In this issue of Blood, Lia et al present a novel mouse model that recapitulates the pathogenic mechanisms of human CLL with deletion 13q14. Their striking finding is that the size of the deletion correlates with disease penetrance and aggressiveness.1
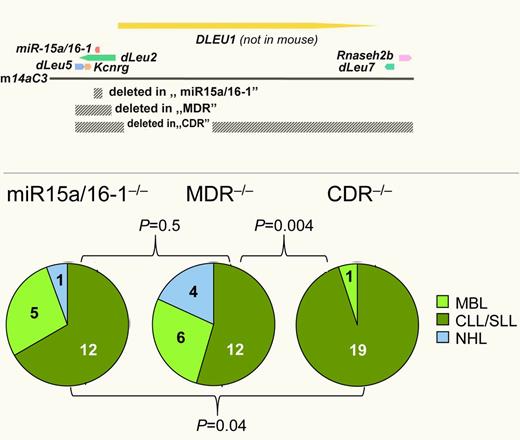
Chromosomal band 13q14.3 is recurrently lost in various hematopoietic and solid tumors. In chronic lymphocytic leukemia (CLL), deletion of at least one allele occurs in more than 50% of cases. Until recently, the underlying mechanism has remained elusive. In a landmark study, Klein and colleagues conclusively show that loss of the 2 microRNA genes Mir15A and Mir16-1, that are localized in this region, is sufficient to cause a late-onset, CLL-like disease in mice (see figure).2 However, a sizeable fraction of mice also developed CD5-negative non-Hodgkin lymphomas (NHLs) and monoclonal B-cell lymphocytosis (MBL), a CLL precursor state. In addition, these malignancies developed with low penetrance (28%) and after a long latency (10 months). In human CLL, deletions in chromosomal band 13q14.3 are usually much larger, and rare CLL cases exist where the deletion does not affect MIR15A and MIR16-1. Recently, an intriguing example of familial CLL was reported where the deletion only affects the DLEU7 gene (see figure).3
The tumor suppressor mechanism in 13q14.3 is multigenic. Three different regions of 13q14.3 were deleted in B cells of mice (top; gray bars, deleted areas; arrows, genes) and led to the development of different proportions of hematopoietic malignancies, with larger deletions resulting in higher disease penetrance (bottom) and more aggressive disease.1,2
The tumor suppressor mechanism in 13q14.3 is multigenic. Three different regions of 13q14.3 were deleted in B cells of mice (top; gray bars, deleted areas; arrows, genes) and led to the development of different proportions of hematopoietic malignancies, with larger deletions resulting in higher disease penetrance (bottom) and more aggressive disease.1,2
Lia and colleagues therefore took mouse modeling one step further. In their previous work, they could show that removal of a “minimally deleted region” (MDR) that was derived from human CLL led to a more aggressive disease compared with deletion of the microRNA genes only. Furthermore, lymphoproliferative disorders occurred with a higher penetrance in MDR−/−, whereas disease latency and distribution of entities (CLL vs MBL vs NHL) remained unchanged (see figure).2 Here, these investigators now present a third mouse strain in which they knocked out a “commonly deleted region” (CDR) that in addition to the MDR also includes DLEU7 and RNASEH2B.1 Strikingly, CDR−/− mice not only developed more aggressive lymphomas, but the phenotype of the resulting malignancies was also shifted almost exclusively toward CLL or small lymphocytic lymphoma (see right panel of figure). This finding strongly suggests that the tumor suppressor activity of band 13q14.3 can be attributed to several genes or genetic elements localized in the CDR.
These exciting observations notwithstanding, some open questions remain: (1) If 13q14.3 is affected by deletions in only approximately 50% of CLL cases, what is the causative genetic aberration in the remaining patients? (2) Are there other tumor suppressor genes in 13q14.3 or are other genetic mechanisms affected? (3) What is the molecular function of the 13q14 tumor suppressor genes?
With regard to the first 2 questions, the low penetrance and long latency of the murine CLL models are of note. It is very likely that genetic lesions in addition to deletion of 13q14.3 have to occur for the development of CLL, and genome-wide sequencing approaches recently identified a number of candidate genes. Similarly, MIR15A and MIR16-1 have been shown to be mutated in CLL patients, resulting in defective precursor transcript processing, but these mutations occur only very rarely (2 of 75 patients).4,5 Another mode of functional inactivation of candidate CLL genes could be transcriptional deregulation. In fact, MIR15A and MIR16-1 are down-regulated in all CLL patients, irrespective of the deletion status of the region.6,7 This suggests that deregulation of 13q14 may have a more general role in CLL pathogenesis, although further work will be required to determine the spectrum of underlying molecular mechanisms. Epigenetic aberrations are prime candidates as drivers of transcriptional deregulation.
The third open question concerns the molecular function of the 13q14.3 genes: if several genes that are localized next to each other are causative for malignant transformation, it is likely that they are also involved in at least cooperative or possibly even the same cancer-related pathway. In fact, for the majority of candidate tumor suppressors encoded by genes in 13q14.3, involvement in the NF-κB signaling pathway has been shown: KPNA3 binds to and transports NF-κB subunits.8 RFP2/DLEU5/TRIM13 has been found to be the strongest inducer of NF-κB in a genome-wide reporter screen.9 The MIR15A and MIR16-1 microRNA family targets NF-κB via TAB3 and TAK1.10 Finally, DLEU7, which is localized together with RNASEH2B in the CDR, functions as a decoy for the TACI receptor and thereby regulates NF-κB activity.11 Thus, it is very likely that the multifaceted tumor suppressor activity of 13q14.3 could, in addition to its role in the cell cycle,2 be involved in regulating the NF-κB signaling pathway. This may open the door for new approaches to the treatment of CLL.
In conclusion, Lia and colleagues describe an exceptional example of a cluster of functionally related genes localized next to each other. This provides an explanation why deletion size influences disease development and course, with more aggressive leukemias developing in mice (and possibly in man) with larger deletions.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal