Abstract
Mutations of Fms-like tyrosine kinase 3 (FLT3) are among the most frequently detected molecular abnormalities in AML patients. Internal tandem duplications (ITDs) are found in approximately 25% and point mutations within the second tyrosine kinase domain (TKD) in approximately 7% of AML patients. Patients carrying the FLT3-ITD but not the FLT3-TKD mutation have a significantly worse prognosis. Therefore, both FLT3 mutations seem to exert different biologic functions. FLT3-ITD but not FLT3-TKD has been shown to induce robust activation of the STAT5 signaling pathway. In the present study, we investigated the mechanisms leading to differential STAT5 activation and show that FLT3-ITD but not FLT3-TKD uses SRC to activate STAT5. Coimmunoprecipitation and pull-down experiments revealed an exclusive interaction between SRC but not other Src family kinases and FLT3-ITD, which is mediated by the SRC SH2 domain. We identified tyrosines 589 and 591 of FLT3-ITD to be essential for SRC binding and subsequent STAT5 activation. Using site-specific Abs, we found that both residues were significantly more strongly phosphorylated in FLT3-ITD compared with FLT3-TKD. SRC inhibition and knock-down blocked STAT5 activation and proliferation induced by FLT3-ITD but not by FLT3-TKD. We conclude that SRC might be a therapeutic target in FLT3-ITD+ AML.
Introduction
The Fms-like tyrosine kinase-3 (FLT3) receptor tyrosine kinase is expressed on blast cells in most patients with acute myeloid leukemia (AML).1 Activating mutations of FLT3 have been detected in approximately 30% of AML patients.2,3 Two distinct groups of FLT3 mutations have been described: (1) internal tandem duplications (FLT3-ITDs), which are most common in the juxtamembrane or the beta1-sheet of the tyrosine kinase domain-1 coding sequence in approximately 20%-27% of patients with AML; and (2) point mutations within the second tyrosine kinase domain (FLT3-TKDs) in approximately 7% of AML patients.2,4,5 Both types of mutations activate the FLT3 receptor constitutively, leading to activation of downstream signaling proteins and resulting in factor-independent proliferation of murine lymphoid and myeloid cells.6-10
Studies have identified FLT3-ITD as a prognostic factor because patients harboring this type of mutation have elevated peripheral blood and BM blast counts and an increased chance of relapse and inferior overall survival.2,11,12 For FLT3-TKD mutations, a clear association with an inferior outcome could not be demonstrated in all studies and the relevance of FLT3-TKD for clinical prognosis is being investigated further.13
Because FLT3 mutations can be found in approximately 30% of all AML patients and FLT3-ITD is associated with an inferior clinical prognosis, FLT3 is considered as an attractive therapeutic target.14 Therefore, several tyrosine kinase inhibitors (TKIs) targeting FLT3 have been developed and tested in AML patients.15,16 These studies showed that a single application of the inhibitor led to a short-term clinical response in some patients.17-19 Therefore, to achieve long-term remissions, FLT3 TKIs may have to be combined with conventional chemotherapy. However, published results so far have not demonstrated a clear clinical benefit from combining FLT3 TKIs with conventional chemotherapy.20-22 Other clinical trials have investigated the approach of combining FLT3 TKIs with other small-molecule inhibitors.14,23
We and others have shown that mice receiving a transplantation of BM expressing FLT3-ITD develop myeloproliferative disease.6,9 In contrast, mice that were transplanted with FLT3-TKD–infected BM succumbed to lymphoid disease.9 Therefore, these FLT3 mutations seem to exert different biologic functions leading to distinct phenotypes in murine BM transplantation models, and may have a different prognostic impact in AML. Interestingly, FLT3-ITD but not FLT3-TKD or wild-type FLT3 (FLT3-WT) leads to strong activation of the STAT5 signaling pathway. This has been shown in murine and human cell lines, as well as in primary murine and human FLT3-ITD+ leukemic cells.24,25 FLT3-ITD–mediated STAT5 activation might be responsible for the observed differences in biology and conferred prognosis by the 2 groups of FLT3 mutations. In the present study, we investigated the signaling mechanisms responsible for STAT5 activation downstream of FLT3-ITD but not FLT3-TKD. We show that FLT3-ITD uses the Src family kinase SRC to activate STAT5. We identified tyrosines 589 and 591 of FLT3-ITD to be essential for SRC binding and subsequent STAT5 activation. The sequence duplication in FLT3-ITD frequently leads to amplification of these amino acids, and stronger phosphorylation of tyrosines 589 and 591 can be detected on FLT3-ITDs compared with FLT3-TKDs. This may be an explanation for the exclusive activation of STAT5 in FLT3-ITD–expressing cells. Therefore, we suggest SRC inhibition as an effective measure in the treatment of FLT3-ITD+ AML.
Methods
Cell lines and primary cells
Murine 32D, Ba/F3 and human MV4-11 cells were obtained from the German Collection of Microorganisms and Cell Cultures (DMSZ) and grown in RPMI 1640 medium (PAA Laboratories) supplemented with 10% FCS (PAA Laboratories) in the presence or absence of murine IL-3 at a concentration of 2 ng/mL (R&D Systems). Phoenix E helper virus-free ecotropic packaging cells (G. Nolan, Stanford, CA) and NIH/3T3 cells (DSMZ) were maintained in DMEM (PAA Laboratories) supplemented with 10% FCS. Cells were cultured in a humidified incubator at 37°C with 5% CO2.
FLT3-ITD+ or FLT3-TKD+ blast cells were obtained from AML patients from BM aspirates or peripheral blood before therapy. A mononuclear cell fraction was prepared using Ficoll-Hypaque separation with Biocoll Separating Solution (Biochrom). Aliquots of blast cells were rapidly thawed at 37°C, washed with RPMI 1640 medium, resuspended at 500 000 cells/mL in RPMI medium supplemented with 20% FCS, and the cells were subjected to proliferation assays. The studies comply with the rules of the local internal review board and the Declaration of Helsinki.
Immunoprecipitation and immunoblotting
After starvation in serum-free medium for 4 hours, the cells were stimulated with FLT3 ligand (FL; R&D Systems) at a concentration of 100 ng/mL for 10 minutes or left unstimulated. Cells were lysed using buffer containing 10mM Tris-HCl (pH 7.4), 5mM EDTA, 130mM NaCl, 1% Triton X-100, 20mM sodium phosphate (pH 7.5), 10mM sodium pyrophosphate (pH 7.0), 50mM NaF, 1mM sodium orthovanadate, 1mM glycerol phosphate, and protease inhibitors (Roche Diagnostics). After clarification by centrifugation and preclearing with protein A–Sepharose (Amersham/Pharmacia Biotech), 2 μg of anti-FLT3 or anti-SRC Ab was added. Ab-protein complexes were precipitated with protein A–Sepharose. For immunoblotting or immunoprecipitation, whole-cell lysates or bound fractions were subjected to SDS-PAGE and blotting was performed on polyvinylidene fluoride membranes (Immobilon-P; Millipore).
Abs and inhibitors
Detection of pY was performed using a mixture of the anti-pY Abs 4G10 (Millipore) and pY20 (BD Biosciences). For the detection of FLAG and FLT3 (both Millipore), pERK1/2, ERK1/2, pAKT, AKT, pSRC, SRC, and LCK (all Cell Signaling Technology), ACTIN (Sigma-Aldrich), and STAT5 (Santa Cruz Biotechnology), the Abs were used according to the manufacturers' recommendations. The mAb against pSTAT5 was kindly provided by Tom Wheelers and Henry B. Sadowski (Hamilton, New Zealand). Bands were visualized using an enhanced chemiluminescence system (Amersham). The phosphorylation analysis of tyrosines 589 and 591 in FLT3-WT, FLT3-ITD, and FLT3-TKD was conducted as described previously by Razumovskaya et al.26
Midostaurin (PKC412) was a kind gift from Novartis Pharma and dasatinib (BMS-354-825) from the Bristol-Myers Squibb Pharmaceutical Research Institute. PD166-326 was synthesized and kindly provided by Darren R. Veach and Wiliam G. Bornmann (Memorial Sloan-Kettering Cancer Center, New York, NY). Each compound was dissolved at 10mM in DMSO and stored at −20°C.
Expression vectors
cDNA of the human/murine chimeric FLT3-WT and FLT3-ITD was kindly provided by Hubert Serve (Frankfurt, Germany) and has been described in detail previously.10 The FLT3-ITD mutant contains an insert of 6 amino acids (YEYDLK) between amino acids 596 and 597 corresponding to human FLT3. We performed site-directed mutagenesis to generate D835Y previously.27 All constructs were confirmed by sequencing. The retroviral vector was constructed by cloning the FLT3 cDNA into the MigRI retroviral vector coexpressing the enhanced green fluorescent protein (EGFP; a kind gift from W. Pear, Philadelphia, PA). FLT3-ITD and FLT3-TKD were cloned into the FLAG-tagged pCMV vector. The FLT3-ITD589/591 mutant constructs were kindly provided by K.S. and the cloning has been described previously.28
shRNA design and cloning
shRNA against SRC and LCK were designed using biopredsi Version 1 software (www.biopredsi.org, developed by the Novartis Institutes for BioMedical Research and provided by the Friedrich Miescher Institute for Biomedical Research) and cloned into the pSUPER.retro.puro vector (OligoEngine) according to the manufacturer's recommendations. Target cells were retrovirally transfected with the shRNA and selected with puromycin at a concentration of 2 mg/mL (Merck) in the presence of IL-3 for 48-96 hours before being used for further experiments. shRNA against luciferase was used as a control.
Retrovirus preparation and generation of cell lines
Phoenix E cells (2 × 106) were plated onto 60-mm dishes and transiently transfected using Lipofectamine 2000 (Invitrogen) 18 hours later according to manufacturer's instructions, as described previously.29,30 The medium was replaced after 12 hours and retroviral stocks were collected at 12-hour intervals starting 24 hours after transfection. To generate stably retroviral infected cell lines derived from a 32D or Ba/F3 cell line origin, 5 × 104 cells were transduced by 2-4 rounds of spin infection (1200g at 32°C for 90 minutes) every 12 hours in retroviral supernatant supplemented with 2 ng/mL of IL-3, 4 μg/mL of polybrene (Sigma-Aldrich), and 20% RPMI 1640 medium. After 48 hours, the FLT3-ITD– and FLT3-TKD–expressing cells were washed and resuspended in cytokine-free RPMI medium supplemented with 10% FCS. To generate FLT3-WT cells, the cells were washed after 48 hours and resuspended in RPMI medium with FL. For further details, please see our previous publications.30,31
Proliferation and viability assays
Proliferation was measured using a 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium–based method by absorption of formazan at 490 nm (CellTiter 96; Promega). Apoptosis was examined using the annexin V–FITC staining kit (BioVision) according to the manufacturer's recommendations. Measures were taken as triplicates after 0, 24, 48, and 72 hours of culture without IL-3.
In vivo tumorigenesis assay
32D syngenic C3H/N mice were IV injected with 1 × 106 32D cells expressing FLT3-ITD control shRNA or 32D FLT3-ITD SRC shRNA. Mice were monitored over time and killed when moribund. The animals were caged in a special caging system with autoclaved food and acidified water at the Technical University of Munich in accordance with national and institutional guidelines for animal care.
Statistical analysis
P values were determined by Student t test except those for survival analysis, which were determined by Wilcoxon test.
Results
SRC SH2 domain binds to FLT3-ITD but not to FLT3-TKD
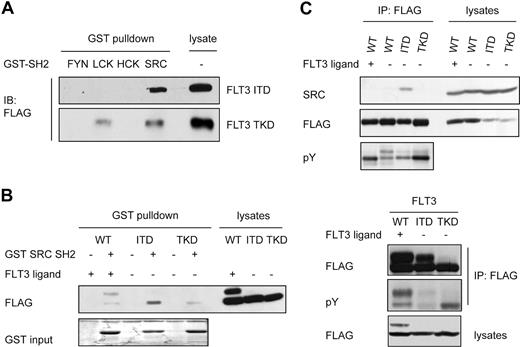
FLT3-ITD but not FLT3-TKD strongly activates STAT5, but the mechanisms for this differential activation are unclear. It has been shown that FLT3-ITD does not activate the JAKs and does not bind to the STAT5 SH2 domains directly.9,24,28 Receptor tyrosine kinases are also able to activate STAT5 via SFKs.32,33 Therefore, we investigated the possible interaction of FLT3-ITD and members of the SFK family. Cells expressing FLAG-tagged FLT3-ITD or FLT3-TKD were subjected to a pull-down experiment with GST fusion proteins containing the SH2 domains of several SFKs, as indicated in Figure 1A. We detected a robust interaction between FLT3-ITD and the SH2 domain of SRC, whereas the SH2 domains of other SFKs did not interact with FLT3-ITD. In contrast, FLT3-TKD only showed weak interactions with the SRC SH2 and LCK SH2 domains (Figure 1A). A direct comparison of the binding of the SRC SH2 domain to FLT3-ITD, FLT3-TKD, and FLT3-WT stimulated with ligand revealed the strongest interaction between FLT3-ITD and the SRC SH2 domain (Figure 1B). We also performed coimmunoprecipitation assays in cells expressing FLT3-WT, FLT3-ITD, or FLT3-TKD with endogenous SRC. Cells were starved for 4 hours, stimulated with FL or left untreated. Coimmunoprecipitation of endogenous SRC could be demonstrated with FLT3-ITD, but not with FLT3-TKD or FLT3-WT with or without ligand stimulation (Figure 1C). FLAG and pY blots confirmed comparable amounts and the anticipated phosphorylation of the precipitated FLT3 constructs. These data indicate that SRC interacts via its SH2 domain with FLT3-ITD and to a lesser extent with FLT3-WT or FLT3-TKD.
SRC physically interacts with FLT3-ITD but not FLT3-TKD. (A) HEK293 cells expressing FLAG-tagged FLT3-ITD or FLT3-TKD were serum starved for 4 hours, followed by lysis and pull-down with a GST fusion protein containing the SH2 domain of FYN, LCK, HCK, and SRC. Samples were separated by SDS-PAGE and analyzed by immunoblotting using an anti-FLAG Ab. (B) GST pull-down experiments were performed as described in panel A using HEK293 cells transiently transfected with FLT3-WT or the indicated mutants and GST SRC SH2 as bait. Cells were starved for 4 hours and stimulated with FL for 10 minutes or left untreated before lysis. Interaction was analyzed by immunoblotting with a FLAG Ab. FLT3 immunoprecipitation followed by immunoblotting with pY and FLAG Ab was carried out to confirm the phosphorylation status of the expressed proteins. (C) HEK293 cells transiently expressing FLAG-tagged FLT3-WT, FLT3-ITD, or FLT3-TKD were serum starved for 4 hours. FLT3-WT–expressing cells were either untreated or stimulated with FL. FLT3 was immunoprecipitated from whole-cell lysates using a FLAG Ab and immunoblotted with a SRC Ab, stripped, and reprobed with anti-FLAG Ab to confirm equal precipitation. In parallel, an aliquot of each cell lysate was immunoblotted and probed with an Ab against pY and FLAG to verify the proper phosphorylation and expression of the proteins in the cells.
SRC physically interacts with FLT3-ITD but not FLT3-TKD. (A) HEK293 cells expressing FLAG-tagged FLT3-ITD or FLT3-TKD were serum starved for 4 hours, followed by lysis and pull-down with a GST fusion protein containing the SH2 domain of FYN, LCK, HCK, and SRC. Samples were separated by SDS-PAGE and analyzed by immunoblotting using an anti-FLAG Ab. (B) GST pull-down experiments were performed as described in panel A using HEK293 cells transiently transfected with FLT3-WT or the indicated mutants and GST SRC SH2 as bait. Cells were starved for 4 hours and stimulated with FL for 10 minutes or left untreated before lysis. Interaction was analyzed by immunoblotting with a FLAG Ab. FLT3 immunoprecipitation followed by immunoblotting with pY and FLAG Ab was carried out to confirm the phosphorylation status of the expressed proteins. (C) HEK293 cells transiently expressing FLAG-tagged FLT3-WT, FLT3-ITD, or FLT3-TKD were serum starved for 4 hours. FLT3-WT–expressing cells were either untreated or stimulated with FL. FLT3 was immunoprecipitated from whole-cell lysates using a FLAG Ab and immunoblotted with a SRC Ab, stripped, and reprobed with anti-FLAG Ab to confirm equal precipitation. In parallel, an aliquot of each cell lysate was immunoblotted and probed with an Ab against pY and FLAG to verify the proper phosphorylation and expression of the proteins in the cells.
Tyrosine residues 589 and 591 in FLT3-ITD are crucial for the interaction of FLT3-ITD and SRC
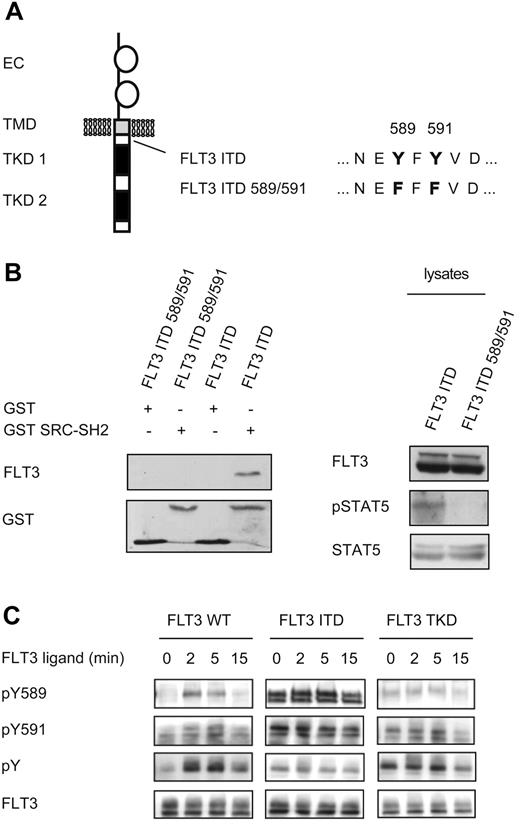
Having demonstrated that SRC interacts with FLT3-ITD, we set out to determine the specific site of interaction. Two potential SRC SH2-binding sites are located within the juxtamembrane region, which is frequently duplicated in FLT3-ITD+ AML (Figure 2A).34,35 To determine the importance of these tyrosine residues in FLT3-ITD for SRC binding, we used a FLT3-ITD mutant in which these tyrosine residues in the ITD sequence were substituted with phenylalanine (FLT3-ITD589/591). Pull-down experiments with GST SRC SH2 in cell extracts containing FLT3-ITD or FLT3-ITD589/591 demonstrated that after substitution of tyrosine 589/591, the interaction of FLT3-ITD and SRC was abolished (Figure 2B left panel). Furthermore, pSTAT5 levels in cells expressing FLT3-ITD589/591 were greatly reduced compared with FLT3-ITD (Figure 2B right panel). This result agrees with the assumption that SRC binding to FLT3-ITD is essential for robust STAT5 activation and with the results of previous studies demonstrating that tyrosine residues 589/591 in FLT3-ITD are essential for strong STAT5 activation and for the induction of myeloproliferative disease in mice.28,35,36
Identification of the interaction site of FLT3-ITD and SRC. (A) Overview of the mutations created in FLT3-ITD. (B) HEK293 cells transiently expressing either FLT3-ITD or a mutated FLT3-ITD589/591 were starved for 4 hours in serum-free medium and then lysed (right). A pull-down assay was performed using GST alone or the SH2 domain of SRC fused to GST as indicated. Precipitated FLT3 was visualized by immunoblotting using an FLT3 Ab. To confirm equal loading, the membrane was stripped and reprobed with a GST Ab (left). In parallel, lysates were analyzed by immunoblotting for FLT3 expression and STAT5 activation. The membrane was then stripped and reprobed with STAT5 Ab to confirm equal loading (right). (C) Cells stably transfected with FLT3-WT, FLT3-ITD, or FLT3-TKD were starved for 4 hours before stimulation with FL (100 ng/mL) for the indicated times. The lysates were subjected to immunoblotting and the membranes were probed with pY589 and pY591, pan-pY (4G10), and total FLT3 Abs, as described previously.26
Identification of the interaction site of FLT3-ITD and SRC. (A) Overview of the mutations created in FLT3-ITD. (B) HEK293 cells transiently expressing either FLT3-ITD or a mutated FLT3-ITD589/591 were starved for 4 hours in serum-free medium and then lysed (right). A pull-down assay was performed using GST alone or the SH2 domain of SRC fused to GST as indicated. Precipitated FLT3 was visualized by immunoblotting using an FLT3 Ab. To confirm equal loading, the membrane was stripped and reprobed with a GST Ab (left). In parallel, lysates were analyzed by immunoblotting for FLT3 expression and STAT5 activation. The membrane was then stripped and reprobed with STAT5 Ab to confirm equal loading (right). (C) Cells stably transfected with FLT3-WT, FLT3-ITD, or FLT3-TKD were starved for 4 hours before stimulation with FL (100 ng/mL) for the indicated times. The lysates were subjected to immunoblotting and the membranes were probed with pY589 and pY591, pan-pY (4G10), and total FLT3 Abs, as described previously.26
SRC SH2-mediated binding to tyrosine 589/591 requires phosphorylation of these sites. To compare the level of phosphorylation at tyrosine 589/591 in FLT3-ITD, FLT3-TKD, and FLT3-WT after ligand stimulation, we used site-specific anti-pY Abs (Figure 2C). Western blotting using phosphospecific Abs against tyrosines 589 and 591 revealed stronger phosphorylation at these sites in FLT3-ITD compared with FLT3-WT after ligand stimulation or FLT3-TKD. Tyrosine phosphorylation of FLT3-WT, FLT3-ITD, and FLT3-TKD was also assessed using Abs detecting pan-pY. These data indicate that phosphorylation at tyrosines 589 and 591 occurs preferentially in FLT3-ITD, whereas in FLT3-WT or FLT3-TKD, phosphorylation at these sites contributes to a lesser extent to the total pY signal. These differences in tyrosine phosphorylation may explain the observed differential SRC binding and STAT5 activation by FLT3 variants.
SRC inhibition reduces STAT5 activation and proliferation of FLT3-ITD– but not FLT3-TKD–expressing cells
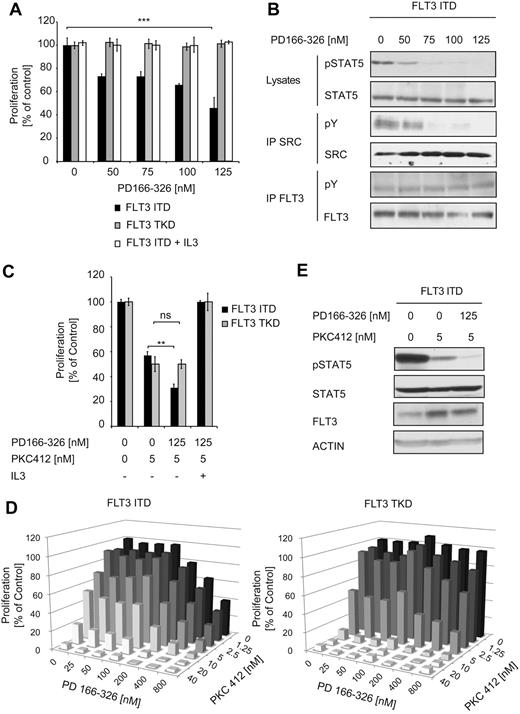
We also sought to determine the effects of SRC kinase inhibition on the proliferation of cells depending on FLT3-ITD or FLT3-TKD signaling. We used the SRC inhibitor PD166-326, which belongs to a group of compounds (pyrido[2,3-days]pyrimidines) that were originally characterized as inhibitors of the fibroblast growth factor receptor, but also potently inhibit SRC.37,38 Cells (32D) proliferating in the absence of growth factors due to stable expression of FLT3-ITD or FLT3-TKD were incubated with increasing concentrations of PD166-326 (Figure 3A).
The effect of SRC inhibition on FLT3 mutant–expressing cells. (A) Cells (32D) stably expressing FLT3-ITD or FLT3-TKD were cultured in the presence of the inhibitor PD166-326 at the indicated concentrations with or without IL-3. The proliferation was measured after 48 hours using an MTT-based assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (B) Cells (32D FLT3-ITD) were treated with PD166-326 in serum-free and cytokine-free medium for 4 hours. The lysates were analyzed for STAT5 activation by immunoblotting. Immunoprecipitation of FLT3-ITD, followed by immunoblotting with pY and FLT3 Abs, was performed. Phosphorylation of SRC was analyzed by immunoprecipitation, followed by immunoblotting with anti-pY Ab. SRC expression was verified by anti-SRC immunoblotting. Activation of STAT5 was investigated by immunoblotting using pSTAT5 Ab. To confirm equal expression of FLT3 and equal loading, the membrane was reprobed with FLT3 and STAT5 Abs. (C-D) Cells (32D) stably expressing FLT3-ITD or FLT3-TKD were incubated for 48 hours with PD166-326 and PKC412 as indicated. The proliferative activity of the cells was determined by MTT assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. The median combinatory index for FLT3-ITD–expressing cells for effective dose 50 and 75 was determined as 0.34 using CalcuSyn Version II software (Biosoft) and indicated synergism. (E) Cells (32D) as described in panel A were starved for 6 hours in serum-free medium and treated with 125nM PD166-326 and 5nM PKC412 or PKC412 alone, and then the cells were lysed and subject to immunoblot analysis. The activation of STAT5 was determined using pSTAT5 Ab. To ensure equal loading and expression, the membranes were stripped and reprobed with FLT3 and ACTIN Abs.
The effect of SRC inhibition on FLT3 mutant–expressing cells. (A) Cells (32D) stably expressing FLT3-ITD or FLT3-TKD were cultured in the presence of the inhibitor PD166-326 at the indicated concentrations with or without IL-3. The proliferation was measured after 48 hours using an MTT-based assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (B) Cells (32D FLT3-ITD) were treated with PD166-326 in serum-free and cytokine-free medium for 4 hours. The lysates were analyzed for STAT5 activation by immunoblotting. Immunoprecipitation of FLT3-ITD, followed by immunoblotting with pY and FLT3 Abs, was performed. Phosphorylation of SRC was analyzed by immunoprecipitation, followed by immunoblotting with anti-pY Ab. SRC expression was verified by anti-SRC immunoblotting. Activation of STAT5 was investigated by immunoblotting using pSTAT5 Ab. To confirm equal expression of FLT3 and equal loading, the membrane was reprobed with FLT3 and STAT5 Abs. (C-D) Cells (32D) stably expressing FLT3-ITD or FLT3-TKD were incubated for 48 hours with PD166-326 and PKC412 as indicated. The proliferative activity of the cells was determined by MTT assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. The median combinatory index for FLT3-ITD–expressing cells for effective dose 50 and 75 was determined as 0.34 using CalcuSyn Version II software (Biosoft) and indicated synergism. (E) Cells (32D) as described in panel A were starved for 6 hours in serum-free medium and treated with 125nM PD166-326 and 5nM PKC412 or PKC412 alone, and then the cells were lysed and subject to immunoblot analysis. The activation of STAT5 was determined using pSTAT5 Ab. To ensure equal loading and expression, the membranes were stripped and reprobed with FLT3 and ACTIN Abs.
Treatment of FLT3-ITD–expressing cells with the SRC inhibitor PD166-326 resulted in a concentration-dependent inhibition of proliferation. Addition of IL-3 to these cells rescued growth inhibition, suggesting that the observed effect was because of specific inhibition of FLT3-ITD signaling (Figure 3A). In contrast, FLT3-TKD–expressing cells were not affected by the treatment with the SRC inhibitor. To confirm inhibition of SRC, we performed immunoprecipitation of SRC, followed by pY immunoblotting (Figure 3B). Inhibition of SRC was accompanied by a significant decrease in the pSTAT5 signal. To rule out any inhibitory effect of PD166-326 on FLT3, we also determined the phosphorylation status of FLT3-ITD, AKT, and ERK, none of which was affected by PD166-326 treatment (Figure 2B and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In addition, IL-3–induced STAT5 activation in FLT3-ITD–expressing cells was not impaired by the SRC inhibitor (supplemental Figure 2). These data indicate that FLT3-ITD– but not IL-3–induced STAT5 activation depends on SRC activity and that SRC is crucial for the proliferation of FLT3-ITD– but not FLT3-TKD–expressing cells.
We also investigateed the effect of simultaneous SRC and FLT3 inhibition in FLT3-ITD and TKD+ cells using PD166-326 in combination with the FLT3 inhibitor PKC412. PKC412 is a potent FLT3 inhibitor currently under clinical investigation in FLT3+ AML.37-39 32D cells expressing FLT3-ITD or FLT3-TKD were kept in culture in the presence of PKC412 alone or in combination with PD166-326 for 48 hours.
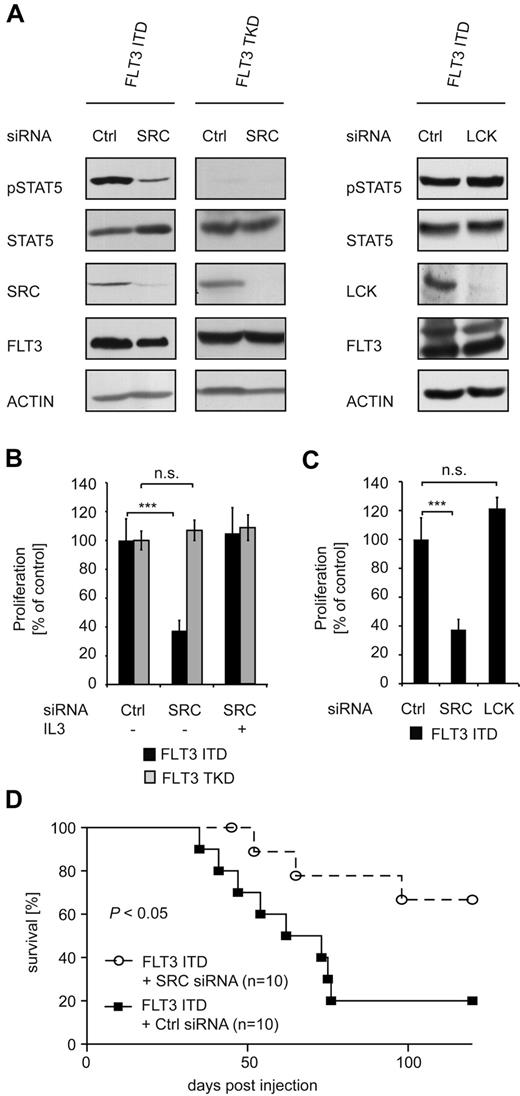
In FLT3-ITD–expressing cells, the addition of PD166-326 to PKC412 led to an additive, statistically significant growth reduction (Figure 3C). We used CalcuSyn Version II software (Biosoft) to determine the combinatory index (Figure 3D). The mean combinatory index for PKC412 and PD166-326 at effective dose 50 and 75 was calculated as 0.34 for FLT3-ITD–expressing cells, indicating a synergistic effect for these 2 drugs. In contrast, in FLT3-TKD–expressing cells, the maximal growth inhibition was observed with PKC412 only, with no further effect by the addition of PD166-326 (Figure 3C-D). IL-3 was able to abrogate growth inhibition by the compounds in both cell lines. Immunoblot analysis for STAT5 activation in FLT3-ITD+ cells showed an additive reduction of STAT5 activation after treatment with both inhibitors (Figure 3E). Having shown that inhibition of SRC only impairs the growth of FLT3-ITD+ cells, we continued to investigate the role of SRC in FLT3-ITD using an siRNA-based approach. Immunoblot analysis revealed efficient down-regulation of SRC in FLT3-ITD– and FLT3-TKD–expressing 32D cells (Figure 4A left panel). STAT5 phosphorylation in FLT3-ITD+ cells with SRC knockdown was strongly reduced compared with FLT3-ITD+ cells transfected with a control siRNA. Interestingly, SRC knock-down did not affect FLT3-ITD–mediated AKT and ERK activation (supplemental Figure 4). The addition of IL-3 restored STAT5 phosphorylation in the SRC knockdown cells (supplemental Figure 3). These data support a specific role of SRC for FLT3-ITD–mediated STAT5 activation. In agreement with this assumption, siRNA-mediated knockdown of another SFK member, LCK, did not effect FLT3-ITD–induced STAT5 phosphorylation (Figure 4A right panel). SRC but not LCK knockdown significantly reduced the proliferative activity of FLT3-ITD–expressing cells, indicating a crucial role of the SRC-STAT5 pathway in the proliferative capacity of this oncogene (Figure 4B-C). In contrast, cells expressing FLT3-TKD were not affected by the down-regulation of SRC. We used a syngenic mouse model to investigate the biologic impact of SRC down-regulation in FLT3-ITD+ myeloid cells in vivo. Cells (32D) stably expressing FLT3-ITD with and without siRNA-mediated SRC knock-down were IV injected into C3H recipient mice. Both cell lines caused a lethal hematopoietic disease, with a significantly prolonged overall survival in the SRC siRNA group (Figure 4D). These results support the conclusion that SRC does play a major role in the signaling of FLT3-ITD, but not in the signaling of FLT3-TKD.
SRC is essential for FLT3-ITD–mediated STAT5 activation and proliferation. (A) Cells (32D) stably expressing FLT3-ITD or FLT3-TKD were retrovirally infected with an SRC, LCK, or control siRNA–encoding vector and selected with puromycin. To ensure the down-regulation of SRC or LCK, immunoblotting of whole-cell lysates was performed using an SRC or LCK Ab, respectively. STAT5 activation was investigated in the same manner using pSTAT5 and STAT5 Abs. FLT3 and ACTIN immunoblotting was conducted to ensure equal loading. (B) Proliferation of 32D cells stably expressing FLT3-ITD or FLT3-TKD in combination with SRC or control siRNA was analyzed after 48 hours using the MTT assay. IL-3 stimulation of the FLT3-ITD + SRC siRNA-expressing cells for 48 hours served as a control. Data were correlated to cells expressing the control siRNA and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (C) Proliferation of FLT3-ITD+ 32D cells transduced with an SRC, LCK, or control siRNA. Analysis was performed as in panel B. (D) Kaplan-Meier plot detailing survival times of C3H mice injected with 32D FLT3-ITD cells stably expressing either control siRNA or SRC siRNA (n = 10). The mice were closely monitored and killed when moribund.
SRC is essential for FLT3-ITD–mediated STAT5 activation and proliferation. (A) Cells (32D) stably expressing FLT3-ITD or FLT3-TKD were retrovirally infected with an SRC, LCK, or control siRNA–encoding vector and selected with puromycin. To ensure the down-regulation of SRC or LCK, immunoblotting of whole-cell lysates was performed using an SRC or LCK Ab, respectively. STAT5 activation was investigated in the same manner using pSTAT5 and STAT5 Abs. FLT3 and ACTIN immunoblotting was conducted to ensure equal loading. (B) Proliferation of 32D cells stably expressing FLT3-ITD or FLT3-TKD in combination with SRC or control siRNA was analyzed after 48 hours using the MTT assay. IL-3 stimulation of the FLT3-ITD + SRC siRNA-expressing cells for 48 hours served as a control. Data were correlated to cells expressing the control siRNA and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (C) Proliferation of FLT3-ITD+ 32D cells transduced with an SRC, LCK, or control siRNA. Analysis was performed as in panel B. (D) Kaplan-Meier plot detailing survival times of C3H mice injected with 32D FLT3-ITD cells stably expressing either control siRNA or SRC siRNA (n = 10). The mice were closely monitored and killed when moribund.
SRC is a potential molecular target in FLT3-ITD+ leukemia
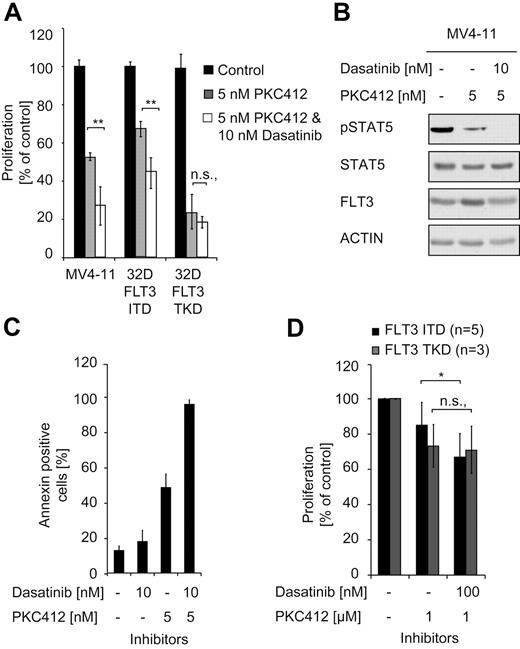
Having demonstrated that SRC is a crucial signaling mediator of FLT3-ITD using murine cell lines, we then also investigated FLT3-ITD+ human cells using MV4-11 cells, a human FLT3-ITD+ AML cell line. Cells were treated with the FLT3 inhibitor PKC412 or in combination with dasatinib, a clinically approved BCR-ABL inhibitor that also inhibits SRC.40,41 Similar to murine cells expressing FLT3-ITD (supplemental Figure 5), an additive effect of SRC inhibition in combination with FLT3 inhibition could be demonstrated in the human FLT3-ITD+ MV4-11 cell line (Figure 5A).
SRC is a signaling mediator in FLT3-ITD+ human AML cells. (A) Cells (32D) positive for FLT3-ITD or FLT3-TKD and FLT3-ITD+ MV4-11 cells were treated with PKC412 alone or in combination with PKC412 and dasatinib. The proliferation was assessed after 48 hours using the MTT assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (B) MV4-11 cells were starved for 6 hours in serum-free medium in the presence of the indicated concentrations of PKC412 and dasatinib. After treatment, the cells were lysed and subjected to immunoblot analysis with regard to the activation of STAT5 (pSTAT5). To control equal loading, the membrane was stripped and reprobed with a STAT5 and ACTIN Ab. (C) MV4-11 cells expressing FLT3-ITD were incubated in the presence of the indicated amounts of PKC412 and dasatinib for 48 hours. Flow cytometric analysis was performed after staining the cells with annexin V/propidium iodide. The total percentage of annexin V/propidium iodide–positive cells is shown. Data represent the values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (D) AML patient samples expressing FLT3-ITD or FLT3-TKD were treated with PKC412 and/or dasatinib for 48 hours. Cell viability was determined using the MTT assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05.
SRC is a signaling mediator in FLT3-ITD+ human AML cells. (A) Cells (32D) positive for FLT3-ITD or FLT3-TKD and FLT3-ITD+ MV4-11 cells were treated with PKC412 alone or in combination with PKC412 and dasatinib. The proliferation was assessed after 48 hours using the MTT assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (B) MV4-11 cells were starved for 6 hours in serum-free medium in the presence of the indicated concentrations of PKC412 and dasatinib. After treatment, the cells were lysed and subjected to immunoblot analysis with regard to the activation of STAT5 (pSTAT5). To control equal loading, the membrane was stripped and reprobed with a STAT5 and ACTIN Ab. (C) MV4-11 cells expressing FLT3-ITD were incubated in the presence of the indicated amounts of PKC412 and dasatinib for 48 hours. Flow cytometric analysis was performed after staining the cells with annexin V/propidium iodide. The total percentage of annexin V/propidium iodide–positive cells is shown. Data represent the values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05. (D) AML patient samples expressing FLT3-ITD or FLT3-TKD were treated with PKC412 and/or dasatinib for 48 hours. Cell viability was determined using the MTT assay. Data are presented as the percentage of DMSO-treated cells and represent values ± SD of triplicates. One representative of at least 3 independent experiments is shown. Statistical analysis was conducted. ***P < .001; **P < .01; *P < .05; and n.s. (no significance), P > .05.
MV4-11 cells were also investigated with regard to the effect of the inhibitors on the activation of STAT5. PKC412 alone induced a reduction in pSTAT5 levels and the combination with dasatinib further reduced pSTAT5 to undetectable levels (Figure 5B). Flow cytometric assays showed an additive increase of apoptosis at doses of 5nM PKC412 and 10nM dasatinib in these cells (Figure 5C).
Finally, we investigated whether our results would hold true with primary human AML cells. Primary cells from 8 AML patients positive for FLT3-ITD or FLT3-TKD (patient characteristics are summarized in Table 1) were treated with PKC412 and dasatinib. Similar to the effects observed in cell lines, the combination of PKC412 and dasatinib had a growth inhibitory effect on cells from FLT3-ITD+ but not FLT3-TKD+ patients (Figure 5D), albeit to a lesser extent than in cell lines because of the variability of growth kinetics of primary human AML cells (supplemental Figure 6).
Summary of patient material
| Patient . | Karyotype . | FAB . | FLT3 mutation . |
|---|---|---|---|
| 1 | 46,XX | M4-M5 | FLT3-ITD |
| 2 | 46,XY | M1 | FLT3-ITD |
| 3 | 46,XY | M1 | FLT3-ITD |
| 4 | 46,XY | M1 | FLT3-TKD |
| 5 | 46,XY | M4 | FLT3-TKD |
| 6 | 46,XX | M1 | FLT3-ITD |
| 7 | 46,XY | M1 | FLT3-ITD |
| 8 | 46,XX | M5b | FLT3-TKD |
| Patient . | Karyotype . | FAB . | FLT3 mutation . |
|---|---|---|---|
| 1 | 46,XX | M4-M5 | FLT3-ITD |
| 2 | 46,XY | M1 | FLT3-ITD |
| 3 | 46,XY | M1 | FLT3-ITD |
| 4 | 46,XY | M1 | FLT3-TKD |
| 5 | 46,XY | M4 | FLT3-TKD |
| 6 | 46,XX | M1 | FLT3-ITD |
| 7 | 46,XY | M1 | FLT3-ITD |
| 8 | 46,XX | M5b | FLT3-TKD |
Discussion
Despite intensive research, the molecular mechanisms of cell transformation by FLT3-ITDs versus FLT3-TKDs are incompletely understood. Therefore, in the present study, we investigated the signaling pathways leading to differential activation of STAT5 by the 2 types of activating FLT3 mutations.
The 2 groups of mutations differ with respect to their clinical outcome and structural features and also show significant disparities with regard to their biologic transforming potential.1,3,42,43 We and others have shown in previous studies that FLT3-ITD induces an MPD-like disease in a murine BM transplantation model.6,9,44 In contrast, FLT3-TKD–transplanted mice succumb to lymphoid hematologic diseases. Biochemical analysis of cell lines stably expressing FLT3-ITD or FLT3-TKD or of human and murine FLT3-ITD or FLT3-TKD primary hematopoietic cells revealed dissimilarities of the downstream signaling pathways activated by these 2 mutations.9,10,25 Most intriguing are the differences with regard to STAT5 activation, which is strongly induced in FLT3-ITD–expressing cells, but only marginally induced in FLT3-TKD+ cells, suggesting that STAT5 activation might be particularly important for FLT3-ITD–mediated AML.7,25 Extensive research was conducted to elucidate the activation mechanisms of STAT5 by FLT3-ITD and led to the identification of tyrosine residues within the juxtamembrane region that are crucial for proliferation and STAT5 activation of FLT3-ITD–expressing cells.28,35,36 Heiss et al suggested that tyrosines 589 and 599 are also involved in the activation of downstream signaling molecules of FLT3-WT.35 In addition, these residues were shown to play a crucial role in the induction of myeloproliferative disease by FLT3-ITD in mice.36 Vempati et al showed that the tyrosines 589 and 591 are also important for growth of Ba/F3 cells expressing FLT3-WT stimulated with FL or FLT3-TKD.28
Previous studies have compared the signaling of FLT3-ITD and FLT3-TKD and demonstrated that the localization of FLT3-ITD and FLT3-TKD significantly alters the transforming capability and activation of downstream signaling molecules. In the case of FLT3-ITD, retention in the endoplasmic reticulum was suggested to be a prerequisite for the activation of STAT5.45,46 Choudhary et al demonstrated that after membrane localization, the activation of STAT5 by FLT3-ITD is reduced, whereas localization at the endoplasmic reticulum up-regulates STAT5 activity in FLT3-ITD–expressing cells.
Downstream signaling molecules involved in the activation of STAT5 by FLT3-ITD have been investigated in previous studies. Choudhary et al showed that neither Src-family kinases nor JAKs are involved in the activation of STAT5, and postulated instead a direct interaction of STAT5 and FLT3-ITD. In contrast, several other groups proposed the involvement of various signaling molecules, including LYN, in the process of STAT5 activation by FLT3-ITD in murine and human cells.28,47,48 Up to now, there have been no investigations of the signaling requirements of FLT3-ITD compared with FLT3-TKD in leading to differential STAT5 activation.
In the present study, we have shown that FLT3-ITD but not FLT3-TKD strongly binds in vitro to the SH2 domain of SRC. No binding of FLT3-ITD to the SH2 domains of other SFK family members such as LCK, HCK, or FYN could be detected. Coimmunoprecipitation revealed an interaction between SRC and FLT3-ITD but not between SRC and FLT3-TKD. Mutation of the putative SH2-binding sites in FLT3-ITD led to the disruption of the FLT3-ITD–SRC complex and to the abolishment of FLT3-ITD–induced STAT5 activation. These data explain and are in agreement with previous studies showing that the loss of tyrosines 589 and 591 abolished STAT5 activation and the transforming potential of FLT3-ITD in a syngenic mouse transplantation model.28,36 Because tyrosines 589 and 591 are also present in FLT3-TKD, it is surprising that FLT3-TKD, although highly autophosphorylated, does not form a complex with SRC that is detectable by coimmunoprecipitation. An obvious explanation could be that in FLT3-ITD, tyrosines 589 and 591 are amplified. However, amplification of these residues is observed in many but not all cases of FLT3-ITD. In approximately 30% of FLT3-ITD+ AML, the ITD is located downstream of the juxtamembrane region in the beta-1 sheet of the kinase domain.5 Using a panel of residue-specific pY Abs, Razumovskaya et al demonstrated significant differences between the autophosphorylation pattern of FLT3-ITD and FLT3-TKD.26 In agreement with these data, we showed significantly enhanced phosphorylation of tyrosines 589 and 591 in FLT3-ITDs compared with FLT3-TKDs. Therefore, structural alterations induced by the ITD may lead to an enhanced autophosphorylation of tyrosines 589 and 591. These data may explain activation of the STAT5 pathway only by FLT3-ITD but not FLT3-TKD, and SRC activation might be the basis for the observed differences of both mutations in terms of biology and clinical impact.
The involvement of SFKs in FLT3-ITD–mediated transformation has been controversial and much discussed in the literature. Whereas Choudhary et al found no evidence that SFKs are involved in the signaling of FLT3-ITD, Okamoto et al demonstrated an involvement of the SFK LYN in FLT3-ITD–mediated STAT5 activation.24,47 To obtain further evidence that SFKs indeed play a role in FLT3-ITD– but not FLT3-TKD–mediated STAT5 activation, we used SFK inhibitors in murine cells artificially overexpressing FLT3-ITD or FLT3-TKD, as well as in human AML cell lines and in primary patient material from FLT3-ITD+ and FLT3-TKD+ patients. In all experiments performed, we found that SRC inhibition blocked STAT5 activation and proliferation in FLT3-ITD+ cells but not in FLT3-TKD+ cells. We used an siRNA-based approach to exclude off-target effects of the compounds used. SRC down-regulation only effected the proliferation of FLT3-ITD+ but not FLT3-TKD+ cells, and this effect was mediated by the specific SFK SRC and was not observed with the SFK LCK. Finally, we confirmed these findings in a syngenic mouse transplantation model, in which we observed a prolonged survival of mice transplanted with cells expressing FLT3-ITD and SRC knockdown compared with mice receiving FLT3-ITD control siRNA cells. SRC has emerged as a critical signaling node in many oncogene-driven malignancies and was also identified recently as a key convergent point for resistance against molecular-targeted therapies.49,50 Therefore, we investigated whether a combination of SRC and FLT3-ITD inhibition may lead to reduced STAT5 activation, proliferation, and induction of apoptosis and found that this could be demonstrated in FLT3-ITD+ but not FLT3-TKD+ cells.
In conclusion, we identified SRC as an essential signal transduction molecule and druggable target for FLT3-ITD– but not FLT3-TKD–mediated transformation. Therefore, the combination of FLT3 and SRC inhibitors warrants further investigation in FLT3-ITD+ but not FLT3-TKD+ AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB684 to J.D.) and the Deutsche Krebshilfe. H.L. was supported by a grant from the Kommission für Klinische Forschungsprojekte of the Technische Universität München.
Authorship
Contribution: H.L., C.A., and J.D. conceived of and designed the experiments; H.L., C.A., and R.G. analyzed the data; H.L., C.A., E.R., and L.R. performed the experiments; K.S., S.B., and K.G. provided materials; R.G. and C.P. contributed to the interpretation of the experimental data and discussed the experiments; and H.L., C.A., and J.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for H.L. is Department of Hematology, Oncology, and Tumor Immunology, HELIOS Klinikum Berlin-Buch, Berlin, Germany.
Correspondence: Justus Duyster, Department of Internal Medicine III, Technical University Munich, Ismaningerstr 22, 81675 Munich, Germany; e-mail: justus.duyster@lrz.tum.de.
References
Author notes
C.A. and R.G. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal