experienced centers, clinically challenging patients such as inhibitor patients tend to be referred to these centers. This may lead to overestimation of the inhibitor incidence and potentially to selection bias. Therefore, we included in the sensitivity analysis only studies that did not include inhibitor patients who were referred to that center because of the inhibitor. Further inclusion criteria that were used for the sensitivity analysis were studies that reported a cumulative incidence and were unlikely to have misdiagnosed inhibitor patients (these studies measured the inhibitor titer at least once a year and whenever there was a clinical suspicion) and studies including patients who had been treated on at least 50 exposure days with FVIII.
Exact 95% CIs were calculated using Episheet 2007 spreadsheets for the analysis of epidemiologic data.6 The SAS Version 9.2 Academic Analysis Suite and the Enterprise Guide 4.1 United Kingdom were used for the meta-analyses.
HADB
To complement our findings, we summarized the data present in the Hemophilia A Mutation Database (HADB, formerly the Hemophilia A Mutation, Structure, Test and Resource Site [HAMSTeRS]) database.10 On October 8, 2010, the data on all 2718 patients were downloaded from the website. Patients with severe hemophilia A were identified as those patients who had a FVIII activity of < 1% or a missing FVIII activity with a severity reported as severe. There were 1276 patients with severe hemophilia A. We excluded 16 patients who were entered twice because they were included in more than one database (combined insertions and deletions were entered in both the insertions and deletions databases; splice-site mutations were entered in both splice site and missense mutations databases). Two patients who had a concurrent intron 1 and 22 inversion were excluded. Four patients who were entered twice (1 patient 3 times) because of additional mutations that were regarded as a polymorphism were excluded. Of the remaining 1253 patients, 922 patients had a known inhibitor status. These patients were used for the summary. Data on patients with intron 1 and 22 inversions were not collected in the HADB. Data of the HADB were not included in the meta-analysis.
Results
Study selection
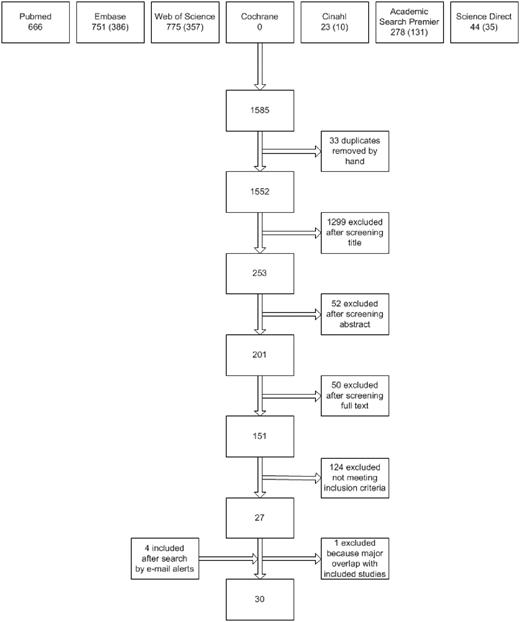
Figure 1 presents the flow chart of the study selection process. Using the above search strategies, 1552 unique references were identified. After screening of the titles and abstracts, 201 unique articles were retrieved. Screening reference lists of selected studies identified no additional studies. In total, 151 studies were identified as being potentially relevant. After strictly applying the inclusion criteria, 26 were eligible for inclusion. Four eligible studies were identified by automatic literature search updates after the first search. After contacting the investigators of 22 studies for additional information, a total of 30 studies were included in the data analysis.
Flow diagram of study selection. Numbers of unique reports are indicated in parentheses.
Flow diagram of study selection. Numbers of unique reports are indicated in parentheses.
Excluded were 24 reviews or meeting abstracts, 17 duplicate reports of studies, 2 studies in a foreign language that did not meet the inclusion criteria (Czech and Portuguese), 14 publications that did not report data on incident inhibitor patients and total numbers of patients for each genotype, 2 studies without data on intron 22 inversions, 1 study without any data on inhibitor development, 44 studies that did not report the full mutation spectrum, 13 studies that did not meet the mutation detection level, 6 studies in which the study cohorts were nearly completely overlapping with other included studies, and 2 studies with insufficient data.
Supplemental Table 2 summarizes the studies that appeared to meet the eligibility criteria but on further inspection did not. One potentially eligible study was excluded because of unavailable data on patients with severe hemophilia.11 One study was excluded during the exclusion of double patients because the majority of study participants were already part of other study cohorts.12 One study was excluded because it did not meet the criterion of a mutation detection rate of at least 80%.13
Included studies
The characteristics of the included studies are summarized in in Table 1. All studies were published in English. Seventeen studies were conducted in Europe, 3 in the United States, 7 in Asia, 1 in South America, 1 in Europe and Northern America, and 1 in Europe and Asia. The studies were published from 1995-2010. One study had families as the unit of observation instead of individual patients. The number of subjects per study varied from 20-971. The mutation detection rate varied from 80%-100%.
Study characteristics
| Source . | N . | Patients or families . | Unrelated patients . | Year . | Country . | Study cohort . | Mutation analyses methods: intron 1 and 22 inversion screening and mutation analysis . | Mutation detection rate, % . | Ethnicity . | Frequency inhibitor screening . | Inhibitor assay . | Treated > 50 ED . | Unselected cohort . | High quality . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahmed14 | 46 | Patients | Yes | 2005 | New Delhi, India | Single | PCR, DHPLC | 92 | NA | NA | Bethesda | NA | NA | − |
| Awidi15 | 117 | Patients | No | 2010 | Jordan | NA | PCR sequencing | NA | NA | NA | Bethesda | Incl < 50 ED | NA | − |
| Boekhorst16 | 664 | Patients | NA | 2008 | Nijmegen, the Netherlands Paris, France Tehran, Iran | Multi | PCR-DGGE/CSGE sequencing | 100 | NA | NA | Bethesda or Nijmegen Bethesda | Incl < 50 ED | No | − |
| Casaña17 | 92 | Families | Yes | 2008 | Valencia, Spain | Single | Southern-blot/PCR sequencing | 100 | NA | NA | Bethesda | NA | NA | − |
| Castaman18 | 27 | Patients | No | 2007 | Albany | Single | PCR, DHPLC sequencing | 100 | NA | 18 patients screened once | NA | Minimally treated | NA | − |
| Chen19 | 88 | Patients | Yes | 2010 | Taipei, Taiwan | Single | PCR, DHPLC sequencing, RNA analysis | 98 | NA | NA | Bethesda | NA | Yes | − |
| David20 | 108 | Patients | Yes | 2006 | Lisboa/Porto, Portugal | Multi | PCR, SSCP/restriction enzyme cleavage/sequencing | 100 | White | NA | Bethesda | NA | NA | − |
| Fernandez-Lopez21 | 55 | Patients | Yes | 2005 | Andalusia, Spain | Single | PCR sequencing | 89 | NA | NA | NA | Na | Yes | − |
| Goodeve22 | 49 | Patients | NA | 2000 | Recombinate Study Group* | Multi | Southern-blot, CSGE sequencing | 93 | 31 white, 7 African American, 4 Hispanic, 1 Asian, 1 other, 1 unknown | At least every 3 months, ≥ 0.6 BU/mL | Bethesda | Incl < 50 ED | No | − |
| Gouw23 | 122 | Patients | No | 2007 | Leuven, Madrid, Stockholm, Malmö, Montreal, Göteborg | Multi | NA | 83 | 151 white, 1 African American, 3 Asian, 7 other, 1 unknown | Various frequency of testing, ≥ 2 positive inhibitor and decreased recovery | Bethesda or Nijmegen Bethesda | All > 50 ED | Yes | + |
| Gouw24 | 318 | Patients | No | 2010 | Utrecht, The Netherlands | Single | Southern blot/PCR sequencing | 95 | NA | ≥ 1/y (1970-1980), ≥ 1/6 m (early 1990s), ≥ 1/3 m (late 1990s onwards) and at clinical indication; ≥ 2 positive inhibitor and decreased recovery | Bethesda or Nijmegen modification | All > 50 ED | Yes | + |
| Green25 | 248 | Patients | No | 2008 | United Kingdom† | Multi | Southern blot/PCR, fluorescent solid-phase CCM/DHPLC sequencing | 99 | NA | Various frequency of testing, clinically relevant inhibitors | NA | NA | Yes | − |
| Hill26 | 28 | Patients | No | 2005 | Nottingham, United Kingdom | Single | PCR, CSGE/DHPLC sequencing | NA | NA | NA | NA | NA | Yes | − |
| Ivaskevicius27 | 50 | Patients | Yes | 2001 | Lithuania | Single | Southern blot, DGGE/CMC/DHPLC sequencing | 98 | NA | NA | Nijmegen Bethesda | NA | Yes | − |
| Jayandharan28 | 92 | Patients | Yes | 2005 | Vellore, India | Single | PCR, CSGE sequencing | 93 | Asian | NA | Bethesda | Many < 50ED | Yes | − |
| Liu29,30 | 708 | Patients | No | 2008 | Seattle, US‡ | Single | Southern blot, heteroduplex screening/restriction enzyme cleavage sequencing | 99 | NA | Before mid1980s, tested if clinical suspicion; later more frequently | Bethesda | Almost all > 50 ED | Yes | + |
| Ma31 | 21 | Patients | Yes | 2008 | Changhua, Taiwan | Single | PCR sequencing | 95 | Asian | NA | NA | NA | Yes | − |
| Margaglione32 | 971 | Patients | Yes | 2008 | Italy§ | Multi | PCR, DHPLC/CSGE sequencing | 89 | NA | NA | NA | NA | Yes | − |
| Miller33 | 407 | Patients | NA | 2010 | 12 US centers¶ | Multi | PCR sequencing | 96 | 35 black non-Hispanic, 32 Hispanic, 321 white non-Hispanic | At study entry, annually, at product switch, clinical indication | Nijmegen Bethesda | NA | NA | + |
| Oldenburg34 | 917 | Patients | NA | 2011 | Bonn, Germany | Multi | CCM/DGGE sequencing | 97 | NA | Regularly, various frequencies | Various | Majority > 50 ED | Yes | + |
| Owaidah35 | 20 | Patients | No | 2009 | Saudi Arabia | Single | PCR sequencing | 100 | Arab | At presentation, at least twice annually | Bethesda | Incl < 50 ED | No | − |
| Pieneman36 | 25 | Patients | Yes | 1995 | Leiden, the Netherlands | Single | PCR, SSCP/DSCP sequencing | 84 | NA | NA | NA | NA | NA | − |
| Reitter37 | 155 | Patients | No | 2010 | Austria# | Multi | PCR sequencing | 99 | NA | NA | NA | Incl < 50 ED | Yes | − |
| Repesse38 | 59 | Patients | No | 2007 | Caen, France | Single | PCR sequencing | 100 | NA | NA | Nijmegen Bethesda | NA | No | − |
| Rosetti39 | 88 | Patients | No | 2007 | Buenos Aires, Argentina | Single | Southern blot/PCR, CSGE sequencing | 95 | NA | NA | NA | NA | NA | − |
| Sanna40 | 108 | Patients | Yes | 2008 | Naples, Isernia, Italy | Single | PCR sequencing | 94 | NA | NA | NA | NA | NA | − |
| Sirocova41 | 29 | Patients | Yes | 2009 | Chisinau, Moldovia | Single | PCR, heteroduplex screening sequencing | 100 | NA | NA | Bethesda | Incl < 50 ED | Yes | − |
| Vidal42 | 38 | Patients | Yes | 2001 | Barcelona, Spain | Single | PCR sequencing | 97 | White | NA | NA | NA | NA | − |
| Viel43 | 48 | Patients | No | 2009 | Atlanta, Birmingham, Augusta, Jackson, US | Multi | PCR sequencing | 96 | African American | Annually | Nijmegen Bethesda, ≥ once > 0.6 BU/mL | NA | No | − |
| Xue44 | 111 | Patients | Yes | 2010 | Tianjin, China | Single | PCR sequencing | 97 | Asian | Once measured | Bethesda | Incl < 50 ED | NA | − |
| Source . | N . | Patients or families . | Unrelated patients . | Year . | Country . | Study cohort . | Mutation analyses methods: intron 1 and 22 inversion screening and mutation analysis . | Mutation detection rate, % . | Ethnicity . | Frequency inhibitor screening . | Inhibitor assay . | Treated > 50 ED . | Unselected cohort . | High quality . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahmed14 | 46 | Patients | Yes | 2005 | New Delhi, India | Single | PCR, DHPLC | 92 | NA | NA | Bethesda | NA | NA | − |
| Awidi15 | 117 | Patients | No | 2010 | Jordan | NA | PCR sequencing | NA | NA | NA | Bethesda | Incl < 50 ED | NA | − |
| Boekhorst16 | 664 | Patients | NA | 2008 | Nijmegen, the Netherlands Paris, France Tehran, Iran | Multi | PCR-DGGE/CSGE sequencing | 100 | NA | NA | Bethesda or Nijmegen Bethesda | Incl < 50 ED | No | − |
| Casaña17 | 92 | Families | Yes | 2008 | Valencia, Spain | Single | Southern-blot/PCR sequencing | 100 | NA | NA | Bethesda | NA | NA | − |
| Castaman18 | 27 | Patients | No | 2007 | Albany | Single | PCR, DHPLC sequencing | 100 | NA | 18 patients screened once | NA | Minimally treated | NA | − |
| Chen19 | 88 | Patients | Yes | 2010 | Taipei, Taiwan | Single | PCR, DHPLC sequencing, RNA analysis | 98 | NA | NA | Bethesda | NA | Yes | − |
| David20 | 108 | Patients | Yes | 2006 | Lisboa/Porto, Portugal | Multi | PCR, SSCP/restriction enzyme cleavage/sequencing | 100 | White | NA | Bethesda | NA | NA | − |
| Fernandez-Lopez21 | 55 | Patients | Yes | 2005 | Andalusia, Spain | Single | PCR sequencing | 89 | NA | NA | NA | Na | Yes | − |
| Goodeve22 | 49 | Patients | NA | 2000 | Recombinate Study Group* | Multi | Southern-blot, CSGE sequencing | 93 | 31 white, 7 African American, 4 Hispanic, 1 Asian, 1 other, 1 unknown | At least every 3 months, ≥ 0.6 BU/mL | Bethesda | Incl < 50 ED | No | − |
| Gouw23 | 122 | Patients | No | 2007 | Leuven, Madrid, Stockholm, Malmö, Montreal, Göteborg | Multi | NA | 83 | 151 white, 1 African American, 3 Asian, 7 other, 1 unknown | Various frequency of testing, ≥ 2 positive inhibitor and decreased recovery | Bethesda or Nijmegen Bethesda | All > 50 ED | Yes | + |
| Gouw24 | 318 | Patients | No | 2010 | Utrecht, The Netherlands | Single | Southern blot/PCR sequencing | 95 | NA | ≥ 1/y (1970-1980), ≥ 1/6 m (early 1990s), ≥ 1/3 m (late 1990s onwards) and at clinical indication; ≥ 2 positive inhibitor and decreased recovery | Bethesda or Nijmegen modification | All > 50 ED | Yes | + |
| Green25 | 248 | Patients | No | 2008 | United Kingdom† | Multi | Southern blot/PCR, fluorescent solid-phase CCM/DHPLC sequencing | 99 | NA | Various frequency of testing, clinically relevant inhibitors | NA | NA | Yes | − |
| Hill26 | 28 | Patients | No | 2005 | Nottingham, United Kingdom | Single | PCR, CSGE/DHPLC sequencing | NA | NA | NA | NA | NA | Yes | − |
| Ivaskevicius27 | 50 | Patients | Yes | 2001 | Lithuania | Single | Southern blot, DGGE/CMC/DHPLC sequencing | 98 | NA | NA | Nijmegen Bethesda | NA | Yes | − |
| Jayandharan28 | 92 | Patients | Yes | 2005 | Vellore, India | Single | PCR, CSGE sequencing | 93 | Asian | NA | Bethesda | Many < 50ED | Yes | − |
| Liu29,30 | 708 | Patients | No | 2008 | Seattle, US‡ | Single | Southern blot, heteroduplex screening/restriction enzyme cleavage sequencing | 99 | NA | Before mid1980s, tested if clinical suspicion; later more frequently | Bethesda | Almost all > 50 ED | Yes | + |
| Ma31 | 21 | Patients | Yes | 2008 | Changhua, Taiwan | Single | PCR sequencing | 95 | Asian | NA | NA | NA | Yes | − |
| Margaglione32 | 971 | Patients | Yes | 2008 | Italy§ | Multi | PCR, DHPLC/CSGE sequencing | 89 | NA | NA | NA | NA | Yes | − |
| Miller33 | 407 | Patients | NA | 2010 | 12 US centers¶ | Multi | PCR sequencing | 96 | 35 black non-Hispanic, 32 Hispanic, 321 white non-Hispanic | At study entry, annually, at product switch, clinical indication | Nijmegen Bethesda | NA | NA | + |
| Oldenburg34 | 917 | Patients | NA | 2011 | Bonn, Germany | Multi | CCM/DGGE sequencing | 97 | NA | Regularly, various frequencies | Various | Majority > 50 ED | Yes | + |
| Owaidah35 | 20 | Patients | No | 2009 | Saudi Arabia | Single | PCR sequencing | 100 | Arab | At presentation, at least twice annually | Bethesda | Incl < 50 ED | No | − |
| Pieneman36 | 25 | Patients | Yes | 1995 | Leiden, the Netherlands | Single | PCR, SSCP/DSCP sequencing | 84 | NA | NA | NA | NA | NA | − |
| Reitter37 | 155 | Patients | No | 2010 | Austria# | Multi | PCR sequencing | 99 | NA | NA | NA | Incl < 50 ED | Yes | − |
| Repesse38 | 59 | Patients | No | 2007 | Caen, France | Single | PCR sequencing | 100 | NA | NA | Nijmegen Bethesda | NA | No | − |
| Rosetti39 | 88 | Patients | No | 2007 | Buenos Aires, Argentina | Single | Southern blot/PCR, CSGE sequencing | 95 | NA | NA | NA | NA | NA | − |
| Sanna40 | 108 | Patients | Yes | 2008 | Naples, Isernia, Italy | Single | PCR sequencing | 94 | NA | NA | NA | NA | NA | − |
| Sirocova41 | 29 | Patients | Yes | 2009 | Chisinau, Moldovia | Single | PCR, heteroduplex screening sequencing | 100 | NA | NA | Bethesda | Incl < 50 ED | Yes | − |
| Vidal42 | 38 | Patients | Yes | 2001 | Barcelona, Spain | Single | PCR sequencing | 97 | White | NA | NA | NA | NA | − |
| Viel43 | 48 | Patients | No | 2009 | Atlanta, Birmingham, Augusta, Jackson, US | Multi | PCR sequencing | 96 | African American | Annually | Nijmegen Bethesda, ≥ once > 0.6 BU/mL | NA | No | − |
| Xue44 | 111 | Patients | Yes | 2010 | Tianjin, China | Single | PCR sequencing | 97 | Asian | Once measured | Bethesda | Incl < 50 ED | NA | − |
ED indicates exposure days; NA, not available; DGGE, denaturing gradient gel electrophoresis; CSGE, conformation sensitive gel electrophoresis; DHPLC, denaturing high performance liquid chromatography; SSCP, single-strand conformation polymorphism analysis; CCM, chemical cleavage of mismatch; CMC, chemical mismatch cleavage; and DSCP, double-strand conformation polymorphism.
Recombinate PUP study group: East Lansing, MI; Cleveland, OH; Morgantown, WV; Chapel Hill, NC; San Juan, PR, Washington, DC; Cincinnati, OH; Denver, CO; New York, NY; New Brunswick, NJ; Las Vegas, NV; Wauwatosa, WI; Louisville, KY; Norfolk, VA; Chicago, IL; Los Angeles, CA; Summit, NJ; Lexington, KY; Iowa City, IA; Paris (Bicetre); Paris (Necker); Milan; Vicenza; Copenhagen; and Aarhus.
United Kingdom National Database: Bangor, Basingstoke, Bath, Bournemouth, Bristol, Cambridge, Canterbury, Cardiff, Colchester, Exeter, Gillingham, Glasgow, Leeds, Liverpool, London (Hammersmith, Lewisham, Royal Free, Royal London, and St Georges), Manchester, Norwich, Oxford, Peterborough, Plymouth, Portsmouth, Southampton, Swansea, Taunton, Truro, and Yeovil.
Alaska, Arizona, California, Colorado, Georgia, Hawaii, Idaho, Indiana, Massachusetts, Michigan, Missouri, Montana, New Hampshire, New Mexico, New York, Oregon, Pennsylvania, Texas, Utah, and Vermont.
AICE (Italian Association of Hemophilia Centers) database: Alessandria, Arezzo, Bari, Bologna, Cagliari, Castelfranco Veneto, Catania, Catanzaro, Cesena, Cosenza, Cremona, Ferrara, Faenza, Florence, Genoa, Ivrea, L'Aquila, Latina, Macerata, Milan, Modena, Naples, Orvieto, Padua, Palermo, Parma, Pavia, Perugia, Pescara, Piacenza, Ravenna, Reggio Calabria, Reggio Emilia, Rome, Sassari, Torino, Turin, Trento, Udine, Vallo della Lucania, Verona, and Vicenza.
Atlanta, GA; Ann Arbor, MI; Worcester, MA; Iowa City, IA; Richmond, VA; Nashville, TN; Indianapolis, IN; Peoria, IL; Kansas City, MO; Aurora, CO; and Phoenix, AZ.
Bregenz, Dornbirn, Graz, Innsbruck, Klagenfurt, Linz, Salzburg, St Poelten, and Vienna.
Participants.
The included studies involved 5383 patients with severe hemophilia A, including 1029 patients with inhibitor development. In 23 studies, data on high-titer-inhibitor development were available, comprising 3686 patients and 510 inhibitor patients. In 4 studies, all patients were at a low risk of subsequent inhibitor development because all patients had received FVIII on at least 50 exposure days.
Types of outcomes.
The inhibitor assays reported were the original Bethesda assay and the Bethesda assay with the Nijmegen modification.45,46 The frequency of inhibitor screening varied markedly between studies. Some studies screened for inhibitors only once, and others tested regularly for the presence of inhibitors.
Results of individual studies
The numbers and proportions of inhibitor development according to the subgroups of F8 genotype and in the total study population are presented in Table 2. Table 3 presents the numbers and proportions of patients with high-responder inhibitor development. The total cumulative incidence of inhibitor development in the individual studies varied from 0%-40%. The total cumulative incidence of high-titer-inhibitor development varied from 0%-26%. No inhibitors were detected in 3 studies with study population sizes of 21, 25, and 27 patients.18,31,36
Summary of studies evaluating the incidence of inhibitor development and inhibitor development according to genotype in patients with severe hemophilia A
| Source . | Large deletions . | Single exon . | Multiple exon . | Nonsense mutations . | Light chain . | Non-light chain . | Intron 1/22 inversions . | Intron 22 inversion . | Intron 1 inversion . | Small deletions/insertions . | in poly-A-runs . | outside poly-A- runs . | Missense mutations . | Light chain . | Non-light chain . | Splice site . | Conserved splice site . | Non-conserved splice site . | Unknown mutation . | Total . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahmed14 | 0/1 (0) | 0/1 (0) | 0/0 (−) | 1/6 (17) | 0/3 (0) | 1/3 (33) | 2/24 (8) | 2/22 (9) | 0/2 (0) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 0/4 (0) | 0/1 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0%) | 0/0 (−) | 0/4 (0) | 3/46 (7) |
| Awidi15 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 10/67 (15) | 10/66 (15) | 0/1 (0) | 1/14 (7) | 1/1 (100) | 0/13 (0) | 6/36 (17) | 6/21 (29) | 0/15 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 17/117 (15) |
| Boekhorst16 | 5/23 (22) | 2/17 (19) | 3/6 (50) | 22/90 (24) | 10/33 (30) | 12/57 (21) | 65/276 (24) | 61/260 (23) | 4/16 (25) | 11/130 (8) | 2/68 (3) | 9/62 (15) | 8/120 (7) | 2/56 (4) | 6/64 (9) | 11/25 (44) | NA | NA | NA | 122/664 (18) |
| Casaña17 | 6/6 (100) | 1/1 (100) | 5/5 (100) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 15/55 (27) | 14/52 (27) | 1/3 (33) | 3/17 (18) | 1/6 (17) | 2/11 (18) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 1/3 (33) | 1/3 (33) | 0/0 (−) | 0/0 (−) | 25/92 (27) |
| Castaman18 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/5 (0) | 0/1(0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/0 (NA) | 0/7 (0) | 0/4 (0) | 0/3 (0) | 0/12 (0) | 0/4 (0) | 0/8 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 0/27 (0) |
| Chen19 | 4/7(57) | 1/2 (50) | 3/5 (60) | 1/13 (8) | 0/9 (0) | 1/4 (25) | 6/40 (15) | 4/34 (12) | 2/6 (33) | 1/15 (7) | 0/0 (−) | 1/15 (7) | 0/7 (0) | 0/3 (0) | 0/4 (0) | 0/4 (0) | 0/3 (0) | 0/0 (−) | 0/2 (0) | 12/88 (14) |
| David20 | 0/1 (0) | 0/1 (0) | 0/0 (−) | 4/6 (67) | 3/3 (100) | 1/3 (33) | 13/76 (17) | 12/73 (16) | 1/3 (33) | 3/18 (17) | 1/9 (11) | 2/9 (22) | 1/6 (17) | 1/3 (33) | 0/3 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 21/108 (19) |
| Fernandez-Lopez21 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 3/11 (27) | 3/9 (33) | 0/2 (0) | 1/23 (4) | 1/20 (5) | 0/3 (0) | 1/8 (0) | 0/3 (0) | 1/5 (20) | 0/6 (0) | 0/4 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/6 (0) | 5/55 (9) |
| Goodeve22 * | 2/2 (100) | NA | NA | 0/2 (0) | 0/2 (−) | 0/0 (−) | 8/23 (35) | 8/23 (35) | NA | 0/10 (0) | 0/5 (0) | 0/5 (0) | 3/7 (43) | 2/4 (50) | 1/3 (33) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 1/4 (25) | 14/49 (29) |
| Gouw23 † | 2/7 (29) | 0/5 (0) | 1/1 (100) | 7/16 (44) | 1/3 (33) | 2/6 (33) | 21/76 (28) | 19/73 (26) | 1/1 (100) | 4/19 (21) | 0/0 (−) | 0/5 (0) | 1/12 (8) | 0/3 (0) | 0/4 (0) | 0/1 (0) | NA | NA | 3/32 (9) | 38/163 (23) |
| Gouw24 | 2/3 (67) | 1/2 (50) | 1/1 (100) | 6/20 (30) | 4/9 (44) | 2/11 (18) | 27/177 (15) | 27/175 (15) | 0/2 (0) | 4/54 (7) | 3/28 (11) | 1/26 (4) | 3/51 (6) | 3/38 (8) | 0/13 (0) | 1/13 (8) | 1/10 (10) | 0/3 (0) | 0/16 (0) | 43/334 (13) |
| Green25 | 2/7 (29) | 0/2 (0) | 2/5 (40) | 8/29 (28) | 6/11 (55) | 2/18 (11) | 19/119 (16) | 18/108 (17) | 1/11 (9) | 4/41 (10) | 0/21 (0) | 4/20 (20) | 4/41 (10) | 3/20 (15) | 1/21 (5) | 0/9 (0) | 0/6 (0) | 0/3 (0) | 0/3 | 37/249 (15) |
| Hill26 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 4/6 (67) | 2/4 (50) | 2/2 (100) | 4/11 (36) | 3/8 (38) | 1/3 (33) | 2/6 (33) | 1/3 (33) | 1/3 (33) | 0/5 (0) | 0/5 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | NA | 10/28 (36) |
| Ivaskevicius27 | 2/4 (50) | 0/1 (0) | 2/3 (67) | 0/6 (0) | 0/0 (−) | 0/6 (0) | 1/24 (4) | 1/24 (4) | 0/0 (−) | 0/8 (0) | 0/4 (0) | 0/4 (0) | 0/6 (0) | 0/4 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 3/50 (6) |
| Jayandharan28 | 1/4 (25) | 0/1 (0) | 1/3 (33) | 1/9 (11) | 0/4 (0) | 1/5 (25) | 4/53 (8) | 4/51 (8) | 0/2 (0) | 0/10 (10) | 0/3 (0) | 0/7 (0) | 0/6 (0) | 0/3 (0) | 0/4 (0) | 0/3 (0) | 0/1 (0) | 0/2 (0) | 1/6 (20) | 7/91 (8) |
| Liu29,30 | 6/9 (67) | 0/3 (0) | 6/6 (100) | 9/28 (32) | 8/13(62) | 1/15 (7) | 25/110 (23) | 24/106 (23) | 1/4 (25) | 7/44 (16) | 1/13 (8) | 6/31 (19) | 5/16 (31) | 3/8 (38) | 2/8 (25) | 3/15 (20) | NA | NA | 0/3 (0) | 55/225 (24) |
| Ma31 | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/13 (0) | 0/12 (0) | 0/1 (0) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/1 (0) | 0/21 (0) |
| Margaglione32 | 8/13 (62) | 1/2 (50) | 7/11 (64) | 27/61 (44) | 11/27 (41) | 16/34 (47) | 119/470 (25) | 113/451 (25) | 6/19 (32) | 22/146 (15) | 4/54 (7) | 18/92 (20) | 8/142 (6) | 5/84 (6) | 3/58 (5) | 7/31 (23) | NA | NA | 15/108 (14) | 206/971 (21) |
| Miller33 | 12/21 (57) | 3/9 (33) | 9/12 (75) | 13/51 (25) | 3/14 (21) | 10/37 (27) | 46/177 (26) | 45/168 (27) | 1/9 (11) | 8/65 (12) | 2/25 (8) | 6/40 (15) | 6/63 (10) | 5/36 (14) | 1/27 (4) | 5/14 (36) | NA | NA | 4/16 (25) | 94/407 (23) |
| Oldenburg34 | 23/45 (51) | 14/33 (42) | 9/12 (75) | 33/123 (27) | 17/49 (35) | 19/85 (22) | 100/408 (25) | 95/387 (25) | 5/21 (24) | 27/151 (18) | 4/37 (11) | 23/114 (20) | 12/130 (9) | 4/55 (7) | 8/75 (11) | 7/30 (23) | 5/16 (31) | 2/14 (14) | 1/25 (4) | 203/912 (22) |
| Owaidah35 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (NA) | 2/10 (20) | 2/10 (20) | 0/0 (−) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/9 (0) | 0/1 (0) | 0/8 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 2/20 (10) |
| Pieneman36 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (NA) | 0/10 (0) | 0/10 (0) | 0/0 (−) | 0/11 (0) | 0/4 (0) | 0/5 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/4 (0) | 0/25 (0) |
| Reitter37 | 1/2 (50) | 0/0 (−) | 1/2 (50) | 1/13 (8) | 1/6 (17) | 0/7 (0) | 14/77 (18) | 13/73 (18) | 1/4 (25) | 5/17 (29) | 1/4 (25) | 4/13 (31) | 3/34 (9) | 2/26 (8) | 1/8 (13) | 1/11 (9) | NA | NA | 0/1 (0) | 25/155 (16) |
| Repesse38 | 1/1 (100) | 1/1 (100) | 0/0 (−) | 3/11 (27) | 1/2 (50) | 2/9 (22) | 7/25 (28) | 7/25 (28) | 0/0 (−) | 0/9 (0) | 0/0 (−) | 0/9 (0) | 0/9 (0) | 0/1 (0) | 0/8 (0) | 1/4 (25) | 1/3 (33) | 0/1 (0) | 0/0 (−) | 12/59 (20) |
| Rosetti39 | 7/11 (64) | 1/4 (25) | 6/7 (86) | 3/9 (33) | 1/3 (33) | 2/6 (33) | 12/40 (30) | 12/39 (31) | 0/1 (0) | 5/15 (33) | 1/2 (50) | 4/13 (31) | 0/8 (0) | 0/6 (0) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/4 (0) | 27/88 (30) |
| Sanna40 | 1/1 (100) | 0/0 (−) | 1/1 (100) | 3/11 (27) | NA | NA | 9/67 (13) | 9/64 (14) | 0/3 (0) | 0/11 (0) | NA | NA | 0/9 (0) | NA | NA | 1/3 (33) | NA | NA | 0/6 (0) | 14/108 (13) |
| Sirocova41 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 2/14 (14) | 2/13 (15) | 0/1 (0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 3/29 (10) |
| Vidal42 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 2/3 (67) | 0/1 (0) | 2/2 (100) | 4/13 (31) | 4/13 (31) | 0/0 (−) | 2/6 (33) | 0/1 (0) | 2/5 (40) | 1/11 (9) | 1/8 (13) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 10/38 (26) |
| Viel43 | 5/7 (71) | 3/5 (60) | 2/2 (100) | 2/5 (40) | 0/1 (0) | 2/4 (50) | 11/17 (65) | 10/13 (77) | 1/4 (25) | 0/5 (0) | 0/3 (0) | 0/2 (0) | 1/12 (8) | 0/1 (0) | 1/11 (9) | 0/0 (0) | 0/0 (−) | 0/0 (−) | 0/2 | 19/48 (40) |
| Xue44 | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 2/60 (3) | 2/57 (4) | 0/3 (0) | 0/24 (0) | 0/11 (0) | 0/13 (0) | 0/12 (0) | 0/3 (0) | 0/9 (0) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/3 (0) | 2/111 (2) |
| Source . | Large deletions . | Single exon . | Multiple exon . | Nonsense mutations . | Light chain . | Non-light chain . | Intron 1/22 inversions . | Intron 22 inversion . | Intron 1 inversion . | Small deletions/insertions . | in poly-A-runs . | outside poly-A- runs . | Missense mutations . | Light chain . | Non-light chain . | Splice site . | Conserved splice site . | Non-conserved splice site . | Unknown mutation . | Total . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahmed14 | 0/1 (0) | 0/1 (0) | 0/0 (−) | 1/6 (17) | 0/3 (0) | 1/3 (33) | 2/24 (8) | 2/22 (9) | 0/2 (0) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 0/4 (0) | 0/1 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0%) | 0/0 (−) | 0/4 (0) | 3/46 (7) |
| Awidi15 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 10/67 (15) | 10/66 (15) | 0/1 (0) | 1/14 (7) | 1/1 (100) | 0/13 (0) | 6/36 (17) | 6/21 (29) | 0/15 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 17/117 (15) |
| Boekhorst16 | 5/23 (22) | 2/17 (19) | 3/6 (50) | 22/90 (24) | 10/33 (30) | 12/57 (21) | 65/276 (24) | 61/260 (23) | 4/16 (25) | 11/130 (8) | 2/68 (3) | 9/62 (15) | 8/120 (7) | 2/56 (4) | 6/64 (9) | 11/25 (44) | NA | NA | NA | 122/664 (18) |
| Casaña17 | 6/6 (100) | 1/1 (100) | 5/5 (100) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 15/55 (27) | 14/52 (27) | 1/3 (33) | 3/17 (18) | 1/6 (17) | 2/11 (18) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 1/3 (33) | 1/3 (33) | 0/0 (−) | 0/0 (−) | 25/92 (27) |
| Castaman18 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/5 (0) | 0/1(0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/0 (NA) | 0/7 (0) | 0/4 (0) | 0/3 (0) | 0/12 (0) | 0/4 (0) | 0/8 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 0/27 (0) |
| Chen19 | 4/7(57) | 1/2 (50) | 3/5 (60) | 1/13 (8) | 0/9 (0) | 1/4 (25) | 6/40 (15) | 4/34 (12) | 2/6 (33) | 1/15 (7) | 0/0 (−) | 1/15 (7) | 0/7 (0) | 0/3 (0) | 0/4 (0) | 0/4 (0) | 0/3 (0) | 0/0 (−) | 0/2 (0) | 12/88 (14) |
| David20 | 0/1 (0) | 0/1 (0) | 0/0 (−) | 4/6 (67) | 3/3 (100) | 1/3 (33) | 13/76 (17) | 12/73 (16) | 1/3 (33) | 3/18 (17) | 1/9 (11) | 2/9 (22) | 1/6 (17) | 1/3 (33) | 0/3 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 21/108 (19) |
| Fernandez-Lopez21 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 3/11 (27) | 3/9 (33) | 0/2 (0) | 1/23 (4) | 1/20 (5) | 0/3 (0) | 1/8 (0) | 0/3 (0) | 1/5 (20) | 0/6 (0) | 0/4 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/6 (0) | 5/55 (9) |
| Goodeve22 * | 2/2 (100) | NA | NA | 0/2 (0) | 0/2 (−) | 0/0 (−) | 8/23 (35) | 8/23 (35) | NA | 0/10 (0) | 0/5 (0) | 0/5 (0) | 3/7 (43) | 2/4 (50) | 1/3 (33) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 1/4 (25) | 14/49 (29) |
| Gouw23 † | 2/7 (29) | 0/5 (0) | 1/1 (100) | 7/16 (44) | 1/3 (33) | 2/6 (33) | 21/76 (28) | 19/73 (26) | 1/1 (100) | 4/19 (21) | 0/0 (−) | 0/5 (0) | 1/12 (8) | 0/3 (0) | 0/4 (0) | 0/1 (0) | NA | NA | 3/32 (9) | 38/163 (23) |
| Gouw24 | 2/3 (67) | 1/2 (50) | 1/1 (100) | 6/20 (30) | 4/9 (44) | 2/11 (18) | 27/177 (15) | 27/175 (15) | 0/2 (0) | 4/54 (7) | 3/28 (11) | 1/26 (4) | 3/51 (6) | 3/38 (8) | 0/13 (0) | 1/13 (8) | 1/10 (10) | 0/3 (0) | 0/16 (0) | 43/334 (13) |
| Green25 | 2/7 (29) | 0/2 (0) | 2/5 (40) | 8/29 (28) | 6/11 (55) | 2/18 (11) | 19/119 (16) | 18/108 (17) | 1/11 (9) | 4/41 (10) | 0/21 (0) | 4/20 (20) | 4/41 (10) | 3/20 (15) | 1/21 (5) | 0/9 (0) | 0/6 (0) | 0/3 (0) | 0/3 | 37/249 (15) |
| Hill26 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 4/6 (67) | 2/4 (50) | 2/2 (100) | 4/11 (36) | 3/8 (38) | 1/3 (33) | 2/6 (33) | 1/3 (33) | 1/3 (33) | 0/5 (0) | 0/5 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | NA | 10/28 (36) |
| Ivaskevicius27 | 2/4 (50) | 0/1 (0) | 2/3 (67) | 0/6 (0) | 0/0 (−) | 0/6 (0) | 1/24 (4) | 1/24 (4) | 0/0 (−) | 0/8 (0) | 0/4 (0) | 0/4 (0) | 0/6 (0) | 0/4 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 3/50 (6) |
| Jayandharan28 | 1/4 (25) | 0/1 (0) | 1/3 (33) | 1/9 (11) | 0/4 (0) | 1/5 (25) | 4/53 (8) | 4/51 (8) | 0/2 (0) | 0/10 (10) | 0/3 (0) | 0/7 (0) | 0/6 (0) | 0/3 (0) | 0/4 (0) | 0/3 (0) | 0/1 (0) | 0/2 (0) | 1/6 (20) | 7/91 (8) |
| Liu29,30 | 6/9 (67) | 0/3 (0) | 6/6 (100) | 9/28 (32) | 8/13(62) | 1/15 (7) | 25/110 (23) | 24/106 (23) | 1/4 (25) | 7/44 (16) | 1/13 (8) | 6/31 (19) | 5/16 (31) | 3/8 (38) | 2/8 (25) | 3/15 (20) | NA | NA | 0/3 (0) | 55/225 (24) |
| Ma31 | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/13 (0) | 0/12 (0) | 0/1 (0) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/1 (0) | 0/21 (0) |
| Margaglione32 | 8/13 (62) | 1/2 (50) | 7/11 (64) | 27/61 (44) | 11/27 (41) | 16/34 (47) | 119/470 (25) | 113/451 (25) | 6/19 (32) | 22/146 (15) | 4/54 (7) | 18/92 (20) | 8/142 (6) | 5/84 (6) | 3/58 (5) | 7/31 (23) | NA | NA | 15/108 (14) | 206/971 (21) |
| Miller33 | 12/21 (57) | 3/9 (33) | 9/12 (75) | 13/51 (25) | 3/14 (21) | 10/37 (27) | 46/177 (26) | 45/168 (27) | 1/9 (11) | 8/65 (12) | 2/25 (8) | 6/40 (15) | 6/63 (10) | 5/36 (14) | 1/27 (4) | 5/14 (36) | NA | NA | 4/16 (25) | 94/407 (23) |
| Oldenburg34 | 23/45 (51) | 14/33 (42) | 9/12 (75) | 33/123 (27) | 17/49 (35) | 19/85 (22) | 100/408 (25) | 95/387 (25) | 5/21 (24) | 27/151 (18) | 4/37 (11) | 23/114 (20) | 12/130 (9) | 4/55 (7) | 8/75 (11) | 7/30 (23) | 5/16 (31) | 2/14 (14) | 1/25 (4) | 203/912 (22) |
| Owaidah35 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (NA) | 2/10 (20) | 2/10 (20) | 0/0 (−) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/9 (0) | 0/1 (0) | 0/8 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 2/20 (10) |
| Pieneman36 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (NA) | 0/10 (0) | 0/10 (0) | 0/0 (−) | 0/11 (0) | 0/4 (0) | 0/5 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/4 (0) | 0/25 (0) |
| Reitter37 | 1/2 (50) | 0/0 (−) | 1/2 (50) | 1/13 (8) | 1/6 (17) | 0/7 (0) | 14/77 (18) | 13/73 (18) | 1/4 (25) | 5/17 (29) | 1/4 (25) | 4/13 (31) | 3/34 (9) | 2/26 (8) | 1/8 (13) | 1/11 (9) | NA | NA | 0/1 (0) | 25/155 (16) |
| Repesse38 | 1/1 (100) | 1/1 (100) | 0/0 (−) | 3/11 (27) | 1/2 (50) | 2/9 (22) | 7/25 (28) | 7/25 (28) | 0/0 (−) | 0/9 (0) | 0/0 (−) | 0/9 (0) | 0/9 (0) | 0/1 (0) | 0/8 (0) | 1/4 (25) | 1/3 (33) | 0/1 (0) | 0/0 (−) | 12/59 (20) |
| Rosetti39 | 7/11 (64) | 1/4 (25) | 6/7 (86) | 3/9 (33) | 1/3 (33) | 2/6 (33) | 12/40 (30) | 12/39 (31) | 0/1 (0) | 5/15 (33) | 1/2 (50) | 4/13 (31) | 0/8 (0) | 0/6 (0) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/4 (0) | 27/88 (30) |
| Sanna40 | 1/1 (100) | 0/0 (−) | 1/1 (100) | 3/11 (27) | NA | NA | 9/67 (13) | 9/64 (14) | 0/3 (0) | 0/11 (0) | NA | NA | 0/9 (0) | NA | NA | 1/3 (33) | NA | NA | 0/6 (0) | 14/108 (13) |
| Sirocova41 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 2/14 (14) | 2/13 (15) | 0/1 (0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 3/29 (10) |
| Vidal42 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 2/3 (67) | 0/1 (0) | 2/2 (100) | 4/13 (31) | 4/13 (31) | 0/0 (−) | 2/6 (33) | 0/1 (0) | 2/5 (40) | 1/11 (9) | 1/8 (13) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 10/38 (26) |
| Viel43 | 5/7 (71) | 3/5 (60) | 2/2 (100) | 2/5 (40) | 0/1 (0) | 2/4 (50) | 11/17 (65) | 10/13 (77) | 1/4 (25) | 0/5 (0) | 0/3 (0) | 0/2 (0) | 1/12 (8) | 0/1 (0) | 1/11 (9) | 0/0 (0) | 0/0 (−) | 0/0 (−) | 0/2 | 19/48 (40) |
| Xue44 | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 2/60 (3) | 2/57 (4) | 0/3 (0) | 0/24 (0) | 0/11 (0) | 0/13 (0) | 0/12 (0) | 0/3 (0) | 0/9 (0) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/3 (0) | 2/111 (2) |
NA indicates not available.
Numbers of patients with inhibitors and total numbers of patients with the specific mutations are reported, with the percentages in parentheses.
Goodeve: 6 patients excluded: 1 patient from Milan and 2 patients from Vincenza (overlap with Margaglione et al32 ), 2 patients from Paris, Bicetre (overlap with Boekhorst et al16 ), and 1 patient from Iowa (overlap with Miller et al33 ).
Restricted to patients with factor VIII:C < 0.01 IU/mL; 169 patients excluded: 41 patients from Milan (overlap Margaglione et al32 ), 35 patients from Utrecht (overlap Gouw et al24 ), 17 patients from Valencia (overlap Casaña et al17 ), 28 patients from London (Royal Free Hospital), and 31 patients from London (Great Ormond Street Hospital; overlap with Green et al25 ).
Summary of studies evaluating the incidence of high titer inhibitor development according to genotype in patients with severe hemophilia A
| Source . | Large deletions . | Single exon . | Multiple exon . | Nonsense mutations . | Light chain . | Non-light chain . | Intron 1/22 inversions . | Intron 22 inversion . | Intron 1 inversion . | Small deletions/insertions . | In poly-A-runs . | Outside poly-A- runs . | Missense mutations . | Light chain . | Non-light chain . | Splice site . | Conserved splice site . | Non-conserved splice site . | Unknown mutation . | Total . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahmed14 | 0/1 (0) | 0/1 (0) | 0/0 (−) | 1/6 (17) | 0/3 (0) | 1/3 (33) | 2/24 (8) | 2/22 (9,1) | 0/2 (0) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 0/4 (0) | 0/1 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/4 (0) | 3/46 (7) |
| Awidi15 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 6/67 (9) | 6/66 (9) | 0/1 (0) | 1/14 (7) | 1/1 (100) | 0/13 (0) | 5/36 (14) | 5/21 (24) | 0/15 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 12/117 (10) |
| Boekhorst16 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Na | NA | NA |
| Casaña17 | 6/6 (100) | 1/1 (100) | 5/5 (100) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 10/55 (18) | 10/52 (19) | 0/3 (0) | 3/17 (18) | 1/6 (17) | 2/11 (18) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/0 (−) | 19/92 (21) |
| Castaman18 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/5 (0) | 0/1(0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/0 (−) | 0/7 (0) | 0/4 (0) | 0/3 (0) | 0/12 (0) | 0/4 (0) | 0/8 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 0/27 (0) |
| Chen19 | 3/7 (43) | 0/2 (0) | 3/5 (60) | 1/13 (8) | 0/9 (0) | 1/4 (25) | 5/40 (13) | 4/34 (12) | 1/6 (17) | 0/15 (0) | 0/0 (−) | 0/15 (0) | 0/7 (0) | 0/3 (0) | 0/4 (0) | 0/4 (0) | 0/3 (0) | 0/0 (−) | 0/2 (0) | 9/88 (10) |
| David20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Fernandez-Lopez21 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Goodeve22 | 1/2 (50) | NA | NA | 0/2 (0) | 0/2 (0) | 0/0 (−) | 4/23 (17) | 4/23 (17) | NA | 0/10 (0) | 0/5 (0) | 0/5 (0) | 0/7 (0) | 0/4 (0) | 0/3 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/4 (0) | 5/49 (10) |
| Gouw Canal23 | 2/7 (29) | 0/5 (0) | 1/1 (100) | 6/16 (38) | 1/3 (33) | 1/6 (17) | 18/76 (24) | 16/73 (22) | 1/1 (100) | 2/19 (11) | 0/0 (−) | 0/5 (0) | 1/12 (8) | 0/3 (0) | 0/4 (0) | 0/1 (0) | NA | NA | 3/32 (9) | 32/163 (20) |
| Gouw VCK24 | 2/3 (67) | 1/2 (50) | 1/1 (100) | 5/20 (25) | 3/9 (33) | 2/11 (18) | 17/177 (10) | 17/175 (10) | 0/2 (0) | 4/54 (7) | 3/28 (11) | 1/26 (4) | 2/51 (4) | 2/38 (5) | 0/13 (0) | 0/13 (0) | 0/10 (0) | 0/3 (0) | 0/16 (0) | 30/334 (9) |
| Green25 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Hill26 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Ivaskevicius27 | 2/4 (50) | 0/1 (0) | 2/3 (67) | 0/6 (0) | 0/0 (−) | 0/6 (0) | 1/24 (4,2) | 1/24 (4) | 0/0 (−) | 0/8 (0) | 0/4 (0) | 0/4 (0) | 0/6 (0) | 0/4 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 3/50 (6) |
| Jayandharan28 | 1/4 (25) | 0/1 (0) | 1/3 (33) | 1/9 (11) | 0/4 (0) | 1/5 (20) | 3/53 (6) | 3/51 (6) | 0/2 (0) | 0/10 (10) | 0/3 (0) | 0/7 (0) | 0/6 (0) | 0/3 (0) | 0/4 (0) | 0/3 (0) | 0/2 (0) | 0/1 (0) | NA | 5/91 (5) |
| Liu29,30 | 6/9 (67) | 0/3 (0) | 6/6 (100) | 4/28 (14) | 4/13 (31) | 0/15 (0) | 15/110 (14) | 15/106 (14) | 0/4 (0) | 7/44 (16) | 1/13 (8) | 6/31 (19) | 2/16 (13) | 1/8 (13) | 1/8 (13) | 2/15 (13) | NA | NA | 0/3 (0) | 36/225 (16) |
| Ma31 | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/13 (0) | 0/12 (0) | 0/1 (0) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/0 | 0/0 (−) | 0/0 (−) | 0/1 (0) | 0/21 (0) |
| Margaglione32 | 8/13 (62) | 1/2 (50) | 7/11 (64) | 25/61 (41) | 9/27 (33) | 16/34 (47) | 100/470 (21) | 95/451 (21) | 5/19 (26) | 19/146 (13) | 4/54 (7) | 15/92 (16) | 8/142 (5) | 5/84 (6) | 3/58 (5) | 7/31 (23) | NA | NA | 14/108 (13) | 181/971 (19) |
| Miller33 | 11/21 (52) | 3/9 (33) | 8/12 (67) | 9/51 (18) | 2/14 (14) | 7/37 (19) | 26/173 (15) | 25/164 (15) | 1/9 (11) | 4/65 (6) | 1/25 (4) | 3/40 (8) | 3/62 (5) | 3/36 (8) | NA | 3/14 (21) | NA | NA | 2/16 (13) | 58/402(14) |
| Oldenburg34 | 6/18 (33) | 3/14 (21) | 3/4 (75) | 10/70 (14) | 8/32 (25) | 2/38 (5) | 35/236 (15) | 34/228 (15) | 1/8 (13) | 4/79 (5) | 0/18 (0) | 4/61 (7) | 3/67 (0) | 0/34 (0) | 0/33 (0) | 2/19 (11) | 1/9 (11) | 1/10 (10) | 0/16 (0) | 57/505 (11) |
| Owaidah35 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 2/10 (20) | 2/10 (20) | 0/0 (−) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/9 (0) | 0/1 (0) | 0/8 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 2/20 (10) |
| Pieneman36 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/10 (0) | 0/10 (0) | 0/0 (−) | 0/11 (0) | 0/4 (0) | 0/5 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/4 (0) | 0/25 (0) |
| Reitter37 | 1/2 (50) | 0/0 (−) | 1/2 (50) | 1/13 (8) | 1/6 (17) | 0/7 (0) | 12/77 (16) | 11/73 (15) | 1/4 (25) | 5/17 (29) | 1/4 (25) | 4/13 (31) | 2/34 (6) | 1/26 (4) | 1/8 (13) | 1/11 (9) | NA | NA | 0/1 (0) | 22/155 (14) |
| Repesse38 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Rosetti39 | 5/11 (45) | 0/4 (0) | 5/7 (71) | 1/9 (11) | 1/3 (33) | 0/6 (0) | 5/40 (13) | 5/39 (13) | 0/1 (0) | 2/15 (13) | 0/2 (0) | 2/13 (15) | 0/8 (0) | 0/6 (0) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/4 (0) | 13/88 (15) |
| Sanna40 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Sirocova41 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 1/14 (7) | 1/13 (8) | 0/1 (0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 2/29 (7) |
| Vidal42 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 2/3 (67) | 0/1 (0) | 2/2 (100) | 4/13 (31) | 4/13 (31) | 0/0 (−) | 2/6 (33) | 0/1 (0) | 2/5 (40) | 1/11 (9) | 1/8 (13) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 10/38 (26) |
| Viel43 | 1/3 (33) | 1/3 (33) | 0/0 (−) | 0/4 (0) | 0/1 (0) | 0/3 (0) | 7/14 (50) | 6/10 (60) | 1/4 (25) | 0/5 (0) | 0/3 (0) | 0/2 (0) | 1/12 (8) | 0/1 (0) | 1/11 (9) | 0/0 (0) | 0/0 (−) | 0/0 (−) | 0/2 (0) | 9/40 (23) |
| Xue44 | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 2/60 (3) | 2/57 (4) | 0/3 (0) | 0/24 (0) | 0/11 (0) | 0/13 (0) | 0/12 (0) | 0/3 (0) | 0/9 (0)) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/3 (0) | 2/111 (2) |
| Source . | Large deletions . | Single exon . | Multiple exon . | Nonsense mutations . | Light chain . | Non-light chain . | Intron 1/22 inversions . | Intron 22 inversion . | Intron 1 inversion . | Small deletions/insertions . | In poly-A-runs . | Outside poly-A- runs . | Missense mutations . | Light chain . | Non-light chain . | Splice site . | Conserved splice site . | Non-conserved splice site . | Unknown mutation . | Total . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahmed14 | 0/1 (0) | 0/1 (0) | 0/0 (−) | 1/6 (17) | 0/3 (0) | 1/3 (33) | 2/24 (8) | 2/22 (9,1) | 0/2 (0) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 0/4 (0) | 0/1 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/4 (0) | 3/46 (7) |
| Awidi15 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 6/67 (9) | 6/66 (9) | 0/1 (0) | 1/14 (7) | 1/1 (100) | 0/13 (0) | 5/36 (14) | 5/21 (24) | 0/15 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 12/117 (10) |
| Boekhorst16 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Na | NA | NA |
| Casaña17 | 6/6 (100) | 1/1 (100) | 5/5 (100) | 0/8 (0) | 0/3 (0) | 0/5 (0) | 10/55 (18) | 10/52 (19) | 0/3 (0) | 3/17 (18) | 1/6 (17) | 2/11 (18) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/0 (−) | 19/92 (21) |
| Castaman18 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/5 (0) | 0/1(0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/0 (−) | 0/7 (0) | 0/4 (0) | 0/3 (0) | 0/12 (0) | 0/4 (0) | 0/8 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 0/27 (0) |
| Chen19 | 3/7 (43) | 0/2 (0) | 3/5 (60) | 1/13 (8) | 0/9 (0) | 1/4 (25) | 5/40 (13) | 4/34 (12) | 1/6 (17) | 0/15 (0) | 0/0 (−) | 0/15 (0) | 0/7 (0) | 0/3 (0) | 0/4 (0) | 0/4 (0) | 0/3 (0) | 0/0 (−) | 0/2 (0) | 9/88 (10) |
| David20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Fernandez-Lopez21 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Goodeve22 | 1/2 (50) | NA | NA | 0/2 (0) | 0/2 (0) | 0/0 (−) | 4/23 (17) | 4/23 (17) | NA | 0/10 (0) | 0/5 (0) | 0/5 (0) | 0/7 (0) | 0/4 (0) | 0/3 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/4 (0) | 5/49 (10) |
| Gouw Canal23 | 2/7 (29) | 0/5 (0) | 1/1 (100) | 6/16 (38) | 1/3 (33) | 1/6 (17) | 18/76 (24) | 16/73 (22) | 1/1 (100) | 2/19 (11) | 0/0 (−) | 0/5 (0) | 1/12 (8) | 0/3 (0) | 0/4 (0) | 0/1 (0) | NA | NA | 3/32 (9) | 32/163 (20) |
| Gouw VCK24 | 2/3 (67) | 1/2 (50) | 1/1 (100) | 5/20 (25) | 3/9 (33) | 2/11 (18) | 17/177 (10) | 17/175 (10) | 0/2 (0) | 4/54 (7) | 3/28 (11) | 1/26 (4) | 2/51 (4) | 2/38 (5) | 0/13 (0) | 0/13 (0) | 0/10 (0) | 0/3 (0) | 0/16 (0) | 30/334 (9) |
| Green25 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Hill26 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Ivaskevicius27 | 2/4 (50) | 0/1 (0) | 2/3 (67) | 0/6 (0) | 0/0 (−) | 0/6 (0) | 1/24 (4,2) | 1/24 (4) | 0/0 (−) | 0/8 (0) | 0/4 (0) | 0/4 (0) | 0/6 (0) | 0/4 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 3/50 (6) |
| Jayandharan28 | 1/4 (25) | 0/1 (0) | 1/3 (33) | 1/9 (11) | 0/4 (0) | 1/5 (20) | 3/53 (6) | 3/51 (6) | 0/2 (0) | 0/10 (10) | 0/3 (0) | 0/7 (0) | 0/6 (0) | 0/3 (0) | 0/4 (0) | 0/3 (0) | 0/2 (0) | 0/1 (0) | NA | 5/91 (5) |
| Liu29,30 | 6/9 (67) | 0/3 (0) | 6/6 (100) | 4/28 (14) | 4/13 (31) | 0/15 (0) | 15/110 (14) | 15/106 (14) | 0/4 (0) | 7/44 (16) | 1/13 (8) | 6/31 (19) | 2/16 (13) | 1/8 (13) | 1/8 (13) | 2/15 (13) | NA | NA | 0/3 (0) | 36/225 (16) |
| Ma31 | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/13 (0) | 0/12 (0) | 0/1 (0) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/0 | 0/0 (−) | 0/0 (−) | 0/1 (0) | 0/21 (0) |
| Margaglione32 | 8/13 (62) | 1/2 (50) | 7/11 (64) | 25/61 (41) | 9/27 (33) | 16/34 (47) | 100/470 (21) | 95/451 (21) | 5/19 (26) | 19/146 (13) | 4/54 (7) | 15/92 (16) | 8/142 (5) | 5/84 (6) | 3/58 (5) | 7/31 (23) | NA | NA | 14/108 (13) | 181/971 (19) |
| Miller33 | 11/21 (52) | 3/9 (33) | 8/12 (67) | 9/51 (18) | 2/14 (14) | 7/37 (19) | 26/173 (15) | 25/164 (15) | 1/9 (11) | 4/65 (6) | 1/25 (4) | 3/40 (8) | 3/62 (5) | 3/36 (8) | NA | 3/14 (21) | NA | NA | 2/16 (13) | 58/402(14) |
| Oldenburg34 | 6/18 (33) | 3/14 (21) | 3/4 (75) | 10/70 (14) | 8/32 (25) | 2/38 (5) | 35/236 (15) | 34/228 (15) | 1/8 (13) | 4/79 (5) | 0/18 (0) | 4/61 (7) | 3/67 (0) | 0/34 (0) | 0/33 (0) | 2/19 (11) | 1/9 (11) | 1/10 (10) | 0/16 (0) | 57/505 (11) |
| Owaidah35 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 2/10 (20) | 2/10 (20) | 0/0 (−) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/9 (0) | 0/1 (0) | 0/8 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 2/20 (10) |
| Pieneman36 | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/10 (0) | 0/10 (0) | 0/0 (−) | 0/11 (0) | 0/4 (0) | 0/5 (0) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/0 (−) | 0/4 (0) | 0/25 (0) |
| Reitter37 | 1/2 (50) | 0/0 (−) | 1/2 (50) | 1/13 (8) | 1/6 (17) | 0/7 (0) | 12/77 (16) | 11/73 (15) | 1/4 (25) | 5/17 (29) | 1/4 (25) | 4/13 (31) | 2/34 (6) | 1/26 (4) | 1/8 (13) | 1/11 (9) | NA | NA | 0/1 (0) | 22/155 (14) |
| Repesse38 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Rosetti39 | 5/11 (45) | 0/4 (0) | 5/7 (71) | 1/9 (11) | 1/3 (33) | 0/6 (0) | 5/40 (13) | 5/39 (13) | 0/1 (0) | 2/15 (13) | 0/2 (0) | 2/13 (15) | 0/8 (0) | 0/6 (0) | 0/2 (0) | 0/1 (0) | 0/0 (−) | 0/1 (0) | 0/4 (0) | 13/88 (15) |
| Sanna40 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Sirocova41 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 0/2 (0) | 0/0 (−) | 0/2 (0) | 1/14 (7) | 1/13 (8) | 0/1 (0) | 0/4 (0) | 0/2 (0) | 0/2 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/0 (−) | 0/0 (−) | 2/29 (7) |
| Vidal42 | 1/2 (50) | 0/1 (0) | 1/1 (100) | 2/3 (67) | 0/1 (0) | 2/2 (100) | 4/13 (31) | 4/13 (31) | 0/0 (−) | 2/6 (33) | 0/1 (0) | 2/5 (40) | 1/11 (9) | 1/8 (13) | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 10/38 (26) |
| Viel43 | 1/3 (33) | 1/3 (33) | 0/0 (−) | 0/4 (0) | 0/1 (0) | 0/3 (0) | 7/14 (50) | 6/10 (60) | 1/4 (25) | 0/5 (0) | 0/3 (0) | 0/2 (0) | 1/12 (8) | 0/1 (0) | 1/11 (9) | 0/0 (0) | 0/0 (−) | 0/0 (−) | 0/2 (0) | 9/40 (23) |
| Xue44 | 0/3 (0) | 0/2 (0) | 0/1 (0) | 0/6 (0) | 0/2 (0) | 0/4 (0) | 2/60 (3) | 2/57 (4) | 0/3 (0) | 0/24 (0) | 0/11 (0) | 0/13 (0) | 0/12 (0) | 0/3 (0) | 0/9 (0)) | 0/3 (0) | 0/3 (0) | 0/0 (−) | 0/3 (0) | 2/111 (2) |
NA indicates not available.
Numbers of patients with inhibitors and total numbers of patients with the specific mutations are reported, with the percentages in parentheses.
Data synthesis
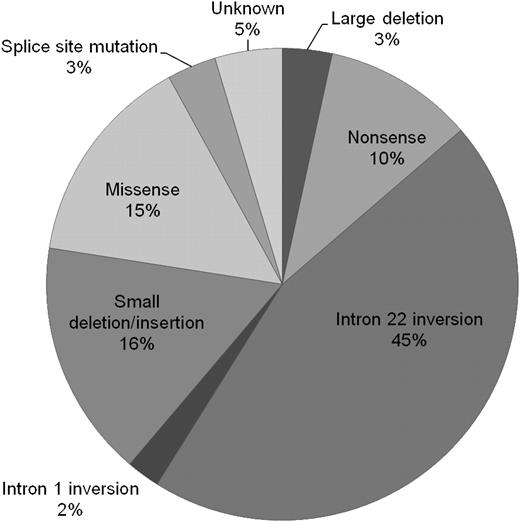
The relative proportions of genotypes are presented in Figure 2. Of all 5383 patients, 3% had large deletions, 10% nonsense mutations, 45% intron 22 inversions, 2% intron 1 inversions, 16% small deletions or insertions, 15% missense mutations, and 3% splice-site mutations. In 4.6% of patients, the mutation was unknown.
All inhibitor development.
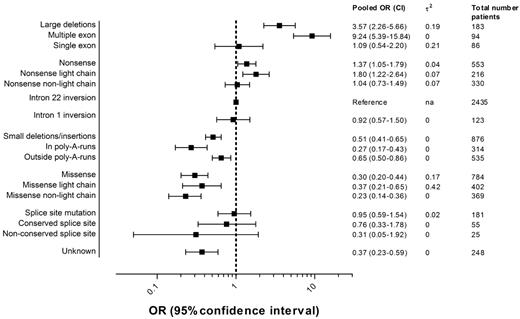
Figure 3 presents the pooled ORs of all inhibitor development and 95% CIs for each F8 genotype including the numbers of patients in each category. Details regarding the individual study ORs for each genotype can be found in the appendix (supplemental Figure 1).
Pooled ORs of inhibitor development according to the F8 genotype.2 indicates the between-study variance of lnOR.
Pooled ORs of inhibitor development according to the F8 genotype.2 indicates the between-study variance of lnOR.
The pooled OR of large deletions compared with intron 22 inversions was 3.6 (95% CI, 2.3-5.7); for nonsense mutations, 1.4 (95% CI, 1.1-1.8); for intron 1 inversions, 0.9 (95% CI, 0.6-1.5); for small deletions and insertions, 0.5 (95% CI, 0.4-0.6); for missense mutations, 0.3 (95% CI, 0.2-0.4); and for splice-site mutations, 1.0 (95% CI, 0.6-1.5).
The risk of inhibitor development in patients with deletions of more than one exon was higher than the risk in patients with deletions of one exon or less. Nonsense mutations and missense mutations in the light chain carried a higher risk than those not located in the light chain. Small deletions, insertions, or combined deletions/insertions located within a poly-A run were associated with a lower risk than those located outside a poly-A run. The location of the nucleotide substitution was only available for few patients with splice-site mutations; therefore, the estimates of the pooled ORs of splice-site mutations at conserved and nonconserved nucleotide positions are imprecise and 95% CIs are wide.
High-titer-inhibitor development.
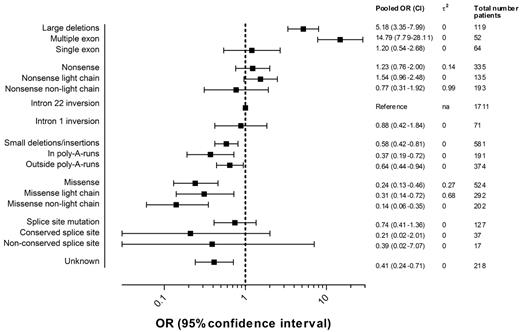
Figure 4 presents the pooled ORs of high-titer-inhibitor development and 95% CIs for each F8 genotype including the numbers of patients in each category. Details regarding the individual study ORs for each genotype can be found in supplemental Figure 1.
Pooled ORs of high-titer-inhibitor development according to the F8 genotype.2 indicates the between-study variance of lnOR.
Pooled ORs of high-titer-inhibitor development according to the F8 genotype.2 indicates the between-study variance of lnOR.
The pooled OR of large deletions relative to intron 22 inversions was 5.2 (95% CI, 3.4-8.0); for nonsense mutations, 1.2 (95% CI, 0.8-2.0); for intron 1 inversions, 0.9 (95% CI, 0.4-1.8); for small deletions and insertions, 0.6 (95% CI, 0.4-0.8); for missense mutations, 0.2 (95% CI, 0.1-0.5); and for splice-site mutations, 0.7 (95% CI, 0.4-1.4). The effect of large deletions of more than one exon on the risk of high-titer-inhibitor development was more pronounced. Too few patients were available in the subgroups of splice-site mutations at conserved and nonconserved nucleotide positions to yield meaningful estimates.
Data evaluation
Small study trends.
To explore the potential presence of small study trends, the forest plots were sorted by study sample size (supplemental Figure 1). No clear small study data trends could be identified.
Sensitivity analysis.
The analysis was repeated in a subgroup of 5 studies meeting the methodological high-quality criteria and resulted in similar ORs (supplemental Table 3).
HADB
In the HADB, the proportion of patients with inhibitor development was 19.5% (180 of 922). The numbers of inhibitor patients in each type of gene mutation are summarized in Table 4. Information on patients with intron 1 and 22 inversions was not collected in the HADB. In large deletions and insertions (greater than 50 base pairs) the incidence of inhibitor development was 45% (≤ 1 exon, 21%; > 1 exon, 67%). In nonsense mutations, the incidence of inhibitor development was 28% (nonsense mutations in light chain (for A3, C1, and C2, 43%; for nonsense mutations outside of the light chain, 12%). In small deletions and insertions (< 50 base pairs), the proportion of inhibitors is 15% (in poly-A runs, 6%; outside poly-A runs, 19%). In missense mutations, the proportion was 8% (for missense mutations in the light chain, 11%; for missense mutations outside of the light chain, 6%). In splice-site mutations, the proportion of inhibitors was 7%.
Type of F8 gene mutation and inhibitor development in HADB
| . | Positive inhibitor . | N . | |
|---|---|---|---|
| % . | n . | ||
| Large deletions | 49 | 45 | 109 |
| > 1 exon | 38 | 67 | 57 |
| < 1 exon | 11 | 21 | 52 |
| Nonsense mutations | 65 | 28 | 230 |
| Light chain | 52 | 43 | 122 |
| Non-light chain | 13 | 12 | 108 |
| Small deletions/insertions | 43 | 15 | 289 |
| Outside poly-A run | 37 | 19 | 191 |
| Poly-A run | 6 | 6 | 98 |
| Missense mutations | 20 | 8 | 246 |
| Light chain | 12 | 11 | 114 |
| Non-light chain | 8 | 6 | 132 |
| Splice site mutations | 3 | 7 | 46 |
| Unknown | 0 | 0 | 2 |
| Total | 180 | 20 | 922 |
| . | Positive inhibitor . | N . | |
|---|---|---|---|
| % . | n . | ||
| Large deletions | 49 | 45 | 109 |
| > 1 exon | 38 | 67 | 57 |
| < 1 exon | 11 | 21 | 52 |
| Nonsense mutations | 65 | 28 | 230 |
| Light chain | 52 | 43 | 122 |
| Non-light chain | 13 | 12 | 108 |
| Small deletions/insertions | 43 | 15 | 289 |
| Outside poly-A run | 37 | 19 | 191 |
| Poly-A run | 6 | 6 | 98 |
| Missense mutations | 20 | 8 | 246 |
| Light chain | 12 | 11 | 114 |
| Non-light chain | 8 | 6 | 132 |
| Splice site mutations | 3 | 7 | 46 |
| Unknown | 0 | 0 | 2 |
| Total | 180 | 20 | 922 |
Data downloaded on October 8, 2010.
Discussion
This meta-analysis merged the data of 5383 severe hemophilia A patients from 30 selected studies to estimate the relative risks of inhibitor development in patients with severe hemophilia A according to specific F8 genotypes with more precision than is possible in single studies. Compared with the risk of inhibitor development in patients with intron 22 inversions, the risk of patients with large deletions and nonsense mutations was higher (pooled OR = 3.6; 95% CI, 2.3-5.7 and OR = 1.4; 95% CI, 1.1-1.8, respectively), the risk of patients with intron 1 inversions and splice-site mutations was equal (pooled OR = 0.9; 95% CI, 0.6-1.5 and OR = 1.0; 95% CI, 0.6-1.5, respectively), and the risk of patients with small deletions and insertions and missense mutations was lower (pooled OR = 0.5; 95% CI, 0.4-0.6 and 0.3; 95% CI, 0.2-0.4, respectively).
The HADB collected data on F8 mutations and information on the patients' inhibitor status.10 The findings from the HADB were similar to our findings. However, the data of the HADB has several limitations when it is used to assess the relationship between F8 genotype and inhibitor development.47 The HADB contains selected patients and the inhibitor status is missing in 26% of patients, which may lead to biased estimates. The relationship between F8 genotype and high-titer-inhibitor development cannot be evaluated because information on the peak inhibitor titer is not collected.
Our study has several limitations. The frequency of screening for the presence of inhibitors in patients varied markedly in the studies. In several studies, significant underdiagnosis of inhibitor development is likely, which will underestimate the cumulative incidence of inhibitors. However, this would predominantly affect the incidence of low-titer inhibitors, because high-titer inhibitors would be more likely to present clinically.
If inhibitor testing occurs more frequently in patients with some genotypes than in patients with other genotypes, this would bias the association. Therefore, we performed a sensitivity analysis in the subgroup of studies that were unlikely to have misdiagnosed inhibitor patients. These studies measured the inhibitor titer at least once a year and whenever there was a clinical suspicion. In this subanalysis, the results were similar to the meta-analysis of all included studies.
Another limitation of the current meta-analysis is that in the majority of the studies, some patients may still be at risk for subsequent inhibitor development. The risk of inhibitor development is highest during the first 50 exposure days to FVIII.2 After 50-75 exposure days, the development of inhibitors becomes rare (approximately 2-5 per 1000 patient-years).48,49 Pooling of studies including patients who were treated on less than 50 exposure days may have underestimated of the inhibitor risk in patients with some genotypes. To address this, we performed a sensitivity analysis among studies that presented patients who were treated on at least 50 exposure days, which showed similar results.
Other genetic and nongenetic factors have been reported to affect the risk of developing inhibitors. Genetic polymorphisms in the HLA complex50,51 and in promoters of cytokine genes (IL-10 and TNFα) and T-cell surface molecules have been associated with the risk of inhibitor development.52-56 Although F8 genotypes are generally believed to be equally distributed across different ethnic populations, this may not be true for other genetic risk factors. To account for geographic differences in treatment practices, we calculated pooled relative effects instead of absolute incidences. Detailed data on ethnic background and other risk factors were unavailable and the impact of these factors could not be evaluated.
Because our analysis was not restricted to unrelated patients, the observations may not be completely independent from each other. Other familial risk factors such as immunologic makeup may account for inhibitor development in several related patients.
A strength of this systematic review is it includes 5383 unique patients with severe hemophilia A, yielding the most precise estimates of relative risk of inhibitor development available to date.
We calculated relative effect measures (ORs) instead of absolute cumulative incidences because the frequency of inhibitor screening, definition of inhibitor development, and FVIII treatment regimen differed among studies, thus affecting the total cumulative incidences of inhibitors. A pooled cumulative risk would include this variation in hemophilia management practices and would not be useful in clinical practice. Because these differences between studies were not expected to be related to F8 genotype, we expect that relative effect measures are similar across studies.
Because of the multitude of F8 genotypes, the total number of patients within the individual genotypes was often small. Therefore, in these genotypes, there may be no or 100% inhibitor patients. Because of these “null cells” in the data, standard methods for meta-analysis give severely biased results. Therefore, we used a meta-analysis model especially suitable for sparse and unbalanced data: the hypergeometric-normal model.8 This model has several advantages in situations when data are sparse; it avoids bias caused by the correlation between estimate and standard error, takes into account the uncertainty in the estimates of the standard errors, and avoids the use of continuity corrections.
A further strength of this study is that it also studied the outcome high-titer-inhibitor development and found similar relative risks. Studying high-titer-inhibitor development avoided potential bias by underdiagnoses of inhibitor development by infrequent inhibitor testing and by known suboptimal specificities in the inhibitor assays.57
Mutations that are expected to cause complete absence of protein were associated with a higher risk of inhibitors, whereas mutations that may result in some protein synthesis were associated with a lower risk of inhibitor development. An immune response against FVIII probably occurs because of lack of central tolerance to FVIII protein to a greater or lesser extent. Patients with F8 mutations that allow the production of some nonfunctional FVIII protein may develop (partial) central tolerance to this altered FVIII protein. In these patients, T lymphocytes specific for fewer FVIII epitopes are present in the periphery, and the generation of FVIII-specific regulatory T cells is possible. In patients with missense mutations and small deletions and insertions, some production of parts of the FVIII protein may occur, and therefore these patients face a lower risk of inhibitor development. Conversely, in patients with F8 mutations that result in complete absence of FVIII protein, central tolerance is lacking. Anti-FVIII–specific T and B cells are not negatively selected, and no FVIII-specific regulatory T cells can be generated. Subsequently, anti–FVIII-specific lymphocytes enter the periphery and may react against infused FVIII product. Large deletions, nonsense mutations, and intron 1 and 22 inversions induce a complete deficit of any production of circulating FVIII polypeptides, and this explains why patients with these mutations have an increased risk for developing inhibitors.58
Based on the recent finding that patients with the intron 22 inversion express the entire FVIII protein as 2 polypeptide chains, it has been suggested that intracellular production of parts of an endogenous FVIII protein may lead to some degree of tolerance toward replacement FVIII proteins and to a reduced risk of inhibitor development.59 When considered together with the findings from the present meta-analysis, which demonstrate that intron 22 inversions confer a lower inhibitor risk as large, multi-exon deletions, this hypothesis may be an explanation for the difference in inhibitor risks among patients with F8 genotypes that do not result in extracellular protein production.
In patients with nonsense and missense mutations, the risk of inhibitor development is higher when the mutation is located in the light chain (ie, in the A3, C1, or C2 domain) than when it is located outside of the light chain (ie, in the A1, A2, or B domain) of the FVIII protein. This may be explained by the presence of a second gene transcript, F8b (HUGO Gene Nomenclature Committee HGNC: F8A2), which runs from a unique first exon within the CpG island in intron 22 through exon 26 at the end of the F8 gene.60 As mentioned previously, the predicted FVIIIB protein was recently found to be expressed within cells from both a hemophilia patient with the intron 22 inversion and from a healthy subject, and the presence of this protein intracellularly may result in some tolerance toward the portion of FVIII encoded by exons 23-26.59 Therefore, mutations that are located outside of this F8b gene may be associated with normal production of this gene product and lead to partial central tolerance toward FVIII.16 Conversely, mutations within this gene may lead to an absent or defective F8b gene product and may be associated with an impaired central tolerance toward this part of the F8 gene. Other investigators hypothesized that mechanisms such as in-frame exon skipping and nonsense-mediated decay of F8 mRNA (and the potential resulting protein production) depend on the location of the nonsense mutation in the coding sequence. However, these hypotheses could not be supported by observational evidence.47,61
Small deletions and insertions usually cause frame shifts and downstream premature stop codons. However, patients with small deletions and insertions have a lower inhibitor risk than patients with nonsense mutations. Small deletions and insertions affecting stretches of adenines (the poly-A run) carry a lower inhibitor risk than those outside of a poly-A run. A milder phenotype and improved thrombo-elastogram parameters have been observed in severe hemophilia A patients with small deletions and insertions in poly-A runs. It is possible that small amounts of FVIII protein could be produced, because the reading frame may be partially restored by errors in DNA replication, RNA transcription, and translation errors in poly-A runs.62-64 The presence of small amounts of FVIII protein may result in a lower inhibitor risk in patients with small deletions and insertions in poly-A runs.
The prevalence of intron 1 inversions was 2%, similar to a previous report.65 The pathogenetic mechanism is similar to that of intron 22 inversions.66,67 In the present study, the inhibitor risk in patients with intron 1 inversions was indeed similar (OR = 0.92; 95% CI, 0.57-1.50) to that in patients with intron 22 inversions.
Most investigators reported low inhibitor incidences in patients with splice-site mutations. However, in one study, splice-site mutations were related to the highest inhibitor risk.16 A higher risk of inhibitor development was found in patients with splice-site mutations affecting conserved nucleotide positions (31%) than in patients with splice-site mutations affecting nonconserved nucleotide positions (13%).34 It may be hypothesized that splice-site mutations in nonconserved positions may not always be recognized and may result in the production of some normal FVIII.68 In splice-site mutations affecting conserved nucleotide positions, the restoration of normal splicing is predicted to occur less often. Altered splicing frequently results in a premature stop codon, and these patients may carry an inhibitor risk similar to that in patients with null mutations. In our study, patients with splice-site mutations had a similar risk as patients with intron 22 inversions. Patients with splice-site mutations affecting conserved nucleotide positions had a higher risk of inhibitor development (OR = 0.76; 95% CI, 0.33-1.78) than those in nonconserved nucleotide positions (OR = 0.31; 95% CI, 0.05-1.92). However, because these data were only available for a few studies, only 55 and 25 patients, respectively, were analyzed, the confidence intervals were wide, and it is difficult to draw any definite conclusions.
In 4.6% of patients, the causative mutation was unknown. This included both patients in whom genetic testing did not reveal a causative mutation and patients who were part of the study cohort and in whom genetic testing was not performed. Because of the variety of studies, these reasons could not be reliably discerned.
This meta-analysis provides more precise estimates of the relative risks for different F8 genotypes relative to intron 22 inversions. In future research on other possible genetic and nongenetic risk factors, the predisposition related to the hemophilic genotype should be taken into account.
Conclusions
The results of this meta-analysis confirm earlier reports that the F8 genotype is an important determinant of inhibitor development in patients with severe hemophilia A. Compared with the risk of inhibitor development in patients with intron 22 inversion, the risk of patients with large deletions and nonsense mutations was higher, the risk of patients with intron 1 inversions and splice-site mutations was equal, and the risk of patients with small deletions and insertions and missense mutations was lower.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank P. Green, P. Casaña, S. Haya, C. Miller, A. Awidi, K. Viel, T. Howard, D. David, F. Xue, C. Mannhalter, L. Rossetti, Y.-C. Chen, E. Chalmers, R. Saxena, and C Altisent for providing additional information; J. W. Schoones of the Walaeus Library (Leiden University Medical Center, Leiden, The Netherlands) for expert support in designing the literature search; and Professor T. Stijnen for indispensable statistical advice.
Authorship
Contribution: S.C.G., S.l.C., and J.G.v.d.B. designed the research; S.C.G., J.O. M.M., A.R.T., W.v.H., J.B., and C.H.M. contributed the data; S.C.G. analyzed the data; S.C.G., H.M.v.d.B., and J.G.v.d.B. and wrote the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: S.C.G. and H.M.v.d.B. have reported receiving unrestricted research support from ZLB Behring, Novo Nordisk, Wyeth, Baxter, and Bayer. J.G.v.d.B. has received unrestricted research/educational funding for various projects from Bayer Schering Pharma, Baxter, ZLB Behring, Novo Nordisk, and Wyeth; has been a consultant to Baxter and Wyeth; and has been a teacher in the educational activities of Bayer Schering Pharma. The remaining authors declare no competing financial interests.
Correspondence: S. C. Gouw, Wilhelmina Children's Hospital, University Medical Center Utrecht, Rm KE 04.133.1, P O Box 85090, 3508 AB Utrecht, The Netherlands; e-mail: s.c.gouw@umcutrecht.nl.