Abstract
Regulator of G-protein signaling 18 (RGS18) is a GTPase-activating protein for the G-α-q and G-α-i subunits of heterotrimeric G-proteins that turns off signaling by G-protein coupled receptors. RGS18 is highly expressed in platelets. In the present study, we show that the 14-3-3γ protein binds to phosphorylated serines 49 and 218 of RGS18. Platelet activation by thrombin, thromboxane A2, or ADP stimulates the association of 14-3-3 and RGS18, probably by increasing the phosphorylation of serine 49. In contrast, treatment of platelets with prostacyclin and nitric oxide, which trigger inhibitory cyclic nucleotide signaling involving cyclic AMP-dependent protein kinase A (PKA) and cyclic GMP-dependent protein kinase I (PKGI), induces the phosphorylation of serine 216 of RGS18 and the detachment of 14-3-3. Serine 216 phosphorylation is able to block 14-3-3 binding to RGS18 even in the presence of thrombin, thromboxane A2, or ADP. 14-3-3–deficient RGS18 is more active compared with 14-3-3–bound RGS18, leading to a more pronounced inhibition of thrombin-induced release of calcium ions from intracellular stores. Therefore, PKA- and PKGI-mediated detachment of 14-3-3 activates RGS18 to block Gq-dependent calcium signaling. These findings indicate cross-talk between platelet activation and inhibition pathways at the level of RGS18 and Gq.
Introduction
In healthy vasculature, endothelial cells lining the blood vessels constantly produce and release prostacyclin (PGI2) and nitric oxide (NO) into the vessel lumen. The interaction of endothelial factors with platelets plays a fundamental role in controlling hemostasis and in maintaining platelets in a resting state. Platelet inhibition by both PGI2 and NO has been well established. The signaling pathways of both molecules result in an elevation of cyclic nucleotides that activate cyclic AMP-dependent protein kinase A (PKA) and cyclic GMP-dependent protein kinase I (PKGI). These in turn phosphorylate an unknown number of substrate proteins, resulting in reduced release of calcium ions (Ca2+) from intracellular stores and reduced activation of G-proteins such as Rap1, ultimately leading to a block of platelet adhesion, granule release, and aggregation. PKA and PKGI have overlapping substrate specificities, which may explain the synergistic role of the 2 pathways. Few substrates have been established in platelets, among them Rap1GAP2, a GTPase-activating protein (GAP) of the small G-protein Rap1, as we have shown previously.1 Other substrates include vasodilator-stimulated phospho-protein (VASP), heat-shock protein 27 (HSP27), and LIM and SH3 domain protein (LASP), all of which regulate actin dynamics.2,3 The IP3-receptor and the IP3-receptor-associated G-kinase substrate (IRAG) are the only PKA and/or PKGI substrates that have been shown to mediate cAMP/cGMP effects on intracellular Ca2+ release.2-4 Limited data are available on the specific substrates and signaling events that translate PKA/G substrate phosphorylation into platelet inhibition.
Conversely, binding of thrombin, ADP, and thromboxane A2 to G-protein coupled receptors (GPCRs) leads to platelet activation. GPCR signaling is terminated by the family of regulator of G-protein signaling (RGS) proteins, the function of which has mainly been attributed to their ability to interact with G-α subunits of heterotrimeric G proteins. RGS proteins are GAPs that enhance the rate of GTP hydrolysis of G-α subunits. Thirty-seven RGS domain–containing proteins have been identified in humans,5 and in a recent transcriptome analysis, at least 16 were shown to be expressed in human platelets.6 The functional significance of RGS proteins in platelets has recently been validated in a mouse model expressing a mutant G-α-i2 (Gi2) that is unable to interact with RGS.7 RGS18 belongs to the subfamily of B/R4 RGS proteins that contain a conserved central RGS domain and a short N and C terminus. These N- and C-terminal regions may be crucial for the association with specific GPCRs but also with other signaling factors.8,9 First identified in 2001, RGS18 is abundantly expressed in platelets and megakaryocytes.10-13 Interaction partners include G-α-q (Gq) and Gi1,2,3, but not Gz, Gs, or G12.10,11 Platelet activation by GPCR ligands requires both Gq and Gi signaling.14 Gq activates PLC-β, leading to the production of inositol-1,4,5-trisphosphate (IP3) and the release of Ca2+ from intracellular stores. Gi inhibits adenylyl cyclase and activates PI3K pathways,15 and RGS18 may be involved in the regulation of these pathways. To date, no other interaction partners of RGS18 have been described.
In the present study, we show that RGS18 is a target of PKA- and PKGI-mediated platelet inhibition, as well as of GPCR-mediated platelet activation. We show that RGS18 interacts with the 14-3-3γ protein via phosphorylated residues S49 and S218. 14-3-3s are abundantly expressed dimeric proteins that operate as serine/threonine–binding modules with a wide range of functions.16 There are 7 highly conserved isoforms described in mammals, 5 of which are expressed in platelets.17 PKA- and PKGI-mediated phosphorylation of RGS18 on serine 216 (S216) has a dominant regulatory function, leading to detachment of 14-3-3γ, which renders RGS18 more efficient in its ability to inhibit Gq-mediated Ca2+ release.
Methods
Abs and materials
A polyclonal Ab against RGS18 was developed using full-length recombinant GST-tagged human RGS18 purified from Escherichia coli BL21 cells. An Ab against phosphorylated S216 of RGS18 was produced using a phosphorylated peptide conjugated to keyhole limpet hemocyanin. Immunization of rabbits and subsequent Ab purification were performed by ImmunoGlobe Antikörpertechnik (Himmelstadt, Germany). The Ab against phosphorylated S7 of Rap1GAP2 has been described previously.18 The following commercially available Abs were used in this study: mouse anti-RGS18 (1H6; Sigma-Aldrich), rabbit pan 14-3-3 (K19; Santa Cruz Biotechnology), mouse 14-3-3γ (3F8; Abcam), mouse 14-3-3β (H8; Santa Cruz Biotechnology), mouse 14-3-3ϵ (8C3; Santa Cruz Biotechnology), mouse 14-3-3ζ (Abcam), FLAG tag (M2; Sigma-Aldrich), Myc tag (9E10; Santa Cruz Biotechnology), and mouse anti-GST (clone GST-2; Sigma-Aldrich). HRP-coupled donkey anti–rabbit and donkey anti–mouse Abs were from Jackson ImmunoResearch Europe and were used as secondary Abs for immunoblot analysis visualized by enhanced chemiluminescence (Thermo Scientific). All statistical analyses were performed using Prism Version 5 software (GraphPad).
Constructs
Full-length human RGS18 cDNA was obtained from the Missouri S&T cDNA Resource Center (Rolla, MO). RGS18 was FLAG-tagged at the N-terminus and expressed using mammalian expression vector pcDNA4/TO (Invitrogen). mCherry-tagged RGS18 was obtained through subcloning into mammalian expression vector pmCherryC1 (Invitrogen), and the GST fusion protein was generated using pGEX-4T3 vector (GE Healthcare). All RGS18 constructs were subcloned using XhoI/BamHI sites of their respective vectors. Site-directed mutagenesis was performed by PCR amplification using mutagenic primer pairs, Pfu DNA polymerase (Fermentas), digestion with DpnI (Fermentas), and transformation into TOP10 bacteria (Invitrogen). Human 14-3-3γ cDNA was a kind gift of James McRedmond (Conway Institute, University College Dublin, Dublin, Ireland). 14-3-3γ was myc-tagged and cloned into mammalian expression vector pcDNA4TO via BamHI and XhoI. For GST-tagged 14-3-3γ, 14-3-3γ was cloned into pGEX-4T3 using EcoRI and XhoI restriction sites. Constructs were verified by DNA sequencing.
Protein purification, 32P labeling, and Phos-Tag gels
GST-14-3-3γ, GST-wt-RGS18, and mutants were expressed in E coli BL21 and affinity purified using glutathione-Sepharose 4B beads (GE Healthcare). The purity of all proteins was examined by SDS-PAGE followed by Coomassie blue staining.
Recombinant GST-RGS18 fusion proteins attached to GSH beads were incubated with the purified C-subunit of PKA (New England Biolabs) in a buffer containing 40μM ATP and 1 μCi (γ-32P) ATP (PerkinElmer) at 30°C for 3 minutes. Reactions were stopped by the addition of SDS-sample buffer, followed by SDS-PAGE, blotting, and exposure of the membrane to film. Equal loading was verified by immunoblotting using anti-GST Ab. For phosphate-binding tag (Phos-tag; Wako Chemicals)–supplemented SDS-PAGE, gels were prepared as described in the manufacturer's protocol. Briefly, 50μM Phos-tag and 100μM MnCl2 (Sigma-Aldrich) were added to the separating gel solution before polymerization. To remove Mn2+ ions before Western blotting, gels were incubated in transfer buffer containing 1mM EDTA for 10 minutes at room temperature.
Cell preparation, transfection, lysis, immunoprecipitation, and pull-down experiments
HEK293T cells were cultured using DMEM supplemented with 10% FCS and 1% penicillin/streptomycin at 37°C and 5% CO2. Cells were transfected using Metafectene (Biontex) or Fugene (Promega) according to manufacturer's instructions. Venous blood was drawn from healthy volunteers taking no medications who gave their informed consent according to the Declaration of Helsinki. Platelet isolation was performed as described previously.19 Platelets were stimulated at 37°C using 0.1 U/mL of thrombin (Roche) for 30 seconds, U-46619 (Cayman Europe) as a thromboxane A2 mimetic at 1μM for 1 minute, ADP (Sigma-Aldrich) at 10μM for 1 minute, forskolin at 10μM for 30 minutes, prostacyclin (Cayman Europe) at 1μM for 5 minutes, sodium nitroprusside (SNP) at 10μM for 10 minutes, the cAMP analog 5,6-di-chlorobenzimidazole riboside-3,5-cyclic monophosphorothioate, Sp-isomer (Biolog) at 0.3mM for 30 minutes or GMP analog 8-(4-chlorophenylthio)guanosine-3,5-cyclic monophosphate (Biolog) at 0.3mM for 20 minutes.
Cell lysis, immunoprecipitation, and pull-down assays were performed as indicated using 5 μL of anti-FLAG M2 Affinity Gel (Sigma-Aldrich), 20 μL of anti-RGS18 polyclonal Ab, or 5 μL of a glutathione Sepharose 4B suspension (GE Healthcare) saturated with GST or GST-14-3-3γ, respectively. For peptide competition assays, lysates were supplemented with 100μM of either TNLRRRSRSFTVN or TNLRRRSR(pS)FTVN acetylated peptides (Schafer-N) before pull-down assays. For dephosphorylation experiments, samples were treated with λ protein phosphatase as described in the manufacturer's protocol (New England Biolabs).
MS
For mass spectrometry (MS) analysis, samples were subjected to SDS-PAGE and subsequently to Coomassie blue staining of gels. Protein bands of interest were excised and subjected to in-gel digestion, as described previously.20 Briefly, gel bands were washed and cysteine residues were reduced and alkylated using dithiothreitol and iodoacetamide. Samples were digested with trypsin and run on a Thermo Scientific LTQ Orbitrap XL mass spectrometer connected to an Eksigent Nano LC.1DPLUS chromatography system. A high-resolution MS scan was performed using the Orbitrap to select the 5 most intense ions before MS/MS analysis using the ion trap. The MS/MS spectra were searched against the UniProt/SwissProt human database (January 2009) using Bioworks Browser 3.3.1 SP1, a proteomics analysis platform. All MS/MS spectra were sequence-database searched using the algorithm TurboSEQUEST. The following search parameters were used: “precursor-ion mass tolerance of < 10 ppm,” “fragment ion tolerance of 0.5 Da,” and “fully tryptic peptide termini,” with “cysteine carboxyamidomethylation” and “methionine oxidation” specified as variable modifications and a maximum of 3 missed cleavages. Protein identifications were accepted if they could be established at greater than 99.0% probability as validated by the Protein Prophet algorithm21 and contained at least 2 identified peptides.
Flow cytometry, Fluo-4 staining, and Ca2+ measurements
HEK293T cells were transfected as described in “Cell preparation, transfection, lysis, immunoprecipitation, and pull-down experiments” using empty pmCherry vector or mCherry-RGS18 constructs. Cells were lifted by incubation in PBS at room temperature for 2 minutes, resuspended in DMEM supplemented with 10% FCS, and subsequently stained with 0.25μM Fluo-4 (Invitrogen) at 37°C for 25 minutes. The cells were then washed twice with serum-free medium. Flow cytometry and Ca2+ measurements were performed using an Accuri C6 flow cytometer, as described previously.22 Fluo-4– and mCherry-positive cells were identified by excitation with a 488-nm laser. The fluorescence was collected using 530 ± 15 (Fluo-4) and 675LP (mCherry) filters. Single cells showing both signals were selected for monitoring intracellular Ca2+ levels after stimulation with 0.1 U/mL of thrombin (Roche). Cells were stimulated after 1 minute in the cytometer and monitored for a total of 3 minutes. Changes in fluorescence, reflecting increases in intracellular Ca2+ concentration, were monitored over time. Samples were run as triplicates. Data from each sample were normalized to Fluo-4 loading according to F/F0, with F being fluorescence collected with the 530 ± 15 band-pass filter at any given time point and F0 being the fluorescence collected with the same filter before the addition of thrombin. Data from each experiment were further normalized to the maximum relative fluorescence after thrombin treatment of control (mCherry) transfected cells to account for variations in thrombin sensitivity of the cells during separate experiments.
Results
Identification of RGS18 as PKA and PKGI substrate
In previous studies, we generated a phospho-specific Ab against the PKA/PKG phosphorylation site (pS7) of Rap1GAP2.18 In platelets with elevated cGMP- or cAMP levels, we observed several bands in addition to Rap1GAP2. For example, treatment with the NO-donor SNP induced the phosphorylation of Rap1GAP2, as described previously.18 Three prominent bands appeared on this blot in addition to Rap1GAP2 at approximately 65, 50, and 30 kDa (Figure 1A). Interestingly, these proteins were not detected in resting platelets and followed similar time dependence as the Rap1GAP2 phosphorylation. We were able to identify the band at 50 kDa as VASP (data not shown) and established the Ab-recognition site as its PKA/G phosphorylation site pS239.23 Therefore, we hypothesized that the additional bands detected might represent genuine PKA and PKGI substrates. Using 2 independent purification approaches, immunoprecipitation with pS7 Ab and lysate fractionation followed by MS, we were able to identify the band at 30 kDa as the RGS18 protein. To determine whether RGS18 is indeed phosphorylated by PKA and recognized by the pS7-Rap1GAP2 specific Ab, cDNA of human RGS18 was cloned into a bacterial expression vector and purified GST-RGS18 was incubated with the catalytic subunit of PKA in the presence or absence of ATP. The pS7 Ab indeed recognized the PKA-phosphorylated form of RGS18 (Figure 1B).
RGS18 is phosphorylated on S216 by both PKA and PKGI. (A) Detection of novel phosphoproteins in platelets. Human platelets were treated with 10μM SNP for the indicated time points. Samples were then lysed, separated by SDS-PAGE, and immunoblotted with pS7-Rap1GAP2 Ab. In addition to pS7-Rap1GAP2, phosphorylated S239 of VASP, a prominent band at approximately 30kDa (P30), and a weaker band at 65 kDa (P65) were detected. (B) Phosphorylation of recombinant RGS18 by PKA. Equal amounts of purified GST-RGS18 were incubated with the catalytic subunit of PKA in the presence or absence of ATP. Samples were separated by SDS-PAGE and Western blots were incubated with pS7-Rap1GAP2 Ab. The Ab recognizes the PKA-phosphorylated form of GST-RGS18 (top panel, anti-pS7Rap1GAP2). Protein amounts of GST-RGS18 were verified using monoclonal RGS18 Ab (bottom panel, total RGS18). (C) Protein sequence alignment of the Rap1GAP2 peptide that served as an Ag in the generation of the phosphospecific Ab with a very similar region around S216 on RGS18. (D) Verification of S216 of RGS18 as the phosphorylation site for PKA. wt-RGS18, S216A-RGS18, and S218A-RGS18 GST constructs were incubated with the catalytic subunit of PKA in the presence of [γ-32P] ATP. Samples were separated by SDS-PAGE and Western blots were exposed to film to detect 32P signal (top panel). Protein amounts were verified by Western blotting using anti-GST Ab (bottom panel, total RGS18). (E) Verification of specificity of new pS216-RGS18 Ab. GST-RGS18 constructs were incubated with the catalytic subunit of PKA in the presence or absence of ATP. Samples were then subjected to SDS-PAGE and immunoblotted using the pS216-specific Ab (top panel, pS216 RGS18) and monoclonal RGS18 Ab as a loading control (bottom panel, total RGS18). (F) Phosphorylation of endogenous RGS18 in intact human platelets. Platelets were stimulated with PGI2 (1μM, 5 minutes), forskolin (10μM, 30 minutes), cAMP analog 5,6-di-chlorobenzimidazole riboside-3,5-cyclic monophosphorothioate Sp-isomer (300μM, 30 minutes), sodium nitroprusside (10μM, 10 minutes), GMP analog 8-(4-chlorophenylthio)guanosine-3,5-cyclic monophosphate (300μM, 20 minutes), thrombin (0.1 U/mL, 30 seconds), thromboxane mimetic U46619 (1μM, 1 minute), or ADP (10μM, 1 minute). Samples were then subjected to SDS-PAGE and immunoblotted using the pS216-specific Ab (top panel) and the monoclonal RGS18 Ab as a loading control (bottom panel). Shown are representative data of independent experiments performed at least 3 times.
RGS18 is phosphorylated on S216 by both PKA and PKGI. (A) Detection of novel phosphoproteins in platelets. Human platelets were treated with 10μM SNP for the indicated time points. Samples were then lysed, separated by SDS-PAGE, and immunoblotted with pS7-Rap1GAP2 Ab. In addition to pS7-Rap1GAP2, phosphorylated S239 of VASP, a prominent band at approximately 30kDa (P30), and a weaker band at 65 kDa (P65) were detected. (B) Phosphorylation of recombinant RGS18 by PKA. Equal amounts of purified GST-RGS18 were incubated with the catalytic subunit of PKA in the presence or absence of ATP. Samples were separated by SDS-PAGE and Western blots were incubated with pS7-Rap1GAP2 Ab. The Ab recognizes the PKA-phosphorylated form of GST-RGS18 (top panel, anti-pS7Rap1GAP2). Protein amounts of GST-RGS18 were verified using monoclonal RGS18 Ab (bottom panel, total RGS18). (C) Protein sequence alignment of the Rap1GAP2 peptide that served as an Ag in the generation of the phosphospecific Ab with a very similar region around S216 on RGS18. (D) Verification of S216 of RGS18 as the phosphorylation site for PKA. wt-RGS18, S216A-RGS18, and S218A-RGS18 GST constructs were incubated with the catalytic subunit of PKA in the presence of [γ-32P] ATP. Samples were separated by SDS-PAGE and Western blots were exposed to film to detect 32P signal (top panel). Protein amounts were verified by Western blotting using anti-GST Ab (bottom panel, total RGS18). (E) Verification of specificity of new pS216-RGS18 Ab. GST-RGS18 constructs were incubated with the catalytic subunit of PKA in the presence or absence of ATP. Samples were then subjected to SDS-PAGE and immunoblotted using the pS216-specific Ab (top panel, pS216 RGS18) and monoclonal RGS18 Ab as a loading control (bottom panel, total RGS18). (F) Phosphorylation of endogenous RGS18 in intact human platelets. Platelets were stimulated with PGI2 (1μM, 5 minutes), forskolin (10μM, 30 minutes), cAMP analog 5,6-di-chlorobenzimidazole riboside-3,5-cyclic monophosphorothioate Sp-isomer (300μM, 30 minutes), sodium nitroprusside (10μM, 10 minutes), GMP analog 8-(4-chlorophenylthio)guanosine-3,5-cyclic monophosphate (300μM, 20 minutes), thrombin (0.1 U/mL, 30 seconds), thromboxane mimetic U46619 (1μM, 1 minute), or ADP (10μM, 1 minute). Samples were then subjected to SDS-PAGE and immunoblotted using the pS216-specific Ab (top panel) and the monoclonal RGS18 Ab as a loading control (bottom panel). Shown are representative data of independent experiments performed at least 3 times.
Identification of RGS18-S216 as the PKA and PKGI phosphorylation site
We next wanted to identify the exact site on RGS18 that is phosphorylated by PKA and PKGI. Comparing the amino acid sequences of RGS18 and Rap1GAP2 revealed a region close to the C-terminus of RGS18 with high similarity to the phosphorylated peptide against which the pS7-Rap1GAP2 Ab was raised (Figure 1C). In addition, the scansite program (http://scansite.mit.edu/)24 predicted S216 on RGS18 as a putative PKA phosphorylation site. To test this prediction, we generated point mutants of RGS18 with S216 mutated to alanine. As a control, we mutated the neighboring S218 residue to alanine. In vitro phosphorylation experiments using purified proteins and isotope labeling confirmed that S216 is indeed the main PKA phosphorylation site on RGS18 (Figure 1D). Next, we generated a phosphospecific Ab against pS216 of RGS18. In vitro phosphorylation using GST-wt-RGS18, S216A, and S218A mutants confirmed the specificity of this Ab (Figure 1E). Treatment of washed platelets with PGI2, forskolin, SNP, a cAMP analog, or a cGMP analog led to S216 phosphorylation of RGS18 (Figure 1F). In contrast to this, thrombin, ADP, or U46619, a thromboxane mimetic, did not alter the phosphorylation state of S216 (Figure 1F). From these data, we conclude that PKA and PKGI phosphorylate endogenous RGS18 on S216 in human platelets.
Interaction of RGS18 and 14-3-3γ
ClustalW2 multiple sequence alignment25 suggests that the S216 PKA phosphorylation site is embedded within a short, highly conserved stretch of 7 amino acids (RRRSRSF; Figure 2A). Furthermore, the second serine residue in this sequence (S218) is predicted to be a mode I 14-3-3–binding site.24 14-3-3 proteins are small, phosphoserine/threonine-binding proteins that function as scaffolds and regulate key signaling components.26 To assess whether RGS18 can bind 14-3-3 at endogenous level in human platelets, we performed immunoprecipitations from human platelet lysate using a rabbit Ab generated against GST-RGS18 and immunoblotted with mouse Abs specific for the γ, β, ζ, and ϵ isoforms (Figure 2B and data not shown), which are believed to be the most abundantly expressed in platelets.17,27 Only 14-3-3γ was observed to interact consistently with RGS18 in thrombin-stimulated human platelets (Figure 2B lane 4). Control immunoprecipitation from thrombin-treated platelets showed the absence of 14-3-3γ (Figure 2B lane 1).
RGS18 interacts with 14-3-3 via S218 in a phosphorylation-dependent manner. (A) Alignment of RGS18 protein sequences from different species using ClustalW2 multiple sequence alignment. A conserved region comprising 7 amino acids is marked with a black border. This region contains both the identified PKA and PKGI phosphorylation site S216 and S218 and a 14-3-3–binding site. Uniprot accession numbers of sequences are: mouse, Q9NS28; rat, Q4L0E8; human, Q9NS28; frog, Q66IM3; and zebrafish, Q08BE2. (B) Endogenous interaction of 14-3-3 with RGS18 in platelets. Washed human platelets were treated without (−) or with either forskolin (10μM, 30 minutes) or thrombin (0.1 U/mL, 30 seconds). Platelets were then lysed and subjected to immunoprecipitation with rabbit anti-RGS18 Ab. As control, platelets were stimulated with thrombin, lysed, and subjected to immunoprecipitation with preimmune serum (ctr thrombin). After SDS-PAGE, Western blots were incubated with mouse anti–14-3-3γ Ab to detect total (middle panel, total 14-3-3γ) and precipitated 14-3-3γ (top panel, 14-3-3γ). Total RGS18 amounts were verified using rabbit anti-RGS18 Ab (bottom panel, total RGS18). Densitometric analysis of the blots of 3 independent experiments confirmed that the difference between control and RGS18 IP in the presence of thrombin was statistically significant (P < .05, data not shown). (C) Interaction of 14-3-3 and RGS18 in transfected cells. FLAG-RGS18 constructs were expressed in HEK293T cells. wt-RGS18, S216A-RGS18, S216E-RGS18, and S218A-RGS18 lysates were subjected to pull-down assays with purified GST-14-3-3γ. As a control, a separate FLAG-wt-RGS18 lysate was subjected to a pull-down assay using GST alone. After SDS-PAGE, Western blots of pull-downs and lysates were incubated with FLAG-Ab to detect precipitated RGS18 (top panel, RGS18) and total RGS18 (bottom panel, total RGS18). (D) Phosphorylation-dependent interaction of RGS18 and 14-3-3 in platelets. Platelets were lysed and subjected to pull-down assays with purified GST-14-3-3γ. Pull-downs were then incubated with λ protein phosphatase for 60 minutes at 30°C according to the manufacturer's protocol. After SDS-PAGE, Western blots of totals and pull-downs were incubated with mouse anti-RGS18 Ab to detect precipitated (top panel, RGS18) and total RGS18 (bottom panel, total RGS18). (E) Phospho-S218–dependent interaction of RGS18 and 14-3-3 in platelets. Platelets were lysed and supplemented with either none or 100μM dephospho (TNLRRRSRSFTVN) or phospho-peptide (TNLRRRSR(pS)FTVN), mimicking the 14-3-3 interaction site. Lysates were subsequently subjected to pull-down assays with purified GST-14-3-3γ. After SDS-PAGE, Western blots of lysates and pull-downs were incubated with mouse anti-RGS18 to detect precipitated (top panel, RGS18) or total (bottom panel, total RGS18) RGS18. Panels B through E are representative data of independent experiments performed at least 3 times.
RGS18 interacts with 14-3-3 via S218 in a phosphorylation-dependent manner. (A) Alignment of RGS18 protein sequences from different species using ClustalW2 multiple sequence alignment. A conserved region comprising 7 amino acids is marked with a black border. This region contains both the identified PKA and PKGI phosphorylation site S216 and S218 and a 14-3-3–binding site. Uniprot accession numbers of sequences are: mouse, Q9NS28; rat, Q4L0E8; human, Q9NS28; frog, Q66IM3; and zebrafish, Q08BE2. (B) Endogenous interaction of 14-3-3 with RGS18 in platelets. Washed human platelets were treated without (−) or with either forskolin (10μM, 30 minutes) or thrombin (0.1 U/mL, 30 seconds). Platelets were then lysed and subjected to immunoprecipitation with rabbit anti-RGS18 Ab. As control, platelets were stimulated with thrombin, lysed, and subjected to immunoprecipitation with preimmune serum (ctr thrombin). After SDS-PAGE, Western blots were incubated with mouse anti–14-3-3γ Ab to detect total (middle panel, total 14-3-3γ) and precipitated 14-3-3γ (top panel, 14-3-3γ). Total RGS18 amounts were verified using rabbit anti-RGS18 Ab (bottom panel, total RGS18). Densitometric analysis of the blots of 3 independent experiments confirmed that the difference between control and RGS18 IP in the presence of thrombin was statistically significant (P < .05, data not shown). (C) Interaction of 14-3-3 and RGS18 in transfected cells. FLAG-RGS18 constructs were expressed in HEK293T cells. wt-RGS18, S216A-RGS18, S216E-RGS18, and S218A-RGS18 lysates were subjected to pull-down assays with purified GST-14-3-3γ. As a control, a separate FLAG-wt-RGS18 lysate was subjected to a pull-down assay using GST alone. After SDS-PAGE, Western blots of pull-downs and lysates were incubated with FLAG-Ab to detect precipitated RGS18 (top panel, RGS18) and total RGS18 (bottom panel, total RGS18). (D) Phosphorylation-dependent interaction of RGS18 and 14-3-3 in platelets. Platelets were lysed and subjected to pull-down assays with purified GST-14-3-3γ. Pull-downs were then incubated with λ protein phosphatase for 60 minutes at 30°C according to the manufacturer's protocol. After SDS-PAGE, Western blots of totals and pull-downs were incubated with mouse anti-RGS18 Ab to detect precipitated (top panel, RGS18) and total RGS18 (bottom panel, total RGS18). (E) Phospho-S218–dependent interaction of RGS18 and 14-3-3 in platelets. Platelets were lysed and supplemented with either none or 100μM dephospho (TNLRRRSRSFTVN) or phospho-peptide (TNLRRRSR(pS)FTVN), mimicking the 14-3-3 interaction site. Lysates were subsequently subjected to pull-down assays with purified GST-14-3-3γ. After SDS-PAGE, Western blots of lysates and pull-downs were incubated with mouse anti-RGS18 to detect precipitated (top panel, RGS18) or total (bottom panel, total RGS18) RGS18. Panels B through E are representative data of independent experiments performed at least 3 times.
To identify the 14-3-3–binding site in RGS18, we expressed FLAG-wt-RGS18 and mutants in HEK293T cells and performed 14-3-3γ pull-down assays. Both wt-RGS18 and S216A-RGS18 strongly bound to GST-14-3-3γ, whereas no binding was detected in GST-only control experiments (Figure 2C). As predicted, mutation of S218 to alanine in RGS18 decreased 14-3-3 binding. Remarkably, mutation of S216 to glutamate, which mimics PKA/G phosphorylation, dramatically decreased binding of RGS18 to 14-3-3γ. To examine whether the interaction of RGS18 with 14-3-3γ is phospho-serine dependent, we performed GST-14-3-3γ pull-down assays from human platelets and subsequently treated samples with λ protein phosphatase. In contrast to the co-immunoprecipitation experiment (Figure 2B), an interaction between GST-14-3-3 and endogenous RGS18 was observed in the absence of thrombin (Figure 2D). This is most likely because of the high sensitivity of GST pull-down assays, which use an excess of GST-fusion protein to probe for binding. In phosphatase-treated samples, the interaction of 14-3-3γ and RGS18 was strongly reduced (Figure 2D). In addition, we performed GST-14-3-3γ pull-down assays from human platelets in the presence of peptides mimicking the phosphorylated or nonphosphorylated S218-binding site on RGS18. Only the phosphorylated peptide reduced the association of RGS18 and 14-3-3γ in pull-down assays (Figure 2E). These data confirm that 14-3-3γ interacts with RGS18 in a phosphorylation-dependent manner and that RGS18 exhibits a basal level of phosphorylation in resting platelets.
To further analyze the regulation of interaction between 14-3-3γ and RGS18, we performed co-immunoprecipitation studies in transfected cells. As observed in pull-down experiments, wt-RGS18 and S216A mutant were strongly associated with 14-3-3γ, whereas mutation of S216 to glutamate again strongly decreased binding (Figure 3A S216E). We did not study mutations of S218 to glutamate or aspartate because these mutations have been well established as not being able to mimic phosphorylated 14-3-3–binding sites.28 Mutation of S218 to alanine greatly decreased binding of 14-3-3 to RGS18; however, there was some residual binding detected, which prompted us to search for a potential secondary 14-3-3–binding site on RGS18. In RGS18, only the sequence surrounding S49 (KRNRLpSLL) displayed convincing evidence as a second potential 14-3-3 site, because amino acids at positions pS −5, −4, −2, and +1 match residues most commonly found in 14-3-3 motifs.28 Furthermore, S49 is highly conserved across species and, most importantly, S49 has already been described as being phosphorylated endogenously in human platelets in response to thrombin receptor–activating peptide stimulation.29 In co-immunoprecipitation studies, S49A mutation alone had a minor but significant effect on 14-3-3γ binding to RGS18 (Figure 3B densitometry from 7 independent experiments). These data indicate that RGS18 contains two 14-3-3–binding sites, one at pS218 and a second one at pS49. To examine the phosphorylation of RGS18 on S218 and S49, we used Phos-tag–supplemented gels.30 Phos-tag, in combination with Mn2+, displays preferential trapping of phosphorylated proteins in SDS-PAGE. This slows down the running velocity of phosphorylated proteins compared with their dephosphorylated counterparts: that is, phosphorylated proteins tend to appear as higher–molecular weight bands in Western blots.30 To obtain the basal, dephosphorylated state, wt-RGS18 expressing HEK293T lysate was treated with λ phosphatase and then Phos-tag SDS-PAGE and Western blotting were performed. Phosphatase treatment resulted in the appearance of a low–molecular weight band of RGS18 (Figure 3C lane 1 b). In contrast, 3 distinct bands of higher apparent molecular weight appeared in untreated wt-RGS18 samples, suggesting phosphorylation-induced shifts (Figure 3C lane 2 s1, s2, and s3). To investigate whether any of these shifts was caused by phosphorylation of the newly identified RGS18 phosphorylation sites, we analyzed lysates from S49A-, S216A-, S218A-, and S49A/S218A mutant-expressing cells. Mutation of S49 resulted in selective loss of the s3 band, whereas mutation of S216 resulted in selective loss of the s2 band (Figure 3C). Mutation of S218 induced a loss of s2 and s3 and the appearance of the dephosphorylated form b, whereas additional mutation of S49 did not result in a different pattern (Figure 3C). For an unequivocal interpretation of these patterns, it will be necessary to develop phosphorylation site–specific Abs against pS49 and the combination of pS216-pS218. However, 3 conclusions can be drawn from our results: (1) S49 phosphorylation contributes to the s3 shift, (2) S216 phosphorylation is involved in the s2 shift, and (3) S218 phosphorylation contributes to the s1 shift of RGS18. Most likely, the s3 band represents a combination of phosphorylated S49 and S218. These data confirm that S49 and S218 are constitutively phosphorylated in HEK293T cells, thus mediating binding of 14-3-3 to RGS18 (Figure 3A-B).
S49 of RGS18 contributes to binding of 14-3-3 to RGS18. (A) Evaluation of the RGS18 and 14-3-3 interaction in co-immunoprecipitation studies. The FLAG-RGS18 constructs (wt-RGS18, S49A-RGS18, S216A-RGS18, S216E-RGS18, S218A-RGS18, and S49A/S218A RGS18) were coexpressed with the myc-14-3-3γ construct in HEK293T cells, lysed, and immunoprecipitated using anti-FLAG–coupled beads. After SDS-PAGE, the Western blots of IPs were incubated with rabbit anti-myc Ab (top panel, 14-3-3) and reincubated with anti-FLAG Ab as a loading control (middle panel, RGS18). Lysate Western blots were incubated with anti-myc Ab as a loading control (bottom panel, total 14-3-3). Shown is a representative blot of independent experiments performed 7 times. A summary of the results obtained is shown in the densitometry panel. (B) Blots of experiments shown in panel A were analyzed by densitometry and data are expressed as means ± SEM representing 7 independent experiments. Statistical significance of relative 14-3-3 binding in relation to wt-RGS18 was detected using 1-way ANOVA in combination with the Tukey posttest (wt to S49A, **P < .01; wt to S216E, S218A, and S218A/49A, ****P < .0001). (C) Phos-tag analysis of overexpressed RGS18. FLAG-RGS18 constructs wt-RGS18, S49A-RGS18, S216A-RGS18, S218A-RGS18, and S49A/S218A RGS18 were expressed in HEK293T cells. Cells were lysed and subjected to Phos-tag–supplemented SDS-PAGE and Western blotting. An aliquot of wt-RGS18 lysate was incubated with λ phosphatase for 1 hour at 30°C before SDS-PAGE and Western blot analysis. Blots were then incubated with mouse anti-RGS18 Ab. Appearing bands were labeled as b, s1, s2, or s3 with increasing apparent molecular weight. (D) Phos-tag analysis of endogenous RGS18. Washed platelets were either unstimulated or incubated with forskolin (10μM, 30 minutes) or thrombin (0.1 U/mL, 30 seconds) before lysis and Phos-tag–supplemented SDS-PAGE and Western blots. An aliquot of nonstimulated platelet lysate was incubated with λ phosphatase for 1 hour at 30°C before SDS-PAGE and Western blot analysis. Blots were then incubated with mouse anti-RGS18 Ab. For appearance of band b in phosphatase-treated samples, a 30-minute exposure is shown (first lane), which is separated by a black line from a shorter exposure (5 minutes) of lanes 2 through 4 of the same Western blot. Blots shown in panels C and D are representative of 4 independent experiments.
S49 of RGS18 contributes to binding of 14-3-3 to RGS18. (A) Evaluation of the RGS18 and 14-3-3 interaction in co-immunoprecipitation studies. The FLAG-RGS18 constructs (wt-RGS18, S49A-RGS18, S216A-RGS18, S216E-RGS18, S218A-RGS18, and S49A/S218A RGS18) were coexpressed with the myc-14-3-3γ construct in HEK293T cells, lysed, and immunoprecipitated using anti-FLAG–coupled beads. After SDS-PAGE, the Western blots of IPs were incubated with rabbit anti-myc Ab (top panel, 14-3-3) and reincubated with anti-FLAG Ab as a loading control (middle panel, RGS18). Lysate Western blots were incubated with anti-myc Ab as a loading control (bottom panel, total 14-3-3). Shown is a representative blot of independent experiments performed 7 times. A summary of the results obtained is shown in the densitometry panel. (B) Blots of experiments shown in panel A were analyzed by densitometry and data are expressed as means ± SEM representing 7 independent experiments. Statistical significance of relative 14-3-3 binding in relation to wt-RGS18 was detected using 1-way ANOVA in combination with the Tukey posttest (wt to S49A, **P < .01; wt to S216E, S218A, and S218A/49A, ****P < .0001). (C) Phos-tag analysis of overexpressed RGS18. FLAG-RGS18 constructs wt-RGS18, S49A-RGS18, S216A-RGS18, S218A-RGS18, and S49A/S218A RGS18 were expressed in HEK293T cells. Cells were lysed and subjected to Phos-tag–supplemented SDS-PAGE and Western blotting. An aliquot of wt-RGS18 lysate was incubated with λ phosphatase for 1 hour at 30°C before SDS-PAGE and Western blot analysis. Blots were then incubated with mouse anti-RGS18 Ab. Appearing bands were labeled as b, s1, s2, or s3 with increasing apparent molecular weight. (D) Phos-tag analysis of endogenous RGS18. Washed platelets were either unstimulated or incubated with forskolin (10μM, 30 minutes) or thrombin (0.1 U/mL, 30 seconds) before lysis and Phos-tag–supplemented SDS-PAGE and Western blots. An aliquot of nonstimulated platelet lysate was incubated with λ phosphatase for 1 hour at 30°C before SDS-PAGE and Western blot analysis. Blots were then incubated with mouse anti-RGS18 Ab. For appearance of band b in phosphatase-treated samples, a 30-minute exposure is shown (first lane), which is separated by a black line from a shorter exposure (5 minutes) of lanes 2 through 4 of the same Western blot. Blots shown in panels C and D are representative of 4 independent experiments.
We next examined lysates of human platelets by Phos-tag SDS-PAGE and Western blotting. Platelets were stimulated with either forskolin or thrombin before lysis. An aliquot of nonstimulated platelet lysate was treated with λ phosphatase. Analogous to HEK293T cells, a low/basal band appeared (Figure 3D lane 1 long exposure b). Nonstimulated samples appeared at higher molecular weights relative to phosphatase-treated samples (Figure 3D lane 2 s1), indicating a phosphorylation-induced shift. Forskolin treatment of platelets resulted in the appearance of an s2 band (Figure 3D lane 3 s2). Thrombin treatment induced an s3 band of the highest apparent molecular weight (Figure 3D lane 4 s3). Together with the data obtained using RGS18 mutants in HEK293T cells (Figure 3C), we conclude that: (1) RGS18 is constitutively phosphorylated on S218 in platelets (the s1 band); (2) PKA activation leads to the phosphorylation of S216 (the s2 band), which confirms previous data shown in Figure 1F; and (3) thrombin treatment results in the phosphorylation of S49 of RGS18 (s3 band), which results in enhanced binding of 14-3-3 to RGS18 (Figures 2B and 4A). We conclude that phosphorylated S218 and S49 are the main 14-3-3–binding sites of RGS18 in platelets.
Regulation of RGS18/14-3-3γ interaction by S216 phosphorylation
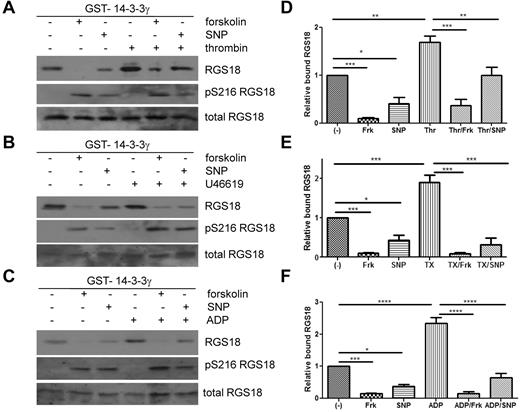
Because the glutamate mutation, mimicking phosphorylation of S216, resulted in decreased binding of RGS18 to 14-3-3γ in pull-down assays and in co-immunoprecipitation studies in HEK293T cells, we sought to determine the role of S216 phosphorylation for 14-3-3γ binding in human platelets. As shown in Figure 1F, S216 on RGS18 can be phosphorylated by stimulation of platelets with PGI2, forskolin, SNP, cAMP analog, or cGMP analog, but not with the GPCR agonists thrombin, ADP, and U46619. We therefore treated platelets with forskolin and SNP to induce S216 phosphorylation (Figure 4A-C middle panels) and performed GST-14-3-3γ pull-down experiments. Indeed, forskolin and SNP strongly reduced binding of RGS18 to 14-3-3γ in human platelets (Figure 4A-C). In contrast, thrombin, ADP, or U46619 potently stimulated the association of RGS18 and 14-3-3γ. When platelets were treated with forskolin or SNP before thrombin, ADP, or U46619 stimulation, 14-3-3γ binding was reduced (Figure 4A-C). Reduced 14-3-3γ binding was correlated with increased phosphorylation of RGS18 on S216. Densitometric analysis of data from repeat experiments confirmed that the effects of forskolin and SNP on basal and thrombin-, U46619-, or ADP-stimulated binding of 14-3-3γ to RGS18 were significant (Figure 4D-F). These data, together with the 14-3-3–binding studies of RGS18 mutants (Figure 3A-B) and Phos-tag analyses (Figure 3C-D), suggest that the increased interaction of RGS18 and 14-3-3γ occurs because of increased phosphorylation of S49. These data also suggest that PKA/PKG-mediated S216 phosphorylation has a dominant inhibitory effect on the interaction of RGS18 and 14-3-3γ.
Cyclic nucleotide signaling removes 14-3-3 from RGS18 in platelets. (A-C) Washed platelets were incubated without or with forskolin (10μM, 30 minutes) or SNP (10μM, 10 minutes), followed by the addition of thrombin (0.1 U/mL, 30 seconds; A), thromboxane mimetic U46619 (1μM, 1 minute (B), or ADP (10μM, 1 minute (C). Platelets were lysed and lysates were subjected to GST-14-3-3γ pull-down. After SDS-PAGE and blotting pull-down assays, samples were incubated with mouse anti-RGS18 Ab (top panel, RGS18), and lysates were either incubated with rabbit anti-RGS18 Ab (bottom panel, total RGS18) or rabbit anti–pS7-Rap1GAP2 Ab, which was shown in Figure 1A and B to recognize phosphorylated S216 of RGS18 (middle panel, pS216 RGS18). Shown are examples of experiments performed at least 3 times. (D-F) Blots of 3-5 independent experiments shown in panels A through C were analyzed by densitometry, and data are expressed as means ± SEM. (D) Densitometry of the thrombin experiment (panel A). (E) Thromboxane mimetic U46619 (panel B). (F) ADP data (panel C). Statistical significance of relative 14-3-3 binding was detected using 1-way ANOVA in combination with the Tukey posttest, as indicated. ****P < .0001; ***P < .001; **P < .01; *P < .05.
Cyclic nucleotide signaling removes 14-3-3 from RGS18 in platelets. (A-C) Washed platelets were incubated without or with forskolin (10μM, 30 minutes) or SNP (10μM, 10 minutes), followed by the addition of thrombin (0.1 U/mL, 30 seconds; A), thromboxane mimetic U46619 (1μM, 1 minute (B), or ADP (10μM, 1 minute (C). Platelets were lysed and lysates were subjected to GST-14-3-3γ pull-down. After SDS-PAGE and blotting pull-down assays, samples were incubated with mouse anti-RGS18 Ab (top panel, RGS18), and lysates were either incubated with rabbit anti-RGS18 Ab (bottom panel, total RGS18) or rabbit anti–pS7-Rap1GAP2 Ab, which was shown in Figure 1A and B to recognize phosphorylated S216 of RGS18 (middle panel, pS216 RGS18). Shown are examples of experiments performed at least 3 times. (D-F) Blots of 3-5 independent experiments shown in panels A through C were analyzed by densitometry, and data are expressed as means ± SEM. (D) Densitometry of the thrombin experiment (panel A). (E) Thromboxane mimetic U46619 (panel B). (F) ADP data (panel C). Statistical significance of relative 14-3-3 binding was detected using 1-way ANOVA in combination with the Tukey posttest, as indicated. ****P < .0001; ***P < .001; **P < .01; *P < .05.
Impact of RGS18/14-3-3γ interaction on Ca2+ signaling
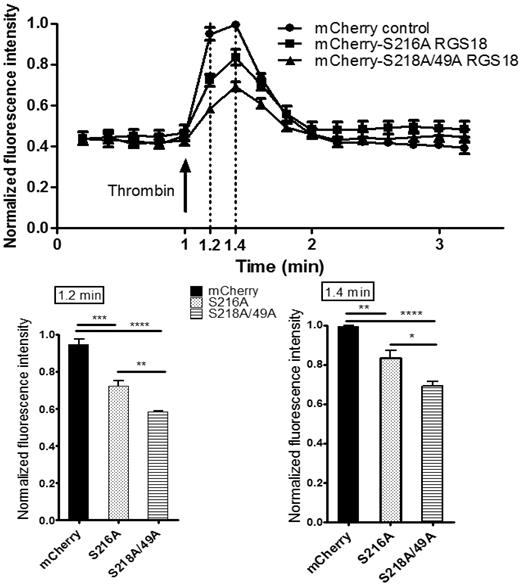
We next assessed the functional consequences of the RGS18-14-3-3γ interaction on downstream signaling events. RGS18 has been shown previously to activate the GTPase function of Gi and Gq.10-13 Gq signaling results in the release of Ca2+ from intracellular stores. To examine RGS18 effects on Gq signaling, we transfected pmCherry-tagged RGS18 constructs into HEK293T cells. We chose 2 different RGS18 constructs, one containing a point mutation of S216 to alanine, which binds 14-3-3 constitutively (Figure 3A-B), and one in which 14-3-3 binding cannot be inhibited by phosphorylation of S216 (Figure 4). The second construct had both 14-3-3–binding sites mutated to alanine (S49A/S218A), corresponding to 14-3-3–deficient RGS18 (Figure 3A-B). Both RGS18 constructs were able to bind Gq in co-immunoprecipitation studies (data not shown). After transfection, cells were stained with Fluo-4, a membrane-permeable dye that greatly increases its fluorescence after Ca2+ binding. Gq-mediated release of Ca2+ from intracellular stores was triggered using thrombin. Thrombin is known to activate Gq-mediated Ca2+ release in platelets via PAR1 receptors,31 and PAR1-dependent intracellular Ca2+ release in HEK293 cells has been described previously.32 The thrombin-induced increase in Fluo-4 fluorescence was monitored by flow cytometry based on the method of Vines et al.22 Only cells that displayed mCherry transfection were considered. As expected, both forms of RGS18 were able to inhibit Gq-mediated Ca2+ release relative to control mCherry-transfected cells. Remarkably, RGS18 deficient in 14-3-3 (the S49A/S218A mutant) was significantly more efficient in inhibiting Gq-mediated Ca2+ release compared with 14-3-3–bound RGS18 (P < .01 by ANOVA and the Bonferroni posttest; Figure 5). These findings suggest that 14-3-3 attenuates the function of RGS18, whereas removal of 14-3-3 enables RGS18 to inhibit Gq-mediated Ca2+ signaling more efficiently.
Removal of 14-3-3 activates RGS18 function. HEK293T cells were transfected with the mCherry-tagged RGS18 constructs S216A or S49A/S218A or the pmCherry vector as a control. Cells were then stained with 0.5μM Fluo-4, a Ca2+ dye, for 25 minutes at 37°C. Ca2+ responses and mCherry transfection efficiency measurements by flow cytometry were performed using an Accuri C6 flow cytometer. Fluo-4–stained single cells that contained Cherry constructs were visualized using a 488-nm laser and 530 ± 15/675LP filter combination. Cells were stimulated with 0.1 U/mL of thrombin (arrow) and intracellular Ca2+ changes were monitored as changes in Fluo-4 fluorescence. Data from 4 independent experiments performed in triplicate were normalized as indicated in “Methods” and are shown as means ± SEM. Thrombin stimulation was carried out at time point 1.0 minutes. Measurements taken at time points 1.2 and 1.4 minutes are shown as separate bar charts (bottom panels). Statistical analyses of data were performed using 1-way ANOVA in combination with the Bonferroni posttest. ****P < .0001; ***P < .001; **P < .01; *P < .05. mCherry-S216A-RGS18– and mCherry-S218/49A-RGS18–transfected cells display a significantly lower increase in Ca2+ compared with mCherry-transfected control cells (time point 1.2 minutes, P < .001 and P < .0001, respectively; time point 1.4 minutes, P < .01 and P < .0001, respectively). Comparing 14-3-3–deficient RGS18 (mCherry-S218/49A-RGS18) with 14-3-3–bound RGS18 (mCherry-S216A-RGS18), Ca2+ peaks are significantly lower in cells transfected with 14-3-3–deficient RGS18 (time point 1.2, P < .01; time point 1.4, P < .05).
Removal of 14-3-3 activates RGS18 function. HEK293T cells were transfected with the mCherry-tagged RGS18 constructs S216A or S49A/S218A or the pmCherry vector as a control. Cells were then stained with 0.5μM Fluo-4, a Ca2+ dye, for 25 minutes at 37°C. Ca2+ responses and mCherry transfection efficiency measurements by flow cytometry were performed using an Accuri C6 flow cytometer. Fluo-4–stained single cells that contained Cherry constructs were visualized using a 488-nm laser and 530 ± 15/675LP filter combination. Cells were stimulated with 0.1 U/mL of thrombin (arrow) and intracellular Ca2+ changes were monitored as changes in Fluo-4 fluorescence. Data from 4 independent experiments performed in triplicate were normalized as indicated in “Methods” and are shown as means ± SEM. Thrombin stimulation was carried out at time point 1.0 minutes. Measurements taken at time points 1.2 and 1.4 minutes are shown as separate bar charts (bottom panels). Statistical analyses of data were performed using 1-way ANOVA in combination with the Bonferroni posttest. ****P < .0001; ***P < .001; **P < .01; *P < .05. mCherry-S216A-RGS18– and mCherry-S218/49A-RGS18–transfected cells display a significantly lower increase in Ca2+ compared with mCherry-transfected control cells (time point 1.2 minutes, P < .001 and P < .0001, respectively; time point 1.4 minutes, P < .01 and P < .0001, respectively). Comparing 14-3-3–deficient RGS18 (mCherry-S218/49A-RGS18) with 14-3-3–bound RGS18 (mCherry-S216A-RGS18), Ca2+ peaks are significantly lower in cells transfected with 14-3-3–deficient RGS18 (time point 1.2, P < .01; time point 1.4, P < .05).
Discussion
The cyclic nucleotide signaling system is an essential regulator of platelet reactivity. Endothelial prostacyclin and nitric oxide trigger the synthesis of cAMP and cGMP in platelets, leading to activation of PKA and PKGI. The phosphorylation of specific PKA and PKGI substrates links cyclic nucleotides to inhibition of platelet function. Only few PKA and PKGI substrate proteins that may be capable of mediating these effects, among them Rap1GAP2, are known to date.18 In the present study, we describe RGS18 as new substrate of PKA and PKGI and establish functional outcomes of RGS18 phosphorylation at the molecular and cellular levels. Cross-reactivity of a phosphorylation site–specific Ab provided initial evidence for a new 30-kDa substrate of PKA and PKGI in platelets, and we were able to identify this protein by MS as RGS18. Mapping studies revealed S216 of RGS18 as the main PKA and PKGI phosphorylation site, and we monitored endogenous phosphorylation of S216 in response to activators of cAMP/cGMP signaling in intact platelets using a newly developed phospho-specific Ab. These experiments clearly establish RGS18 as new target of cAMP/cGMP pathways in platelets and provide another example of the convergence of PKA and PKGI signaling in platelets. The phosphorylated S216 residue is embedded in a highly conserved, RGS18-specific region of 7 amino acids close to the C-terminus, which we show to be involved in binding of 14-3-3γ to RGS18. We confirmed the interaction of RGS18 and 14-3-3γ at the endogenous level in human platelets and in transfected cells and mapped phosphorylated S218 on RGS18 as the primary 14-3-3–binding site. In addition, we identified phosphorylated S49 as a secondary 14-3-3–binding site. Both 14-3-3–binding sites are localized outside of the catalytic RGS domain of RGS18. This is in agreement with other examples of 14-3-3 interactions in which the 14-3-3 dimer straddles either side of a folded functional domain.28 Treatment of platelets with activatory GPCR ligands such as thrombin, ADP, and thromboxane analog further increases binding of 14-3-3 to RGS18, which is most likely mediated by additional phosphorylation of the S49-binding site as indicated by Phos-tag Western blotting (Figures 3C-D and 6). This is supported by the findings of Garcia et al,29 who observed that PAR-1 receptor signaling induces the phosphorylation of S49 of RGS18. Therefore, pS49 might represent the gatekeeper 14-3-3–binding site of RGS18, which does not have a high affinity for 14-3-3 in itself, but significantly augments 14-3-3 binding in the presence of pS218.33,34 The identity of the kinase(s) that phosphorylates S49 and S218 remains to be determined.
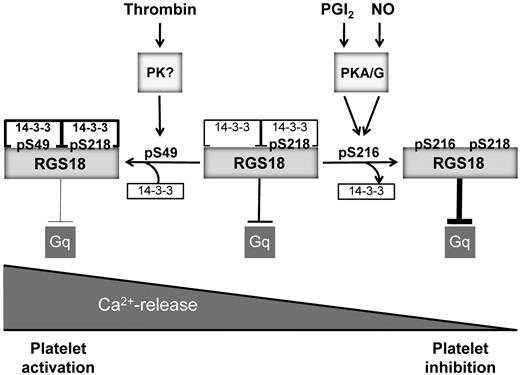
Model of RGS18 regulation during platelet activation and inhibition. RGS18 is a GTPase-activating protein of Gq and is constitutively bound to 14-3-3 via phosphorylated S218. Platelet activation by thrombin leads to the phosphorylation of S49 on RGS18 (pS49) by an unknown protein kinase. This results in increased RGS18 affinity to 14-3-3. RGS18 bound to 14-3-3 is a less efficient GTPase-activator of Gq, resulting in higher levels of active Gq-GTP, which stimulates release of calcium ions from intracellular stores (Ca2+-release), leading to platelet aggregation. Therefore, Gq signaling is facilitated during platelet activation. PGI2 and NO are released from endothelial cells and stimulate the activation of cAMP- and cGMP-dependent protein kinases (PKA/G). PKA and PKG phosphorylate RGS18 on S216 (pS216), leading to the detachment of 14-3-3 from RGS18. Detachment of 14-3-3 causes RGS18 to turn off Gq signaling more efficiently, resulting in decreased intracellular Ca2+ release and platelet inhibition. In this way, RGS18 is able to integrate activating and inhibitory signaling in platelets at the level of Gq.
Model of RGS18 regulation during platelet activation and inhibition. RGS18 is a GTPase-activating protein of Gq and is constitutively bound to 14-3-3 via phosphorylated S218. Platelet activation by thrombin leads to the phosphorylation of S49 on RGS18 (pS49) by an unknown protein kinase. This results in increased RGS18 affinity to 14-3-3. RGS18 bound to 14-3-3 is a less efficient GTPase-activator of Gq, resulting in higher levels of active Gq-GTP, which stimulates release of calcium ions from intracellular stores (Ca2+-release), leading to platelet aggregation. Therefore, Gq signaling is facilitated during platelet activation. PGI2 and NO are released from endothelial cells and stimulate the activation of cAMP- and cGMP-dependent protein kinases (PKA/G). PKA and PKG phosphorylate RGS18 on S216 (pS216), leading to the detachment of 14-3-3 from RGS18. Detachment of 14-3-3 causes RGS18 to turn off Gq signaling more efficiently, resulting in decreased intracellular Ca2+ release and platelet inhibition. In this way, RGS18 is able to integrate activating and inhibitory signaling in platelets at the level of Gq.
PKA and PKGI phosphorylation of S216 on RGS18 has a dominant-negative effect on binding of 14-3-3 to RGS18 in human platelets. Both basal and thrombin-, thromboxane A2-, or ADP-stimulated binding of 14-3-3 to RGS18 are abolished by S216 phosphorylation. As to the direct effect of S216 phosphorylation on 14-3-3 binding, 2 mechanisms can be envisioned: (1) that pS216 interferes directly with the interaction of 14-3-3 and RGS18 or (2) that pS216 induces the dephosphorylation of pS49 or pS218 or both. Preliminary evidence using Phos-tag gels suggests that S216 phosphorylation might induce some decrease in S49 and S218 phosphorylation (data not shown), although the exact consequences of S216 phosphorylation will need to be determined in future studies. PKA/PKG-induced detachment of 14-3-3 from its binding partner has been described previously for the GTPase-activating protein Rap1GAP2 in platelets. PKA- and PKGI-mediated phosphorylation of S7 of Rap1GAP2 triggered the removal of 14-3-3 from its binding site at phosphorylated S9, whereas thrombin and ADP treatment enhanced the phosphorylation of S9 and the association of Rap1GAP2 and 14-3-3.18 Another example of the inhibition of 14-3-3 binding by the phosphorylation of the −2 serine relative to the actual 14-3-3–binding site has been described for Cdc25B35 and Cdc25C36 involving CdkI and Cdc2 kinases.35,36
To understand the consequences of 14-3-3 binding for RGS18 function, we investigated downstream signaling events. In initial studies, we verified the interaction of RGS18 with Gq and Gi (data not shown) and confirmed that RGS18 blocks the Gq-mediated release of Ca2+ from intracellular stores. Removal of 14-3-3 from RGS18 significantly potentiated RGS18-mediated inhibition of Ca2+ release, suggesting increased turnover of Gq-GTP to inactive Gq-GDP. The exact mechanism of inhibition of RGS18 by 14-3-3 remains to be determined. Studies of other RGS family proteins have shown that 14-3-3 binds to RGS3, RGS4, RGS5, RGS7, RGS9-2, and RGS16.37-41 The general consequences of 14-3-3 binding are controversial: either no significant effects or inhibition of RGS activities have been observed.37-40 14-3-3 might interfere with the binding of RGS proteins to their Gα targets, and recent data on RGS3 suggest that 14-3-3 binding induces a conformational change in RGS3 that is likely to affect the interaction of RGS3 and Gα.42,43 Our studies on the interaction between RGS18 and Gi2 and Gq by co-immunoprecipitation from transfected cells showed a trend toward increased binding of the 14-3-3–deficient RGS18 mutant to Gi2 and Gq compared with wt-RGS18; however, this effect was not statistically significant (data not shown). Live microscopy of fluorescently labeled wt-RGS18 and 14-3-3–deficient mutants in transfected cells did not reveal any major difference in subcellular distribution (data not shown). Because RGS proteins are increasingly recognized as having multiple binding partners,44 14-3-3 could potentially regulate RGS18 by interfering with the binding of other currently unknown RGS18-interacting proteins. The PKA and PKGI substrate Rap1GAP2 is regulated in a manner similar to RGS18. Binding of 14-3-3 keeps Rap1GAP2 in an inactive state. Removal of 14-3-3 from Rap1GAP2 by PKA- and PKGI-mediated phosphorylation reduces cell adhesion, implicating increased GAP activity.18 Negative regulation of 14-3-3 binding might represent a common theme of cAMP/cGMP function in platelets that needs to be investigated in other PKA and PKGI substrates.
In summary, the results of the present study suggest that phosphorylation of RGS18 on S216 by PKA and PKGI effects the detachment of 14-3-3, resulting in a potentiation of RGS function (Figure 6). Consequently, Gq signaling is attenuated, leading to reduced receptor-mediated release of Ca2+ from intracellular stores. This mechanism might contribute to the inhibitory actions of cAMP and cGMP in platelets. Furthermore, phosphorylation of RGS18 on S49 during platelet activation stimulates the interaction of RGS18 and 14-3-3, resulting in reduced RGS18 function. Therefore, Gq signaling can be maintained during platelet activation, leading to enhanced release of Ca2+ from intracellular stores. The regulated interaction between RGS18 and 14-3-3 might represent a switch in the control of calcium signaling in platelets.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Access to and use of the University College Dublin Conway Institute Mass Spectrometry Resource instrumentation is gratefully acknowledged.
This work was supported by a Principal Investigator Program grant from Science Foundation Ireland (08/IN.1/B1855 to A.S.) and by the International Society for Advancement of Cytometry Scholar program.
Authorship
Contribution: All authors performed the experiments and analyzed the data; and K.G. and A.S. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Albert Smolenski, UCD Conway Institute, UCD School of Medicine and Medical Science, University College Dublin, Belfield, Dublin 4, Ireland; e-mail: albert.smolenski@ucd.ie.

![Figure 1. RGS18 is phosphorylated on S216 by both PKA and PKGI. (A) Detection of novel phosphoproteins in platelets. Human platelets were treated with 10μM SNP for the indicated time points. Samples were then lysed, separated by SDS-PAGE, and immunoblotted with pS7-Rap1GAP2 Ab. In addition to pS7-Rap1GAP2, phosphorylated S239 of VASP, a prominent band at approximately 30kDa (P30), and a weaker band at 65 kDa (P65) were detected. (B) Phosphorylation of recombinant RGS18 by PKA. Equal amounts of purified GST-RGS18 were incubated with the catalytic subunit of PKA in the presence or absence of ATP. Samples were separated by SDS-PAGE and Western blots were incubated with pS7-Rap1GAP2 Ab. The Ab recognizes the PKA-phosphorylated form of GST-RGS18 (top panel, anti-pS7Rap1GAP2). Protein amounts of GST-RGS18 were verified using monoclonal RGS18 Ab (bottom panel, total RGS18). (C) Protein sequence alignment of the Rap1GAP2 peptide that served as an Ag in the generation of the phosphospecific Ab with a very similar region around S216 on RGS18. (D) Verification of S216 of RGS18 as the phosphorylation site for PKA. wt-RGS18, S216A-RGS18, and S218A-RGS18 GST constructs were incubated with the catalytic subunit of PKA in the presence of [γ-32P] ATP. Samples were separated by SDS-PAGE and Western blots were exposed to film to detect 32P signal (top panel). Protein amounts were verified by Western blotting using anti-GST Ab (bottom panel, total RGS18). (E) Verification of specificity of new pS216-RGS18 Ab. GST-RGS18 constructs were incubated with the catalytic subunit of PKA in the presence or absence of ATP. Samples were then subjected to SDS-PAGE and immunoblotted using the pS216-specific Ab (top panel, pS216 RGS18) and monoclonal RGS18 Ab as a loading control (bottom panel, total RGS18). (F) Phosphorylation of endogenous RGS18 in intact human platelets. Platelets were stimulated with PGI2 (1μM, 5 minutes), forskolin (10μM, 30 minutes), cAMP analog 5,6-di-chlorobenzimidazole riboside-3,5-cyclic monophosphorothioate Sp-isomer (300μM, 30 minutes), sodium nitroprusside (10μM, 10 minutes), GMP analog 8-(4-chlorophenylthio)guanosine-3,5-cyclic monophosphate (300μM, 20 minutes), thrombin (0.1 U/mL, 30 seconds), thromboxane mimetic U46619 (1μM, 1 minute), or ADP (10μM, 1 minute). Samples were then subjected to SDS-PAGE and immunoblotted using the pS216-specific Ab (top panel) and the monoclonal RGS18 Ab as a loading control (bottom panel). Shown are representative data of independent experiments performed at least 3 times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/16/10.1182_blood-2011-11-390369/5/m_zh89991286760001.jpeg?Expires=1767731393&Signature=X4y97DZiC7jCaTqLPQN2Bvx2yvwEsa8E0JeS3egAfPoCIM-W791gzTPdLvCRio5hIQRLV6kavsFXHBo1krfiqIdxlZUC3QC-gQVvg-OpPSMANiFeZvnHozGKGGI-Af1PpePKRQWxiOSeUPaMS8bdKhEpFnxzA43heW88FlP4OktcSjSab6IA-9vEpCWc8GLKjM~9Gb1TPS4499G3qQvQamHUkv38WRDSRGgGFT3dTN47MeihSop-j4WTxh6e~TdhVeC~u6FlRFJmWW6kKCPwRXa~5eexW7DO03KMsS2lMtF9iTSD7Ocgp0-khwkqgMrIjxBbXyJVDtwfHiSX3R~v9g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal