Abstract
The β-globin locus control region (LCR) is necessary for high-level β-globin gene transcription and differentiation-dependent relocation of the β-globin locus from the nuclear periphery to the central nucleoplasm and to foci of hyperphosphorylated Pol II “transcription factories” (TFys). To determine the contribution of individual LCR DNaseI hypersensitive sites (HSs) to transcription and nuclear location, in the present study, we compared β-globin gene activity and location in erythroid cells derived from mice with deletions of individual HSs, deletions of 2 HSs, and deletion of the whole LCR and found all of the HSs had a similar spectrum of activities, albeit to different degrees. Each HS acts as an independent module to activate expression in an additive manner, and this is correlated with relocation away from the nuclear periphery. In contrast, HSs have redundant activities with respect to association with TFys and the probability that an allele is actively transcribed, as measured by primary RNA transcript FISH. The limiting effect on RNA levels occurs after β-globin genes associate with TFys, at which time HSs contribute to the amount of RNA arising from each burst of transcription by stimulating transcriptional elongation.
Introduction
Analysis of naturally occurring and targeted deletions of the endogenous β-globin locus control region (LCR) and of transgenic mice has revealed that the LCR is required for high-level β-globin transcription in erythroid cells.1-3 The LCR DNase1 hypersensitive sites (HSs) contain similar, albeit distinct sequence motifs, and vary in their activities in vitro and in transgenic mouse assays.2-10 Therefore, each HS may play a unique role in LCR function or may contribute a similar spectrum of activities. Transgenic mice bearing individual LCR HSs, combinations of HSs, or an intact LCR linked to the human β-globin locus have led to models in which the LCR HSs interact to form a holocomplex in which the individual HSs 1-4 act synergistically to ensure a permissive chromatin environment and activate expression, whereas 5′HSs 5 and 6 act as a chromatin barrier.5,11-19 In addition, it has been suggested that 5′HS2 has a dominant or unique activity compared with other LCR HSs. In contrast to these transgenic studies, our analysis of the transcriptional phenotypes of mice in which each LCR HS was deleted individually from the endogenous locus suggested that each site might contribute additively to LCR-mediated gene activation.20-22
The LCR has also been implicated in the localization of the β-globin locus during erythroid differentiation. During differentiation, the β-globin locus relocates away from the nuclear periphery and increasingly associates with foci of hyperphosphorylated Pol II “transcription factories” (TFys), where high-level transcription is activated.23 Deletion of the HSs that comprise the endogenous mouse β-globin LCR results in both decreased relocation of the locus to the nucleoplasm and association with TFys and less efficient Pol II elongation, with a reduction in mRNA accumulation to 1%-4% of normal.1,23,24 Whether relocation away from the periphery and/or association with TFys is directly dependent on LCR sequence or is a consequence of LCR-activated transcription is unknown.
We have generated a series of mice with variable transcriptional phenotypes by deleting combinations of 2 HSs from the endogenous LCR. Using primary erythroid cells derived from these mice, in the present study, we investigated the mechanisms by which the HSs contribute to the positioning of the locus and activation of β-globin gene expression.
Methods
Generation of mice with LCR deletions
Mice with LCR-targeted deletions of 2 HSs were generated using standard procedures on a Fred Hutchinson Cancer Research Center institutional animal care and use committee–approved protocol (Figure 1). Deletion of 5′HS1 and 5′HS2 (Δ1-2) and 5′HS2 and 5′HS3 (Δ2-3) were performed in a single step, whereas HS1 was deleted from mice containing a deletion of 5′HS4,20 resulting in mice with 5′HS1 and 4′HS1 deleted (Δ1,4) and 5′HS2 and 5′HS3 maintained. In each case, selectable markers were flanked by LoxP sites and excised before analysis by breeding with mice constitutively expressing Cre recombinase, followed by breeding out the Cre gene. Deletions were made on the HbbD allele of 129 mice. Because this is not the genome project reference strain, coordinates of the deletions were defined relative to the Ey cap site: Δ1-2, −4675 to −10 414; Δ2-3, −9259 to −17 702; and Δ1,4, −4675 to −6999 and −19 849 to −22 561.
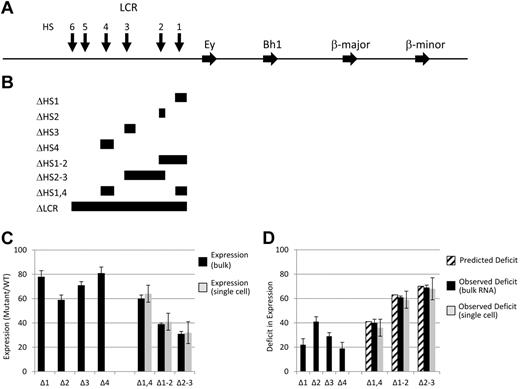
Map of LCR deletions obtained by homologous recombination and the expression analysis of the resulting mutant mice. (A) Map of the mouse β-globin locus. Blocks with arrowheads represent genes. Vertical arrows represent LCR HSs. (B) Diagrammatic representation of the targeted deletions generated and discussed in the text. Each block demonstrates the extent of the deletion listed on the left of the panel. Note that the Δ1-2 and Δ2-3 deletions remove all sequences from the 5′ border to the 3′ border of the corresponding single HS deletions, thus removing sequences between the 2 HSs. In contrast, the Δ1,4 deletion only removes the sequences present in the 2 single HS deletions. (C) Adult β-globin expression resulting from LCR HS deletions. The level of expression from a mutant allele relative to that of a WT allele in heterozygotic mice is presented as a percentage with the SD denoted (see “The β-globin LCR HSs contribute to transcription in an addictive, not synergistic manner” and Table 1 for details). Black bars are data from tissue RNA preparations and light bars are data from single-cell analyses. (D) Deficit in adult β-globin expression resulting from LCR HS deletions. The deficit in expression of a mutant allele compared with a WT allele in heterozygotic mice was calculated by subtracting the expression level (Figure 1C) from 100% and is presented with the SD (see “The β-globin LCR HSs contribute to transcription in an addictive, not synergistic manner” and Table 2 for details). Striped bars are the calculated deficit that would be observed in double HS deletion mice if HSs contribute to expression in an additive manner. Black bars are data from tissue RNA preparations and light bars are data from single-cell analyses.
Map of LCR deletions obtained by homologous recombination and the expression analysis of the resulting mutant mice. (A) Map of the mouse β-globin locus. Blocks with arrowheads represent genes. Vertical arrows represent LCR HSs. (B) Diagrammatic representation of the targeted deletions generated and discussed in the text. Each block demonstrates the extent of the deletion listed on the left of the panel. Note that the Δ1-2 and Δ2-3 deletions remove all sequences from the 5′ border to the 3′ border of the corresponding single HS deletions, thus removing sequences between the 2 HSs. In contrast, the Δ1,4 deletion only removes the sequences present in the 2 single HS deletions. (C) Adult β-globin expression resulting from LCR HS deletions. The level of expression from a mutant allele relative to that of a WT allele in heterozygotic mice is presented as a percentage with the SD denoted (see “The β-globin LCR HSs contribute to transcription in an addictive, not synergistic manner” and Table 1 for details). Black bars are data from tissue RNA preparations and light bars are data from single-cell analyses. (D) Deficit in adult β-globin expression resulting from LCR HS deletions. The deficit in expression of a mutant allele compared with a WT allele in heterozygotic mice was calculated by subtracting the expression level (Figure 1C) from 100% and is presented with the SD (see “The β-globin LCR HSs contribute to transcription in an addictive, not synergistic manner” and Table 2 for details). Striped bars are the calculated deficit that would be observed in double HS deletion mice if HSs contribute to expression in an additive manner. Black bars are data from tissue RNA preparations and light bars are data from single-cell analyses.
mRNA quantitation studies
mRNA quantitation studies were as described previously, including bulk and single-cell quantitative RT-PCR assays.1,25 Fetal livers were obtained at 13.5 days post coitum (dpc). For bulk RNA analyses, 6 animals were assayed. A restriction fragment length polymorphism–based assay was used to compare expression levels of the highly conserved mouse β-globin genes on the HbbD and HbbS alleles quantitatively.1,21
ChIP studies
ChIP studies and quantitation were performed as described previously using 13.5 dpc fetal livers.26 Antibodies (Abs) used were to SPT16 and to SPT5 (sc-28734 and sc-28678; Santa Cruz Biotechnology) and RNA polymerase II CTD of Pol II phosphorylated at serine-2 (CTD repeat YSPTSPS) (ab5095; Abcam). Samples were normalized to a necdin or actin control. ChIP and microscopy studies were performed using Abs to serine-2 and serine-5 phosphorylated CTD, respectively. The ChIPs were performed with the serine-2 Ab because in our experience, it is much more reproducible in ChIP experiments than the serine-5 Ab. The microscopy work was performed using the serine-5 Ab because it gives a more distinctive and reproducible staining pattern when combined with DNA FISH than the serine-2 Ab.
Imaging studies and flow cytometry
In all cases, mice homozygous for wild-type (WT) or LCR mutant loci were used. Fetal liver cells from at least 6 dpc 13.5 embryos were pooled and fractionated as described previously (with the exception of using CD-71hi staining instead of Ery-1).23 Costaining with CD-71 and Ery-1 yields a linear pattern with higher signal intensity of CD-71, which is consistent with CD-71 and Ery-1 both recognizing CD-71 but with varying affinities or titers. Four fractions of cells were isolated based on the following attributes: fraction 1, CD-117+CD-71−TER-119−; fraction 2, CD-117+CD-71+TER-119−; fraction 3, CD-117−CD-71+TER-119+; and fraction 4, CD-117−CD-71−TER-119+. Specific primary Abs used for immunostaining assays include goat anti–LaminB1 (sc-6217; Santa Cruz Biotechnology) and a rabbit polyclonal Ab to the CTD of Pol II phosphorylated at serine-5 (ab5131; Abcam). One to 3 independent hybridizations were performed on each sample. Primary transcript RNA FISH was carried out as described previously,23 except that α- and β-major transcripts were detected using single-stranded digoxin- and biotin-labeled DNA probes complementary to intron 2 of α-globin and β-major,27 followed by signal amplification with secondary Abs. DNA immuno-FISH was carried out as described previously.23 Secondary detection was performed with FITC-, Cy5-, or Cy3-conjugated anti–mouse, anti–goat, or anti–rabbit Abs (Jackson ImmunoResearch Laboratories and Invitrogen). Slides were mounted in 90% glycerol and PBS, pH 8.6, containing 2.5% wt/vol 1,4-diazabicyclo-[2,2,2]-octane (Sigma-Aldrich). Samples were imaged at room temperature on an inverted IX71 microscope (Olympus) with an Olympus 100×/1.40, Plan S Apo oil objective and equipped with a cooled Photometrics CoolSNAP HQ scientific grade CCD camera using Deltavision softWoRx software Version 3.5.1 (Applied Precision). After deconvolution of the image stacks, RNA FISH spots were counted on 2D projections, and DNA FISH results were analyzed in 3D (softWoRx Version 3.5.1). For the localization analysis, β-globin loci were considered to be associated with the lamina if a FISH spot overlapped at least partially with LaminB1 signal. Conversely, β-globin loci were scored as associated with TFys if FISH spots overlapped with Pol II foci that displayed a signal intensity of at least 20% of the brightest TFy in each analyzed cell.
Results
The β-globin LCR HSs contribute to transcription in an additive, not synergistic, manner
If LCR HSs contribute to β-globin expression additively, then the transcriptional phenotypes of the single HS deletions would predict the effect of deleting multiple HSs. For example, deletions of 5′HS2 and 5′HS3 led to decreases in adult expression to 59% and 71% of WT levels, respectively (ie, deficits of 41% and 29%, respectively; Figure 1C-D and Tables 1 and 2). If an allele contained a deletion of 5′HS2 through 5′HS3, we would predict a deficit of 70% or a decrease in expression to 30% compared with a WT allele in heterozygotic mice. In contrast, synergism between HSs would be reflected by a larger decrease in expression, whereas redundancy would be reflected by a lesser effect. Three lines of mice were generated, each with deletions of 2 HSs: 5′HS1 and 5′HS2 (Δ1-2), 5′HS2 and 5′HS3 (Δ2-3), and 5′HS1 and 5′HS4 (Δ1,4). In each case, the 5′ and 3′ extent of the deletions were identical to those of the single-site deletions described previously and selectable markers were excised, leaving a single LoxP site at the junction (Figure 1A-B).20-22 This set of deletions was chosen to reflect the reported hierarchy of HS activity on transcription (5′HS2 > 5′HS3 > 5′HS4 > 5′HS1). Therefore, Δ2-3 removes the 2 most active HSs, whereas these sites are the only active HSs retained in Δ1,4. In addition, Δ1,2 retains 5′HS3 and 5′HS4, permitting comparison of the influence of a strong (5′HS2) or relatively weak (5′HS4) HS on the function of another active HS (5′HS3). To quantify the ratio of expression from each mutant allele to that from a WT allele precisely, we generated adult mice heterozygotic for each deletion on a HbbD allele and a WT HbbS allele. Blood was analyzed using a quantitative RT-PCR assay that exploits a restriction fragment length polymorphism between the HbbD and HbbS cDNA.1,21 The observed ratio of expression from the mutant HbbD allele to that of the internal control WT HbbS allele is shown in Figure 1C and D and Table 2. Strikingly, the Δ2-3 deletion leads to a decrease in adult β-gene activity to 32% of WT levels, indistinguishable from the 30% predicted if individual LCR HSs acted additively. Similarly, the values observed with the Δ1-2 and Δ1,4 mice are indistinguishable from that predicted if each HS contributed in an additive manor.
Effect of single HS deletions on expression
| LCR HS deleted . | Mutant/WT expression . | Deficit in expression . |
|---|---|---|
| Δ1 | 0.78 ± 0.05 | 0.22 |
| Δ2 | 0.59 ± 0.04 | 0.41 |
| Δ3 | 0.71 ± 0.03 | 0.29 |
| Δ4 | 0.81 ± 0.05 | 0.19 |
| Δ5-6 | 0.97 ± 0.09 | 0.03 |
| LCR HS deleted . | Mutant/WT expression . | Deficit in expression . |
|---|---|---|
| Δ1 | 0.78 ± 0.05 | 0.22 |
| Δ2 | 0.59 ± 0.04 | 0.41 |
| Δ3 | 0.71 ± 0.03 | 0.29 |
| Δ4 | 0.81 ± 0.05 | 0.19 |
| Δ5-6 | 0.97 ± 0.09 | 0.03 |
Values are expressed as a percentage of each messenger RNA relative to WT values followed by the SD.
Predicted and observed ratio of mutant to WT expression in double HS deletion mice
| Double HS mutation . | Predicted deficit . | Predicted ratio . | Observed ratio, bulk RNA . | Observed ratio, single cells . |
|---|---|---|---|---|
| Δ1,4 | 0.41 | 0.59 | 0.60 ± 0.03 | 0.64 ± 0.07 |
| Δ1-2 | 0.63 | 0.37 | 0.39 ± 0.01 | 0.41 ± 0.07 |
| Δ2-3 | 0.70 | 0.30 | 0.31 ± 0.02 | 0.32 ± 0.09 |
| Double HS mutation . | Predicted deficit . | Predicted ratio . | Observed ratio, bulk RNA . | Observed ratio, single cells . |
|---|---|---|---|---|
| Δ1,4 | 0.41 | 0.59 | 0.60 ± 0.03 | 0.64 ± 0.07 |
| Δ1-2 | 0.63 | 0.37 | 0.39 ± 0.01 | 0.41 ± 0.07 |
| Δ2-3 | 0.70 | 0.30 | 0.31 ± 0.02 | 0.32 ± 0.09 |
The predicted deficit is the sum of the deficits of the two individual HSs deleted (from Table 1). The predicted ratio is calculated from the predicted deficit. The observed ratio (bulk RNA) is the experimentally determined average ratio of expression of the mutant allele to the WT allele from 6 mice, followed by the SD. The observed ratio (single cells) is the ratio of expression of the mutant allele to the WT allele, followed by the SD.
The LCR HSs increase β-globin gene expression uniformly rather than in a binary manner
We have determined previously that deletion of the LCR (ΔLCR) resulted in each allele being expressed at a low, basal level, in contrast to WT alleles, which uniformly expressed at a high level.1 The intermediate levels of expression obtained in bulk populations of reticulocytes from the double HS deletion mice could be explained by either model or a combination of 2 models. In a binary model, HSs would increase the probability that an allele was activated to a full extent. Therefore, in the Δ2-3 deletion mice, approximately 30% of alleles would be active at WT levels (jackpot cells), with the remainder expressing at basal levels similar to the ΔLCR allele (null cells). In contrast, in a rheostat model, the presence of HSs would lead to a uniform increase in expression of each allele above the basal levels observed with the LCR deletion and to approximately 30% of WT levels. Individual reticulocytes were analyzed to distinguish between these models.25 For each mutation, all individual cells examined expressed the mutant allele at the same level as observed in our analysis of bulk populations (Figure 1C-D and Table 2), and no null or jackpot cells were observed. Therefore, the HSs do not affect the number of alleles permissive for activated expression. Rather, all alleles are in a permissive state and the LCR HSs act to “ramp up” the level of accumulated mRNA from each to a homogeneous level intermediate between WT and ΔLCR.
LCR HSs increase the probability that the β-globin gene is transcribed and the amount of mRNA per burst of transcription
Whereas the single-cell RT-PCR assays quantitate output from individual alleles, the long half-life of β-globin RNA (hours to days) precludes assessment of transcriptional output at a specific point in time.28,29 In contrast, studies of nascent transcripts, with half-lives of minutes, have determined that most genes are not expressed continuously but rather in intermittent bursts of transcription.30,31 The probability that a gene is actively transcribed can be determined by primary transcript FISH, in which each foci of staining identifies a burst of ongoing transcription. To determine whether the intermediate levels of stable mRNA expression observed in the double HS deletions were due to a decreased probability of the β-globin locus being actively transcribed, primary transcript FISH was performed on fractions of mouse fetal liver cells that vary in their degree of erythroid maturation. The ratio of actively transcribing WT or mutant β-globin alleles to WT α-globin alleles was determined in homozygous WT and homozygous mutant LCR mice. Analysis was performed on CD-117−CD-71+TER-119+ cells (fraction 3), a mixture of proerythroblasts and basophilic erythroblasts making up 50%-80% of fetal liver cells, and CD-117−CD-71−TER-119+ cells (fraction 4), a mixture of normoblasts and polychromatic erythrocytes making up 5%-10% of fetal liver cells (Figure 2A-B). Fractions of more immature cells were not studied due the low number of active alleles.
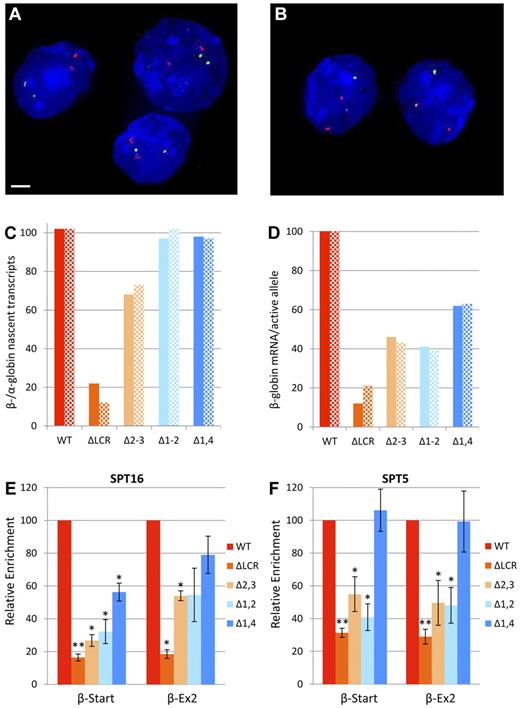
Expression and elongation studies in cells homozygous for WT or LCR mutant β-globin loci. (A-D) Expression of α- and β-globin from WT and mutant alleles during erythroid differentiation by primary transcript RNA FISH. Fetal liver cells from at least 6 homozygous WT or homozygous mutant fetuses were fractionated by flow cytometry and probed for α-globin (red) and β-major primary transcripts (green) by RNA FISH, followed by counterstaining of nuclei by DAPI staining (blue). Scale bar indicates 2 μm. Representative images from WT mice of fraction 3 (A) and fraction 4 (B) are shown. (C) Ratio of β-globin to α-globin nascent transcripts. Values are the ratio of β-globin to α-globin alleles with bursts of active transcription (foci of nascent transcripts) shown as a percentage. Solid bars represent values for fraction 3 and hatched bars are fraction 4. More than 100 cells were analyzed for each data point. (D) β-globin mRNA per active allele. To obtain an estimate of the relative mRNA production from each burst of active transcription, we compared the effect of each mutation on mRNA production (Table 2) with its effect on bursts of transcription (Figure 2C). Values are the ratio of the percentage of WT β-globin mRNA accumulation for each mutation to the percentage of WT alleles associated with foci of nascent transcripts. Solid bars represent data using fraction 3 and hatched bars are fraction 4. Values are divided by 1.02 so that WT yields 100%. (E-F) Recruitment of elongation components to the β-globin locus in erythroid cells from mice homozygous for LCR HS deletions normalized to WT. ChIP was performed on WT and LCR mutant chromatin using Abs to FACT component SPT16 (E) and DSIF component SPT5 (F). Three different chromatin preparations were analyzed by quantitative RT-PCR of the β-major globin start site (β-Start) and exon-2 of β-major (β-Ex2) regions and normalized to necdin, which is not transcribed in these cells. Values for each mutant line were normalized to the WT line (*P < .05 and **P < .001 relative to WT). The same data without normalization to WT is shown in supplemental Figure 1A and B. Error bars represent the SEM.
Expression and elongation studies in cells homozygous for WT or LCR mutant β-globin loci. (A-D) Expression of α- and β-globin from WT and mutant alleles during erythroid differentiation by primary transcript RNA FISH. Fetal liver cells from at least 6 homozygous WT or homozygous mutant fetuses were fractionated by flow cytometry and probed for α-globin (red) and β-major primary transcripts (green) by RNA FISH, followed by counterstaining of nuclei by DAPI staining (blue). Scale bar indicates 2 μm. Representative images from WT mice of fraction 3 (A) and fraction 4 (B) are shown. (C) Ratio of β-globin to α-globin nascent transcripts. Values are the ratio of β-globin to α-globin alleles with bursts of active transcription (foci of nascent transcripts) shown as a percentage. Solid bars represent values for fraction 3 and hatched bars are fraction 4. More than 100 cells were analyzed for each data point. (D) β-globin mRNA per active allele. To obtain an estimate of the relative mRNA production from each burst of active transcription, we compared the effect of each mutation on mRNA production (Table 2) with its effect on bursts of transcription (Figure 2C). Values are the ratio of the percentage of WT β-globin mRNA accumulation for each mutation to the percentage of WT alleles associated with foci of nascent transcripts. Solid bars represent data using fraction 3 and hatched bars are fraction 4. Values are divided by 1.02 so that WT yields 100%. (E-F) Recruitment of elongation components to the β-globin locus in erythroid cells from mice homozygous for LCR HS deletions normalized to WT. ChIP was performed on WT and LCR mutant chromatin using Abs to FACT component SPT16 (E) and DSIF component SPT5 (F). Three different chromatin preparations were analyzed by quantitative RT-PCR of the β-major globin start site (β-Start) and exon-2 of β-major (β-Ex2) regions and normalized to necdin, which is not transcribed in these cells. Values for each mutant line were normalized to the WT line (*P < .05 and **P < .001 relative to WT). The same data without normalization to WT is shown in supplemental Figure 1A and B. Error bars represent the SEM.
In cells actively transcribing at least one α- or β-globin allele, α-globin and WT β-globin alleles were actively transcribed to the same extent (Figure 2C). In contrast, only 12%-22% of ΔLCR alleles had detectable β-globin transcripts. Whereas the Δ1-2 and Δ1,4 deletions had no detectable effect on the number of actively transcribed alleles, the Δ2-3 deletion decreased the number of active bursts to 68%-73% of WT. For each mutation, the reduction in stable mRNA was substantially larger than the reduction in actively transcribing β-globin genes, suggesting that LCR deletions influence the amount of mRNA emanating from each burst of transcription. To obtain an estimate of this decrease, the relative level of accumulated β-globin transcripts compared with WT (Table 2) and the relative number of alleles with bursts of nascent transcripts (Figure 2C) were compared. For each mutation, the decrease in stable RNA accumulation is large compared with the decrease in the number of alleles actively expressing (Figure 2D). Alhough indirect, this result strongly suggests that each of the LCR mutations results in decreased expression per burst of transcription. A direct comparison of the signal intensity of nascent transcripts between WT and mutant alleles was attempted but was not possible because, in control studies, the signal between 2 WT alleles in single cells varied more than 2-fold in half of the cells analyzed (T.R., unpublished observation). Therefore, the single-cell RT-PCR experiments demonstrate that all double deletion HS alleles are permissive for expression over time, and primary transcript FISH reveals that the probability of the β-globin gene being actively transcribed is relatively insensitive to these deletions. In contrast, the decrease in accumulated mRNA observed in the LCR mutants is primarily because of the amount of primary transcripts generated during bursts of active transcription.
LCR HSs recruit factors essential for efficient elongation
We have shown previously that deletion of the entire LCR resulted in a significant decrease in transcription, but a minor decrease in pre-initiation complex formation, suggesting the primary role of the LCR is downstream of pre-initiation complex formation.24 Consistent with this, loss of the LCR results in a profound decrease in elongation complexes, implicating the LCR in releasing paused polymerases and increasing the efficiency of elongation.24,26 To determine whether the variable amount of mRNA emanating from a burst of transcription is correlated with the presence of factors necessary for efficient elongation, ChIP was performed, followed by quantitative RT-PCR. The facilitates chromatin transcription (FACT) complex acts as a histone chaperone during elongation and facilitates p-TEFb recruitment.32,33 Enrichment of the FACT component SPT16 parallels the levels of mRNA for each mutant, with the Δ1,4 mutation having the least effect and the 2 5′HS2–containing deletions having greater effects (Figure 2E). The 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole sensitivity inducing factor complex (DSIF) facilitates polymerase pausing, but, after phosphorylation of its SPT5 component by P-TEFb, switches and promotes elongation.34-36 Therefore, SPT5 enrichment in the body of the gene is a marker of efficient elongation. In the Δ1,4 mutation, which has the least effect on mRNA accumulation, DSIF enrichment was similar to that of WT (Figure 2F). In contrast, the 2 5′HS2–containing double mutations, Δ1-2 and Δ2-3, decreased enrichment to levels similar to that observed with deletion of the full LCR. In both WT and LCR mutant mice, the absolute levels of SPT5 occupancy decrease with progression from the β-globin start to exon 2, which is consistent with accumulation of DSIF-mediated paused polymerases at the 5′ end of the gene and DSIF positively influencing elongation downstream (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In contrast, SPT16 occupancy is similar at the start site and exon 2, which is consistent with its sole role as a facilitator of elongation (supplemental Figure 1B). The reduction in components of 2 different elongation complexes in the LCR mutants with the amount of mRNA per burst of nascent transcripts suggests that essential LCR HS functions include increasing elongation efficiency through the recruitment of DSIF and FACT.
Relocalization away from the nuclear periphery is correlated with level of transcription, whereas association with transcription factories occurs at any level of HS-dependent transcription
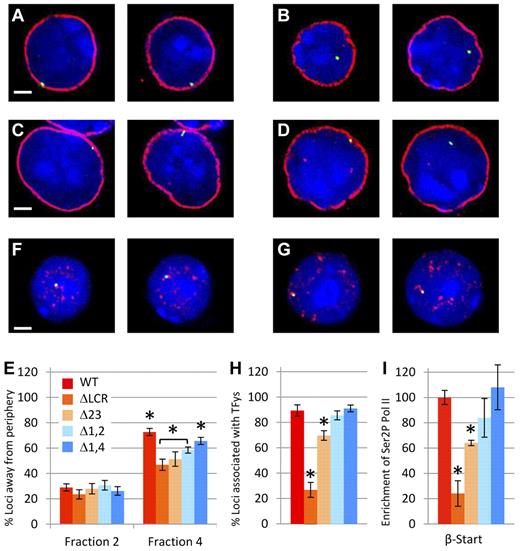
During erythroid maturation, the LCR is essential for both efficient localization of the β-globin locus from the nuclear periphery and to foci of serine-5–phosphorylated RNA polymerase II (TFys).23 The latter may entail relocalization to a preexisting TFy or the nucleation of a new TFy. We sought to distinguish between 2 models linking the LCR to nuclear positioning. In the first, the presence of any LCR HS or any degree of LCR-mediated activation is sufficient for efficient association with TFys and/or localization away from the nuclear periphery. Alternatively, increasing numbers of HSs or expression is directly correlated with increasing localization to the nucleoplasm and TFys. To distinguish between these models, fractionated fetal liver cells from homozygous WT and homozygous mutant mice were analyzed by immuno-FISH (Figure 3A-D and F-G). We showed previously that fraction 2 consisted of cells in which relocation away from the nuclear periphery and activation of expression is rarely detectable.23 Figure 3E demonstrates that before the initiation of β-globin expression, the 4 mutant lines show the same lack of localization away from the periphery as WT in fraction 2. In contrast, in fraction 4, which contains the most mature erythroid cells, the degree of relocalization of each mutant β-globin locus to the nucleoplasm is decreased compared with WT and is correlated with level of mRNA accumulation. Whereas relocalization away from the nuclear periphery is sensitive to loss of LCR HSs, association with TFys is relatively resistant. The Δ1,4 and Δ1-2 mutations do not affect association with TFys despite decreases in expression to 40% of normal (Figure 3H). Although the Δ2-3 mutation does decrease association with TFys, it is to a small extent compared with the reduction in expression.
Nuclear localization studies in cells homozygous for WT or LCR mutant β-globin loci. (A-E) Relocalization of the WT and mutant β-globin loci away from the nuclear periphery during erythroid differentiation. Fetal liver cells from at least 6 homozygous WT or homozygous mutant fetuses were fractionated by flow cytometry and probed by DNA FISH (green), followed by immunostaining of LaminB1 (red) to define the nuclear periphery and counterstaining of nuclei by DAPI staining (blue). Scale bar indicates 2 μm. Representative images from WT and Δ1-2 cells of fraction 2 (A,C) and fraction 4 (B,D) are shown. Images are single z-sections, so the diameter of the nucleus varies depending on the level of the slice. (E) Percentage of β-globin loci located away from the nuclear periphery in immature (fraction 2) and mature (fraction 4) erythroid cells. In fraction 2, no significant differences between WT and LCR HS mutants are present. In contrast, in fraction 4, significantly more WT alleles are located away from the nuclear periphery than for each of LCR HS-deleted lines (*P < .05). Values also vary significantly between Δ1,4 and all other mutants (P < .05) and between the ΔLCR and Δ1-2 mutations (bracketed and shown by asterisks). More than 100 loci were analyzed for each data point. Error bars represent the SEM. (F-I) Association of WT and β-globin loci to foci of hyperphosphorylated Pol II. (F-H) Fetal liver cells from at least 6 homozygous WT or homozygous mutant fetuses were fractionated by flow cytometry and probed by DNA FISH (green), followed by immunostaining of phospho-Pol II (red) to define transcription factories and counterstaining of nuclei by DAPI staining (blue). Scale bar indicates 2 μm. Representative images from WT (F) and Δ1-2 (G) cells are shown. Both alleles are shown in their respective z-sections. (H) Percentage of β-globin loci overlapping with TFys in mature erythroid cells (fraction 4). Association frequency of the ΔLCR and Δ2-3 mutant alleles with TFys differs significantly from all other genotypes (*P < .05). WT, Δ1-2, and Δ1,4 did not differ significantly from each other. More than 50 loci were analyzed for each data point. Error bars represent the SEM. (I) Recruitment of hyperphosphorylated Pol II to the β-globin gene in erythroid cells from mice homozygous for LCR HS deletions normalized to WT. ChIP was performed on WT and LCR mutant chromatin using a phosphorylated form of PolII. Three different chromatin preparations were analyzed by quantitative RT-PCR of the β-major globin start site (β-Start) and normalized to actin. Values for each mutant line were normalized to the WT line (*P < .05 relative to WT).
Nuclear localization studies in cells homozygous for WT or LCR mutant β-globin loci. (A-E) Relocalization of the WT and mutant β-globin loci away from the nuclear periphery during erythroid differentiation. Fetal liver cells from at least 6 homozygous WT or homozygous mutant fetuses were fractionated by flow cytometry and probed by DNA FISH (green), followed by immunostaining of LaminB1 (red) to define the nuclear periphery and counterstaining of nuclei by DAPI staining (blue). Scale bar indicates 2 μm. Representative images from WT and Δ1-2 cells of fraction 2 (A,C) and fraction 4 (B,D) are shown. Images are single z-sections, so the diameter of the nucleus varies depending on the level of the slice. (E) Percentage of β-globin loci located away from the nuclear periphery in immature (fraction 2) and mature (fraction 4) erythroid cells. In fraction 2, no significant differences between WT and LCR HS mutants are present. In contrast, in fraction 4, significantly more WT alleles are located away from the nuclear periphery than for each of LCR HS-deleted lines (*P < .05). Values also vary significantly between Δ1,4 and all other mutants (P < .05) and between the ΔLCR and Δ1-2 mutations (bracketed and shown by asterisks). More than 100 loci were analyzed for each data point. Error bars represent the SEM. (F-I) Association of WT and β-globin loci to foci of hyperphosphorylated Pol II. (F-H) Fetal liver cells from at least 6 homozygous WT or homozygous mutant fetuses were fractionated by flow cytometry and probed by DNA FISH (green), followed by immunostaining of phospho-Pol II (red) to define transcription factories and counterstaining of nuclei by DAPI staining (blue). Scale bar indicates 2 μm. Representative images from WT (F) and Δ1-2 (G) cells are shown. Both alleles are shown in their respective z-sections. (H) Percentage of β-globin loci overlapping with TFys in mature erythroid cells (fraction 4). Association frequency of the ΔLCR and Δ2-3 mutant alleles with TFys differs significantly from all other genotypes (*P < .05). WT, Δ1-2, and Δ1,4 did not differ significantly from each other. More than 50 loci were analyzed for each data point. Error bars represent the SEM. (I) Recruitment of hyperphosphorylated Pol II to the β-globin gene in erythroid cells from mice homozygous for LCR HS deletions normalized to WT. ChIP was performed on WT and LCR mutant chromatin using a phosphorylated form of PolII. Three different chromatin preparations were analyzed by quantitative RT-PCR of the β-major globin start site (β-Start) and normalized to actin. Values for each mutant line were normalized to the WT line (*P < .05 relative to WT).
Because these FISH studies used a 150-kb bacterial artificial chromosome probe, association with a TFy may not fairly represent association with the β-globin gene itself. Therefore, we performed ChIP with a serine-2–phosphorylated PolII Ab to analyze association of the β-globin gene (start site probe) with TFys directly. Similar to our microscopy studies, the ChIP analysis (Figure 3I) revealed that the Δ1,4 and Δ1-2 mutations do not significantly affect phosphorylated PolII at the β-globin gene, whereas the Δ2-3 mutation has a mild but significant effect and the ΔLCR allele is more profoundly affected (compare Figure 3H and I).
In summary, our immuno-FISH studies reveal that relocalization of the β-globin locus away from the nuclear periphery is correlated with the level of transcription, whereas association with TFys occurs at any level of HS-dependent transcription. These results suggest that different mechanisms underlie movement of the β-globin locus away from the periphery and its association with TFys.
Discussion
HS contributions to LCR function
Our previous analysis of mice with single HS deletions of the endogenous mouse LCR and the analysis presented here of mice with double LCR HS deletions suggest that each HS has a similar spectrum of activities affecting the expression and localization of the β-globin genes away from the nuclear periphery and in association with TFys, albeit to quantitatively different degrees. This is consistent with each HS being bound by a similar but nonidentical profile of transcription factors in erythroid cells.6-10 That the transcriptional phenotype of each double HS deletion is accurately predicted from the single HS deletions suggests that the LCR is an array of independently acting modules with the same mechanisms of action, which therefore interact in an additive not a synergistic manner, as was proposed previously.5,11-13,15-19 This is consistent with transgenic mouse studies suggesting that enhancers function as autonomous modules,37 but differs from studies suggesting that regions between LCR HSs contribute to full LCR function.16,17 Our results and conclusions differ from several analyses of human LCR HS function in transgenic mice and question the functional importance of the proposed holocomplex model of LCR function. Some of these disparities are likely because of the endogenous mouse β-globin locus being in a DNaseI-accessible state in erythroid tissue even in the absence of the entire LCR.1 A proposed role of the holocomplex was recruitment of a “chromatin-opening activity”; therefore, studies at the endogenous locus are not suitable for assaying such activity. This may explain why small deletions of LCR core regions in transgenic models have a devastating effect on transcription compared with larger deletions,4,11,12,38-40 but that no such difference is observed at the endogenous locus.41
HSs contribute to the association of the β-globin locus with TFys and stimulation of actively transcribed alleles via different mechanisms
In maturing primary erythroid cells, virtually all WT β-globin loci are associated with TFys and are actively transcribed, as measured by primary transcript FISH.23 Whereas LCR HSs are required for efficient association with TFys and the synthesis of nascent β-globin transcripts, these processes are relatively insensitive to the deletion of up to 2 individual HSs. The Δ1,4 and Δ1-2 mutations do not influence the association of the β-globin locus with TFys or the detection of bursts of nascent transcripts despite up to a 60% decrease in accumulated mRNA. Similarly, in the Δ2-3 mutation, association with TFys and foci of nascent transcripts are reduced only 20% and 30% respectively, despite a 70% reduction in stable message. Therefore, independent of the level of transcription, the presence of 2 of 4 LCR HSs with enhancer activity is sufficient for near WT levels of association of the β-globin locus with TFys and a WT frequency of bursts of nascent transcripts. The former is consistent with the observation that enhancerless genes can associate with TFys.42 Several mechanisms, including promoter-bound factor(s), nucleation of a TFy at the site of active transcription, or transcription leading to the retention of the locus in a TFy after a random association, could explain these results. However, regardless of mechanism, association of the β-globin locus with TFys and the frequency of bursts of nascent transcripts are relatively insensitive to the loss of multiple LCR HSs.
In contrast to the relatively mild effect that loss of LCR HSs have on the probability of the β-globin gene being actively transcribed, the loss of any HS has a significant effect on the level of β-globin mRNA. This suggests that LCR HS deletions lead to a decrease in mRNA emanating from each burst or foci of active transcription. Although nascent transcript FISH studies were performed on fractionated fetal liver cells and mRNA quantitation was performed on peripheral blood, it was intriguing that the ratio of mRNA expression to bursts of active transcription was significantly reduced in all of the LCR HS mutants analyzed (Figure 2D). This strongly suggests that once a β-globin gene associates with a TFy, each LCR HS contributes to the activation of mRNA arising from each foci or burst of transcription. Enrichment of both FACT and DSIF complexes in exon 2 of each of the double HS deletions was below that observed for WT alleles and paralleled transcriptional output. These observations suggest that the major effect of deleting 2 LCR HSs on mRNA accumulation occurs at the level of recruitment of factors required for efficient elongation. Comparison of the double HS deletions suggests that 5′HS2 has the largest effect on both DSIF and FACT enrichment, and is consistent with 5′HS2 having the largest effect on transcription in several systems.
Whereas deletion of the LCR results in a profound decrease in relocation of the β-globin locus away from the nuclear periphery during differentiation, each of the double LCR HS deletions has an intermediate phenotype, which is correlated with their effect on transcription. Consistent with our prior findings, WT and double LCR HS mutant loci can associate with TFys and be transcribed at the nuclear periphery (Ragoczy et al23 and data not shown). Our previous experiments also revealed that after erythroid differentiation, TFys redistribute, moving away from the periphery, suggesting that the β-globin locus may associate with a TFy in the periphery and then move centrally because of its physical interaction with the TFy. The difference in the efficiency of relocalization between mutants is not fully explained by differential association with TFys, raising the possibility that the mutant loci are more likely to associate with a different population of TFys.23,42 Regardless, association of the β-globin locus with TFys and relocation away from the nuclear periphery likely occur via different mechanisms, because the former occurs independently of the level of HS-dependent transcription, whereas the latter is correlated with the level of transcription.
LCR HSs act independently at multiple steps of gene activation
Systematic dissection of the LCR has resulted in a range of transcriptional phenotypes, allowing us to define several steps involved in activation and providing insight into regulation of genes with less complex structures. Tissue-specific genes show sporadic bursts of expression in progenitor cells.43 We propose that enhancers facilitate localization to TFys either by nucleation of a TFy via increasing polymerase density or by recruitment of a factor(s) involved in enhancer-mediated expression that directly facilitates migration to a TFy. This activity of LCR HSs is redundant, because the presence of 2 LCR HSs can be sufficient for localization of the locus to a TFy as efficiently as the intact LCR. Therefore, enhancerless genes would be expected to associate with TFys less frequently and for shorter durations. The LCR and many enhancers mediate the activation of transcription by reducing Pol II promoter proximal pausing and increasing the efficiency of transcriptional elongation.44 Therefore, an attractive hypothesis is that, once a gene associates with a TFy, each additional HS acts independently to increase the efficiency of elongation, resulting in significant increases in mRNA accumulation. This is consistent with models in which release of paused Pol II is not a binary process, but rather is dynamically adjusted in response to differentiation cues and environmental changes.45 Activation of β-globin expression is therefore an extreme example in which the LCR, comprising 4 enhancers (5′HS1-5′HS4), ensures near continuous release from Pol II pausing and efficient elongation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jennie Close and Jessica Halow for expert technical assistance and the Fred Hutchison Cancer Research Center flow cytometry, image analysis, and animal health resources core facilities for assistance.
This work was supported by the National Institutes of Health (grants DK44746 and HL065440 to M.G.), the intramural program of the National Diabetes and Digestive and Kidney Diseases, National Institutes of Health (to A.D.), and the Cooley's Anemia Foundation (to M.A.B).
National Institutes of Health
Authorship
Contribution: M.A.B. designed and performed the research, analyzed the data, and wrote the manuscript; T.R., J.L., R.B., and A.T. performed the research and analyzed the data; and A.D. and M.G. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark Groudine, Fred Hutchinson Cancer Research Center, Mailstop A3-025, 1100 Fairview Ave N, Seattle, WA 98109; e-mail: markg@fhcrc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal