Abstract
Oncogenic mutations leading to persistent kinase activities are associated with malignancies. Therefore, deciphering the signaling networks downstream of these oncogenic stimuli remains a challenge to gather insights into targeted therapy. To elucidate the biochemical networks connecting the Kit mutant to leukemogenesis, in the present study, we performed a global profiling of tyrosine-phosphorylated proteins from mutant Kit-driven murine leukemia proerythroblasts and identified Shp2 and Stat5 as proximal effectors of Kit. Shp2 or Stat5 gene depletion by sh-RNA, combined with pharmacologic inhibition of PI3kinase or Mek/Erk activities, revealed 2 distinct and independent signaling pathways contributing to malignancy. We demonstrate that cell survival is driven by the Kit/Shp2/Ras/Mek/Erk1/2 pathway, whereas the G1/S transition during the cell cycle is accelerated by both the Kit/Stat5 and Kit/PI3K/Akt pathways. The combined use of the clinically relevant drugs NVP-BEZ235, which targets the cell cycle, and Obatoclax, which targets survival, demonstrated synergistic effects to inhibit leukemia cell growth. This synergy was confirmed with a human mast leukemia cell line (HMC-1.2) that expresses mutant Kit. The results of the present study using liquid chromatography/tandem mass spectrometry analysis have elucidated signaling networks downstream of an oncogenic kinase, providing a molecular rationale for pathway-targeted therapy to treat cancer cells refractory to tyrosine kinase inhibitors.
Introduction
Receptor tyrosine kinases (RTKs) are known to generate growth and survival signals once activated, therefore playing a pivotal role in the development of normal and neoplastic cells. Several diseases and neoplasia are related to mutations in RTK leading to constitutive receptor activation and signaling.1 The discovery that Kit, the RTK for SCF, was constitutively activated by mutations in a variety of human malignancies, including hematopoietic disorders, systemic mastocytosis, gastrointestinal stromal tumors, melanoma, and testicular germ cell tumors,2 made Kit activity an attractive target for drug development.3 Although drugs inhibiting tyrosine kinase activity represent the frontline treatment for Kit-driven malignancies, the treatment response varies highly depending on the Kit mutation status and the cellular context.3 In addition, the clinical response to treatment may evolve overtime, ultimately leading to resistance.4 Therefore, it is critical to design alternative therapeutic strategies to Kit inhibitors and for that we need a detailed understanding of: (1) the proximal effectors of the Kit mutant, (2) the biochemical pathways associated with these proximal effectors, (3) their functional contributions in cancer development, and (4) their cooperation in cancer.
There are considerable data about individual molecular changes that underlie leukemic development in acute myeloid leukemia (AML) associated with oncogenic mutated Kit, but knowledge about the overall signaling network promoting the leukemic growth remains rather basic. In the present study, we took advantage of the erythroleukemia that develops in spi-1/PU.1-transgenic (spi-1/PU.1-TG) mice5 to explore the set of molecular pathways downstream of oncogenic Kit. In this model, the founding oncogenic event is the inappropriate overexpression of Spi-1/PU.1, a master transcriptional regulator of B lymphopoiesis and myelopoiesis6,7 that leads to inhibition of both terminal differentiation and apoptosis in the erythroid lineage.8 As a result, spi-1/PU.1-TG mice develop severe anemia and a proliferation of proerythroblastic cells (HS1 preleukemic cells). Later, a clonal population of proerythroblastic cells (HS2 leukemic cells) emerges in these mice characterized by growth factor autonomy and tumorigenicity. The tumorigenic stage is correlated with the acquisition of mutations targeting Kit at residues D814 or D818 in the phosphotransferase domain,9 which confer ligand-independent tyrosine kinase activity to Kit. These residues are homologous to human residues D816 and D820, respectively, which are mutated in patients with mastocytosis or AML. We have demonstrated previously that constitutive mutant Kit activity was sufficient to drive leukemic transformation.9 To elucidate the biochemical networks connecting mutant Kit to leukemogenesis, we started from a global phosphoproteomic study of leukemic cells and identified the tyrosine phosphatase Shp2 and Stat5 as proximal effectors mediating Kit oncogenic signaling. Functional studies allowed assessment of the relationship between each one of these effectors and leukemic cell behavior. We have shown herein that the survival of leukemic cells is controlled by the Kit/Shp2/Ras/Mek/Erk1/2 pathway, whereas the G1/S transition during the cell cycle is accelerated by both the Kit/Stat5 and Kit/PI3K/Akt pathways with no biochemical or biologic evidence for cross-talk interference between these pathways. These data were confirmed with a human Kit-driven mast leukemia cell line. Most importantly, we have demonstrated that a combination of drugs targeting signaling pathways that promote survival and the cell cycle is efficient for successful eradication of leukemia cells.
Methods
Cell lines, growth kinetics, colony assays, and pharmacologic inhibitors
HS1 and HS2 spi-1–transgenic cell lines were established and cultured as described previously.5,9 The number of living cells was measured by trypan blue exclusion using a Vi-Cell analyzer (Beckman Coulter). PP1 (Enzo Life Sciences), PP2 (Calbiochem), imatinib mesylate (IM; Novartis-Pharma), LY294002 (Sigma-Aldrich), NVP-BEZ235 (Novartis Pharma), Jak I (Calbiochem), SU6656 (Calbiochem), Obatoclax (GX15-070; Gemin X Pharmaceuticals), and UO126 (Promega) were added in culture medium at the indicated concentrations. Clonogenic assays were performed in methylcellulose, as described previously.10
LC-MS/MS analysis
Phosphopeptide immunoprecipitation was performed using the PhosphoScan Kit (PY-100) from Cell Signaling Technology. The extracted peptides were concentrated and separated with an Ultimate3000 (Dionex) series HPLC, spotted on-line on a matrix-assisted laser desorption/ionization target with a Probot (Dionex), and analyzed using a 4800 Plus MALDI TOF/TOF analyzer (Applied Biosystems) as described previously13 but phosphorylation on tyrosine (+80 Da) was selected as a partial modification. The liquid chromatography tandem mass spectrometry (LC-MS/MS) data were searched twice using MASCOT software (Matrix Science) against the National Center for Biotechnology Information nonredundant Mus musculus database (National Library of Medicine). Spectra were checked manually for proper matching and the result was accordingly rejected or accepted. All data were validated manually using myProMS.14
Flow cytometric analysis of cell cycle and caspase 3 labeling
Cell-cycle distribution of ethanol-fixed cells stained with propidium iodide (Invitrogen) was analyzed by flow cytometry. For caspase 3 labeling, cells were fixed with paraformaldehyde 1%, permeabilized with 70% cold ethanol, and stained with PE-conjugated anti–active caspase 3 Abs (BD Biosciences). Flow cytometric analysis was performed using a FACSsort flow cytometer (BD Biosciences). Data were analyzed using ModfitLT Version 3.2.1 software (Verity) for cell cycle and FlowJo Version 9.2 (TreeStar) for caspase 3 labeling.
Lentiviral vector production
pLKO.1-Puro vectors transducing either shRNA targeting Shp2 or a nonsilencing (ns) scrambled shRNA were purchased from the MISSION shRNA lentivirus library (Sigma-Aldrich). Selected shRNA sequences against Stat5a/b were inserted down the H1 promoter into the pTRIPΔU3 lentiviral vector encoding green fluorescent protein as a reporter gene. A lentiviral vector encoding an antiluciferase shRNA was used as a control. Lentiviral particles were generated as described previously.15 Freshly prepared supernatants were used to transduce HS2 cells. Forty-eight hours after infection, infected HS2 cells were sorted according to green fluorescent protein expression using a FACSVantage cell sorter (BD Biosciences) or were selected by puromycin resistance (1 mg/mL; CAYLA). Selected cells were used for further investigation 3 days after infection.
Abs
Abs raised against phosphorylated forms of Akt(S473), Stat5a/b(Y694/699), Shp2(Y542) or (Y580), Erk1/Erk2(T202-Y204), and Kit(Y719) and against constitutive forms of Akt, Erk1/Erk2, and Kit were from Cell Signaling Technology. Anti-p85 was described previously.16 Anti-Stat5 and anti-Shp2 Abs were from Santa Cruz Biotechnology and anti-β actin Ab was from Sigma-Aldrich. For immunoprecipitation, an anti-Kit from R&D Systems was used.
Immunoprecipitation and immunoblotting
Ras activation assay
The activity of Ras was measured by detecting the levels of Ras-GTP using a Ras activation kit (Upstate Biotechnology). Briefly, 106 cells were lysed and Ras-GTP was immunoprecipitated using the Ras-GTP–binding domain of Raf1 before Western blot analysis.
Tumorigenicity
Cells (3 × 106 in 0.5 mL of α-MEM medium containing 2% FBS) were injected subcutaneously into 8-week-old female nude mice as described previously.9 Tumor nodules were removed after 3 weeks and weighed.
Results
Shp2 and Stat5 are proximal effectors of the Kit mutant in leukemic cells
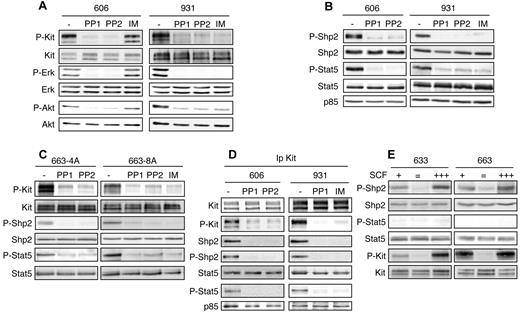
The erythroleukemia in spi-1-TG mice is characterized by an early phase in which preleukemic cells (HS1) that are arrested at the proerythroblast stage and strictly dependent on Epo or SCF for survival and proliferation, accumulate in blood-forming tissues.5 During disease progression, a clonal population of leukemic proerythroblasts (HS2) emerges that had acquired in vivo and in vitro growth factor independency as a consequence of gained somatic mutations in the RTK Kit. These mutations, KitD814Y and KitD818Y, confer ligand-independent tyrosine kinase activity to Kit and lead to persistent activation of the MAPK and PI3K/Akt pathways.9 The constitutive signaling can be abolished by treatment with drugs inhibiting Kit kinase activity, such as IM (Gleevec), PP1, and PP2 in 931HS2 cells expressing KitD818Y and PP1 and PP2, but not IM, in 606HS2 cells expressing mutant KitD814Y (Figure 1A and Kosmider et al9 ). To identify signaling targets of the Kit mutant tyrosine kinase, we made a global mapping of tyrosine-phosphorylated (PY) proteins of leukemic cells. After trypsin digestion of total proteins, PY peptides were enriched by immunoaffinity with anti-phosphotyrosine Abs and then analyzed by LC-MS/MS. A list of PY peptides identified from cells harboring mutant KitD814Y (606 HS2 cells) and KitD818Y (931 HS2 cells) is given in Table 1. Kit was one of the PY proteins found (5 peptides), providing validation of the method. We found literature-documented Kit interactors as p85, Shp1, and Dok1.2 We also identified proteins described to be associated with proliferating cells as RPS27, GSK3, Enolase1, PRPF4B, Dyrk 1, and Cdk217 and signaling players reported previously to be phosphorylated in HS2 cells as Gab1 and Erk1/2.9,18 To discriminate PY proteins likely to be initiators of signaling downstream of Kit, we retained proteins for which phosphorylation on the tyrosine residue known to participate in function was dependent on Kit activity and that were associated with Kit in coimmunoprecipitation assays. Using these criteria, Shp2 and Stat5 were selected. The phosphoproteome data showed that Shp2 was phosphorylated on Y580, Stat5a on Y694, and Stat5b on Y699 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Shp2 is a positive signaling effector of receptors for a variety of growth factors and cytokines.19 Shp2 function results from a combination between its phosphatase activity and its ability to recruit docking proteins through 2 Src homology-2 (SH2) domains. It has been demonstrated that PY580-Shp2 serves to recruit signaling adapters or to stimulate Shp2 phosphatase activity.19 Stat5 belongs to a family of proteins playing a dual role as signal transducers and transcription factors.20 PY694/699-Stat5 allows Stat5 dimerization, followed by translocation to the nucleus, where it drives gene transcription.20
Tyrosine phosphorylation of Shp2 and Stat5 is induced by mutant Kit in HS2 leukemic cells. (A-B) Whole-cell lysates from 606HS2 and 931HS2 cells treated for 4 hours with IM (0.75μM), PP1 (4μM), or PP2 (4μM) or not treated (−; DMSO) were subjected to immunoblotting with the indicated Abs. (C) 663-4A cells overexpressing KitD814Y and 663-8A cells overexpressing KitD818Y were treated for 4 hours with IM, PP1, or PP2 or not treated (−; DMSO). Whole-cell extracts were subjected to immunoblotting with the indicated Abs. (D) Cell extracts from 606HS2 and 931HS2 cells treated for 4 hours with IM, PP1, or PP2 or not treated (−; DMSO) were immunoprecipitated with anti-Kit Abs. Immunoprecipitates were analyzed by Western blotting with the indicated Abs. Data are representative of 5 independent experiments. (E) HS1 preleukemic cells (633HS1 and 663HS1) were cultured routinely in the presence of SCF (50 ng/mL; +) or starved of SCF in the presence of serum for 4 hours (=) before being stimulated for 10 minutes with SCF (500 ng/mL; +++). Whole cell extracts were subjected to immunoblotting with the indicated Abs.
Tyrosine phosphorylation of Shp2 and Stat5 is induced by mutant Kit in HS2 leukemic cells. (A-B) Whole-cell lysates from 606HS2 and 931HS2 cells treated for 4 hours with IM (0.75μM), PP1 (4μM), or PP2 (4μM) or not treated (−; DMSO) were subjected to immunoblotting with the indicated Abs. (C) 663-4A cells overexpressing KitD814Y and 663-8A cells overexpressing KitD818Y were treated for 4 hours with IM, PP1, or PP2 or not treated (−; DMSO). Whole-cell extracts were subjected to immunoblotting with the indicated Abs. (D) Cell extracts from 606HS2 and 931HS2 cells treated for 4 hours with IM, PP1, or PP2 or not treated (−; DMSO) were immunoprecipitated with anti-Kit Abs. Immunoprecipitates were analyzed by Western blotting with the indicated Abs. Data are representative of 5 independent experiments. (E) HS1 preleukemic cells (633HS1 and 663HS1) were cultured routinely in the presence of SCF (50 ng/mL; +) or starved of SCF in the presence of serum for 4 hours (=) before being stimulated for 10 minutes with SCF (500 ng/mL; +++). Whole cell extracts were subjected to immunoblotting with the indicated Abs.
List of tryptic PY peptides purified with the PhosphoScan kit from 606 and 931HS2 cell extracts
| Gene symbol . | NCBI query ID . | Sequence . | PY . | Description . |
|---|---|---|---|---|
| Abi1 | gi116089341 | TLEPVKPPTVPNDYMTSPAR | Y213 | Abl interactor 1 |
| Ata2 | gi37360348 | SHYADVDPENQNFLLESNLGK | Y41 | Solute carrier family38, member 2 |
| Bam32 | gi5733602 | EVEEPCIYESVR | Y139 | Dual adaptor for phosphotyrosine and 3-phosphoinositides1 (DAPP1) |
| Blk | gi198939 | SLDNGGYYISPR | Y181 | Tyrosine-protein kinase Blk |
| Btk | gi2507603 | KVVALYDYMPMNANDLQLR | Y223 | Bruton tyrosine kinase |
| HYVVCSTPQSQYYLAEK | Y344 | |||
| HLFSTIPELINYHQHNSAGLISR | Y361 | |||
| Calml2 | gi4502549 | VFDKDGNGYISAAELR | Y100 | Calmodulin 2 |
| c-Cbl | gi80978932 | IKPSSSANAIYSLAARPLPMPK | Y672 | Casitas B-lineage lymphoma |
| Cdc2 | gi1575290 | IEKIGEGTYGVVYK | Y15 | p34 Cdc2 kinase |
| IGEGTYGVVYKGR | Y15 | |||
| Cdebp | gi1086521 | MQNHGYENPTYK | Y750 | CDE1-binding protein |
| Cdk2 | gi34556205 | VEKIGEGTYGVVYK | Y11 | Cyclin-dependent kinase 2 |
| Cdv3B | gi28461294 | KTPQGPPEIYSDTQFPSLQSTAK | Y213 | Carnitine deficiency-associated gene expressed in ventricle 3 |
| c-Kit | gi42768812 | VVEATAYGLIK | Y608 | Stem cell factor receptor |
| NLLHSTEPSCDSSNEYMDMKPGVSYVVPTKTDK | Y728 | |||
| IDSYIER | Y745 | |||
| MVSPEHAPAEMYDVMK | Y898 | |||
| HIYSNLANCNPNPENPVVVDHSVR | Y934 | |||
| Csk | gi729888 | VMEGTVAAQDEFYRSGWALNMK | Y184 | Tyrosine-protein kinase CSK (p50CSK) |
| Dok1 | gi27805460 | GFSSDTALYSQVQK | Y450 | Docking protein 1 |
| Dyrk1A | gi24418935 | IYQYIQSR | Y321 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a |
| Emd | gi6679641 | LIYGQDSAYQSIAHYRPISNVSR | Y167 | Emerin |
| Eno1 | gi12963491 | AAVPSGASTGIYEALELR | Y43 | Enolase 1 |
| Esyt1 | gi33859650 | QLLDDEERITAETLYMSHR | Y111 | Extended synaptotagmin-like protein 1 |
| ITAETLYMSHR | Y111 | |||
| Fer | gi118572319 | VQENDGKEPPPVVNYEEDAR | Y402 | Tyrosine-protein kinase FER |
| Fyb | gi33469119 | TTAVEIDYDSLKR | Y559 | FYN binding protein |
| Gab1 | gi6165415 | SSGSGSSMADERVDYVVVDQQK | Y660 | GRB2-associated binding protein 1 |
| Gsk3a | gi72384361 | GEPNVSYICSR | Y279 | Glycogen synthase kinase 3 alpha |
| Hgs | gi31982286 | YKVVQDTYQIMK | Y132 | HGF-regulated tyrosine kinase substrate |
| Hlb301 | gi547841 | SQDGYTYPSR | Y849 | Low-density lipoprotein receptor precursor (LDL receptor) |
| Itsn2 | gi20138801 | GEPEALYAAVTK | Y922 | Intersectin-2 |
| Jmjd1c | gi149260924 | MKEDKYDCVSR | Y150 | PREDICTED: jumonji domain containing 1C |
| Jnk2 | gi18254507 | TACTNFMMTPYVVTR | Y185 | c-Jun N-terminal kinase 2 |
| Jnk3 | gi30578163 | TAGTSFMMTPYVVTR | Y185 | c-Jun N-terminal kinase 3 |
| Jtk1 | gi133922607 | AVPEGHEYYR | Y1053 | Tyrosine kinase 2 (TYK2) |
| Lair1 | gi81913091 | AVGAVTSQSTDMAESSTYAAIIRH | Y257 | Leukocyte-associated immunoglobulin-like receptor 1 |
| Lamtor1 | gi81903514 | ALNGAEPNYHSLPSAR | Y40 | Regulator complex protein |
| IAAYAYSALSQIR | Y140 | |||
| Lrrc25 | gi23346481 | SVDQDSQPVYCNLESLGR | Y289 | Leucine-rich repeat-containing protein 25 precursor |
| Mapk1 | gi6754632 | VADPDHDHTGFLTEYVATR | Y185 | Mitogen activated protein kinase 1, Erk2 |
| Mapk14 | gi187465730 | HTDDEMTGYVATR | Y182 | Mitogen activated protein kinase 14, p38 |
| Mapk3 | gi21489933 | IADPEHDHTGFLTEYVATR | Y205 | Mitogen activated protein kinase 3, Erk1 |
| Muc-13 | gi148665438 | TGVPSQTPNPYANQR | Y562 | Mucin 13, epithelial transmembrane |
| p85 a | gi117320524 | DQYLMWLTQK | Y580 | Phosphatidylinositol 3-kinase regulatory subunit, p85 |
| Pctk3 | gi6679233 | LGEGTYATVFKGR | Y132 | Serine/threonine-protein kinase PCTAIRE-3 |
| Prpf4B | gi148708948 | LCDFGSASHVADNDITPYLVSR | Y849 | Pre-mRNA processing factor 4 homolog B |
| Ptpn6 | gi118130785 | GQESEYGNITYPPAVR | Y536 | Protein tyrosine phosphatase, non-receptor type 6 (SHP1) |
| Ptpn11 | gi6755228 | VYENVGLMQQQR | Y580 | Protein tyrosine phosphatase, non-receptor type 11 (SHP2) |
| Ptpn18 | gi113199761 | APTSTDTPIYSQVAPR | Y381 | Protein tyrosine phosphatase, non-receptor type 18 |
| ADTYAVVQKR | Y354 | |||
| Rab2A | gi10946940 | AYAYLFK | Y3 | Ras-related protein Rab-2A |
| Rps27 | gi4506711 | LVQSPNSYFMDVK | Y31 | Ribosomal protein S27 |
| Serpb1 | gi13385280 | SSFSHYSGLKHEDKR | Y207 | SERPINE1 mRNA binding protein 1 |
| Slc1A6 | gi6678003 | QIKYFSFPGELLMR | Y85 | Solute carrier family 1 (high affinity aspartate/glutamate transporter), number 6 |
| Snap23 | gi148696058 | ITNGQPQQTTGAASGGYIKR | Y138 | Synaptosomal associated protein 23 |
| Stat5a | gi6755672 | AVDGYVKPQIK | Y694 | Signal transducer and activator of transcription 5A |
| Stat5B | gi1174465 | AADGYVKPQIK | Y699 | Signal transducer and activator of transcription 5B |
| Stx4 | gi30354363 | NILSSADYVERGQEHVK | Y251 | Syntaxin 4A (placental) |
| Tcte-1 | gi201911 | SGLSYLLSK | Y93 | t-Complex-associated testis expressed 1 |
| TfR1 | gi11596855 | SAFSNLFGGEPLSYTR | Y20 | Transferrin receptor protein 1 |
| Tk1 | gi82919367 | SYINLPTVLPSSPSK | Y3 | Thymidine kinase 1 |
| Tln1 | gi49022858 | KSTVLQQQYNR | Y436 | Talin |
| Tmem 127 | gi28201995 | MYAPGGAGLPGGR | Y2 | Transmembrane protein 127 |
| Zdhhc7 | gi19527186 | EYMESLQLKPGEVIYKCPK | Y130 | Zinc finger, DHHC domain containing 7 |
| Gene symbol . | NCBI query ID . | Sequence . | PY . | Description . |
|---|---|---|---|---|
| Abi1 | gi116089341 | TLEPVKPPTVPNDYMTSPAR | Y213 | Abl interactor 1 |
| Ata2 | gi37360348 | SHYADVDPENQNFLLESNLGK | Y41 | Solute carrier family38, member 2 |
| Bam32 | gi5733602 | EVEEPCIYESVR | Y139 | Dual adaptor for phosphotyrosine and 3-phosphoinositides1 (DAPP1) |
| Blk | gi198939 | SLDNGGYYISPR | Y181 | Tyrosine-protein kinase Blk |
| Btk | gi2507603 | KVVALYDYMPMNANDLQLR | Y223 | Bruton tyrosine kinase |
| HYVVCSTPQSQYYLAEK | Y344 | |||
| HLFSTIPELINYHQHNSAGLISR | Y361 | |||
| Calml2 | gi4502549 | VFDKDGNGYISAAELR | Y100 | Calmodulin 2 |
| c-Cbl | gi80978932 | IKPSSSANAIYSLAARPLPMPK | Y672 | Casitas B-lineage lymphoma |
| Cdc2 | gi1575290 | IEKIGEGTYGVVYK | Y15 | p34 Cdc2 kinase |
| IGEGTYGVVYKGR | Y15 | |||
| Cdebp | gi1086521 | MQNHGYENPTYK | Y750 | CDE1-binding protein |
| Cdk2 | gi34556205 | VEKIGEGTYGVVYK | Y11 | Cyclin-dependent kinase 2 |
| Cdv3B | gi28461294 | KTPQGPPEIYSDTQFPSLQSTAK | Y213 | Carnitine deficiency-associated gene expressed in ventricle 3 |
| c-Kit | gi42768812 | VVEATAYGLIK | Y608 | Stem cell factor receptor |
| NLLHSTEPSCDSSNEYMDMKPGVSYVVPTKTDK | Y728 | |||
| IDSYIER | Y745 | |||
| MVSPEHAPAEMYDVMK | Y898 | |||
| HIYSNLANCNPNPENPVVVDHSVR | Y934 | |||
| Csk | gi729888 | VMEGTVAAQDEFYRSGWALNMK | Y184 | Tyrosine-protein kinase CSK (p50CSK) |
| Dok1 | gi27805460 | GFSSDTALYSQVQK | Y450 | Docking protein 1 |
| Dyrk1A | gi24418935 | IYQYIQSR | Y321 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a |
| Emd | gi6679641 | LIYGQDSAYQSIAHYRPISNVSR | Y167 | Emerin |
| Eno1 | gi12963491 | AAVPSGASTGIYEALELR | Y43 | Enolase 1 |
| Esyt1 | gi33859650 | QLLDDEERITAETLYMSHR | Y111 | Extended synaptotagmin-like protein 1 |
| ITAETLYMSHR | Y111 | |||
| Fer | gi118572319 | VQENDGKEPPPVVNYEEDAR | Y402 | Tyrosine-protein kinase FER |
| Fyb | gi33469119 | TTAVEIDYDSLKR | Y559 | FYN binding protein |
| Gab1 | gi6165415 | SSGSGSSMADERVDYVVVDQQK | Y660 | GRB2-associated binding protein 1 |
| Gsk3a | gi72384361 | GEPNVSYICSR | Y279 | Glycogen synthase kinase 3 alpha |
| Hgs | gi31982286 | YKVVQDTYQIMK | Y132 | HGF-regulated tyrosine kinase substrate |
| Hlb301 | gi547841 | SQDGYTYPSR | Y849 | Low-density lipoprotein receptor precursor (LDL receptor) |
| Itsn2 | gi20138801 | GEPEALYAAVTK | Y922 | Intersectin-2 |
| Jmjd1c | gi149260924 | MKEDKYDCVSR | Y150 | PREDICTED: jumonji domain containing 1C |
| Jnk2 | gi18254507 | TACTNFMMTPYVVTR | Y185 | c-Jun N-terminal kinase 2 |
| Jnk3 | gi30578163 | TAGTSFMMTPYVVTR | Y185 | c-Jun N-terminal kinase 3 |
| Jtk1 | gi133922607 | AVPEGHEYYR | Y1053 | Tyrosine kinase 2 (TYK2) |
| Lair1 | gi81913091 | AVGAVTSQSTDMAESSTYAAIIRH | Y257 | Leukocyte-associated immunoglobulin-like receptor 1 |
| Lamtor1 | gi81903514 | ALNGAEPNYHSLPSAR | Y40 | Regulator complex protein |
| IAAYAYSALSQIR | Y140 | |||
| Lrrc25 | gi23346481 | SVDQDSQPVYCNLESLGR | Y289 | Leucine-rich repeat-containing protein 25 precursor |
| Mapk1 | gi6754632 | VADPDHDHTGFLTEYVATR | Y185 | Mitogen activated protein kinase 1, Erk2 |
| Mapk14 | gi187465730 | HTDDEMTGYVATR | Y182 | Mitogen activated protein kinase 14, p38 |
| Mapk3 | gi21489933 | IADPEHDHTGFLTEYVATR | Y205 | Mitogen activated protein kinase 3, Erk1 |
| Muc-13 | gi148665438 | TGVPSQTPNPYANQR | Y562 | Mucin 13, epithelial transmembrane |
| p85 a | gi117320524 | DQYLMWLTQK | Y580 | Phosphatidylinositol 3-kinase regulatory subunit, p85 |
| Pctk3 | gi6679233 | LGEGTYATVFKGR | Y132 | Serine/threonine-protein kinase PCTAIRE-3 |
| Prpf4B | gi148708948 | LCDFGSASHVADNDITPYLVSR | Y849 | Pre-mRNA processing factor 4 homolog B |
| Ptpn6 | gi118130785 | GQESEYGNITYPPAVR | Y536 | Protein tyrosine phosphatase, non-receptor type 6 (SHP1) |
| Ptpn11 | gi6755228 | VYENVGLMQQQR | Y580 | Protein tyrosine phosphatase, non-receptor type 11 (SHP2) |
| Ptpn18 | gi113199761 | APTSTDTPIYSQVAPR | Y381 | Protein tyrosine phosphatase, non-receptor type 18 |
| ADTYAVVQKR | Y354 | |||
| Rab2A | gi10946940 | AYAYLFK | Y3 | Ras-related protein Rab-2A |
| Rps27 | gi4506711 | LVQSPNSYFMDVK | Y31 | Ribosomal protein S27 |
| Serpb1 | gi13385280 | SSFSHYSGLKHEDKR | Y207 | SERPINE1 mRNA binding protein 1 |
| Slc1A6 | gi6678003 | QIKYFSFPGELLMR | Y85 | Solute carrier family 1 (high affinity aspartate/glutamate transporter), number 6 |
| Snap23 | gi148696058 | ITNGQPQQTTGAASGGYIKR | Y138 | Synaptosomal associated protein 23 |
| Stat5a | gi6755672 | AVDGYVKPQIK | Y694 | Signal transducer and activator of transcription 5A |
| Stat5B | gi1174465 | AADGYVKPQIK | Y699 | Signal transducer and activator of transcription 5B |
| Stx4 | gi30354363 | NILSSADYVERGQEHVK | Y251 | Syntaxin 4A (placental) |
| Tcte-1 | gi201911 | SGLSYLLSK | Y93 | t-Complex-associated testis expressed 1 |
| TfR1 | gi11596855 | SAFSNLFGGEPLSYTR | Y20 | Transferrin receptor protein 1 |
| Tk1 | gi82919367 | SYINLPTVLPSSPSK | Y3 | Thymidine kinase 1 |
| Tln1 | gi49022858 | KSTVLQQQYNR | Y436 | Talin |
| Tmem 127 | gi28201995 | MYAPGGAGLPGGR | Y2 | Transmembrane protein 127 |
| Zdhhc7 | gi19527186 | EYMESLQLKPGEVIYKCPK | Y130 | Zinc finger, DHHC domain containing 7 |
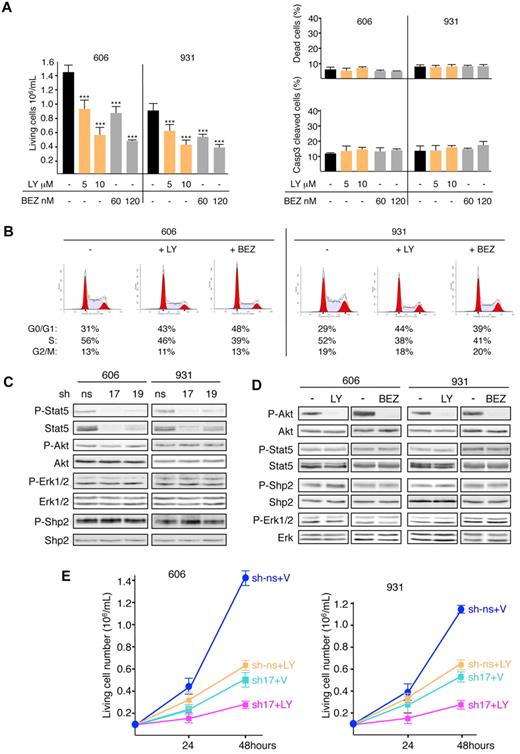
We confirmed the phosphoproteome data by Western blotting using phosphospecific Abs against PY580-Shp2 and PY694/699-Stat5. As illustrated in Figure 1B, Shp2 and Stat5 were tyrosine phosphorylated in 606HS2 and 931HS2 cells; in addition, treatment with Kit inhibitors (PP1, PP2, or IM) abolished Shp2 and Stat5 tyrosine phosphorylation, whereas Shp2 and Stat5 levels were not affected, indicating that the 2 types of Kit mutants were responsible for Shp2 and Stat5 phosphorylation. These findings were confirmed in a model of preleukemic 663HS1 cells in which enforced expression of Kit mutants (663-4A overexpressing KitD814Y and 663-8A overexpressing KitD818Y) led to the acquisition of an HS2 phenotype.9 PY580-Shp2 and PY694/699-Stat5 were seen in these cells in the absence of any cytokine, and their levels were dramatically reduced after treatment with Kit inhibitors, whereas Shp2 and Stat5 levels were not affected (Figure 1C). Therefore, expression of the Kit mutant in preleukemic cells was sufficient to promote constitutive Shp2 and Stat5 tyrosine phosphorylation.
Coimmunoprecipitation assays with Kit Abs of 606 and 931HS2 cell lysates demonstrated that Shp2 and Stat5 belong to Kit-signaling complexes (Figure 1D). The association between Shp2 and Kit was abolished when cells were treated with PP1, PP2, or IM, indicating that the Kit kinase activity determines the interaction of Shp2 with Kit. In contrast, Kit inhibitors did not modify the association between Stat5 and Kit, but PY-Stat5 forms were not detectable. Therefore, Stat5 was constitutively associated with Kit, but its phosphorylation was dependent on Kit activity. This association suggested that Shp2 and Stat5 were substrates of Kit kinase. However, the role of a nonreceptor tyrosine kinase linking Kit mutant with Shp2 and Stat5 cannot be excluded, especially because PP1 and PP2 are also inhibitors of Src kinases. The contribution of Jak and Src kinases in Kit signaling has been reported previously,21 and our proteomics screen identified phosphopeptides corresponding to members of Jak and Src kinase family. Nevertheless, the participation of Jak and Src kinases was excluded because neither Shp2 and Stat5 phosphorylation nor Kit signaling was altered when HS2 cells were treated with a panJak inhibitor22 or the Src-selective kinase inhibitor SU665623 (supplemental Figure 2).
Finally, we addressed the specificity of Shp2 and Stat5 phosphorylation with regard to the oncogenic status of Kit using preleukemic cells (HS1) from spi-1–TG mice. These cells made possible the analysis of signaling downstream of wild-type Kit in an erythroid cell context. In HS1 cells (633 and 663HS1) cultured with SCF, Shp2 was tyrosine phosphorylated (Figure 1E). When cells were SCF starved, both Kit and Shp2 phosphorylation levels were abolished, but phosphorylation of both proteins rapidly increased after SCF hyperstimulation. In contrast, no phosphorylated forms of Stat5 could be detected in SCF-stimulated HS1 cells. This indicates that Shp2 phosphorylation occurs downstream of wild-type and mutant Kit, whereas Stat5 is specifically phosphorylated downstream of mutant Kit.
These data strongly suggest that Shp2 and Stat5 are proximal effectors of Kit mutant kinase. To determine their pathogenic role in Kit-mediated transformation, we analyzed the biologic responses of HS2 leukemic cells after Shp2 and Stat5 knockdown by RNA interference.
Depletion of Shp2 induces apoptosis in leukemic cells
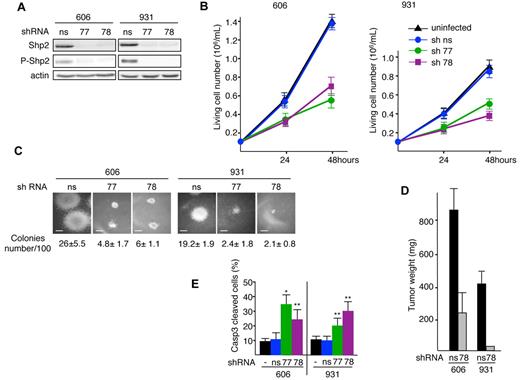
Two lentiviruses encoding independent shRNAs for Shp2 (sh77 or sh78) were used. Both shRNAs induced a major decrease (90%-95%) in Shp2 protein level compared with a nonsilencing shRNA (ns-shRNA; Figure 2A). The growth of Shp2-shRNA77– or sh78-transduced 606HS2 and 931HS2 cells was reduced by more than 50% over 48 hours compared with the proliferation rate of cells transduced with ns-shRNA or uninfected cells (Figure 2B). This was confirmed in methylcellulose clonogenic assays. Transduction of Shp2, shRNA77, or sh78 in 606HS2 and 931HS2 cells resulted in reductions of both the cloning efficiency (approximately 80%) and colony size (Figure 2C).
Knockdown of Shp2 reduces proliferation and induces apoptosis of leukemic cells. Cells (606HS2 and 931HS2) were infected with a lentiviral vector transducing Shp2-shRNA77 or Shp2-shRNA78 or a control ns-shRNA and selected for puromycin resistance. Experiments were performed 72 hours after infection. (A) Whole-cell extracts were immunoblotted with the indicated Abs and anti–β-actin Ab as a loading control. (B) Cells were plated at 1 × 105 cells/mL. Viable cells were scored by trypan blue exclusion at 24 and 48 hours. Data are means ± SEM (n = 3, performed in duplicate). (C) Representative colonies grown into methylcellulose for 6 days. Scale bars indicate 80 μm. The average number of colonies per 100 cells seeded ± SEM is indicated (n = 3, performed in duplicate). (D) Subcutaneous tumors in nude mice engrafted with 606HS2 and 931HS2 cells transduced with Shp2-shRNA78 or control ns-shRNA were taken 3 weeks after injection and weighed. Bar represents the mean weight ± SEM of 8 tumors for Shp2-shRNA78-606HS2 cells, ns-shRNA-606HS2 cells, and ns-shRNA-931HS2 cells. For Shp2-shRNA78-931HS2 cells, only 2 tumors were obtained. (E) Cells were plated at 1 × 105 cells/mL and the percentage of apoptotic cells was evaluated 48 hours later by flow cytometry using anti-active caspase 3 Abs. Bars represent means ± SEM (n = 3). Statistical differences from the value of the control (ns) are indicated as follows: *P < .05; **P < .01 (by Student t test).
Knockdown of Shp2 reduces proliferation and induces apoptosis of leukemic cells. Cells (606HS2 and 931HS2) were infected with a lentiviral vector transducing Shp2-shRNA77 or Shp2-shRNA78 or a control ns-shRNA and selected for puromycin resistance. Experiments were performed 72 hours after infection. (A) Whole-cell extracts were immunoblotted with the indicated Abs and anti–β-actin Ab as a loading control. (B) Cells were plated at 1 × 105 cells/mL. Viable cells were scored by trypan blue exclusion at 24 and 48 hours. Data are means ± SEM (n = 3, performed in duplicate). (C) Representative colonies grown into methylcellulose for 6 days. Scale bars indicate 80 μm. The average number of colonies per 100 cells seeded ± SEM is indicated (n = 3, performed in duplicate). (D) Subcutaneous tumors in nude mice engrafted with 606HS2 and 931HS2 cells transduced with Shp2-shRNA78 or control ns-shRNA were taken 3 weeks after injection and weighed. Bar represents the mean weight ± SEM of 8 tumors for Shp2-shRNA78-606HS2 cells, ns-shRNA-606HS2 cells, and ns-shRNA-931HS2 cells. For Shp2-shRNA78-931HS2 cells, only 2 tumors were obtained. (E) Cells were plated at 1 × 105 cells/mL and the percentage of apoptotic cells was evaluated 48 hours later by flow cytometry using anti-active caspase 3 Abs. Bars represent means ± SEM (n = 3). Statistical differences from the value of the control (ns) are indicated as follows: *P < .05; **P < .01 (by Student t test).
We next investigated whether extinction of Shp2 expression would affect tumor development in vivo. 606HS2 and 931HS2 cells were transduced with either Shp2-shRNA78 or ns-shRNA and subcutaneously implanted into nude mice. After 3 weeks, all (8 of 8) recipients engrafted with Shp2-shRNA78–606HS2 cells developed tumors that were 72% reduced in size compared with tumors generated by cells transduced with ns-shRNA. When Shp2-shRNA78–931HS2 cells were tested, very small tumors (25 and 30 mg) were found in only 2 of 6 recipients, whereas tumors weighing 350-510 mg were seen in all (8 of 8) recipients engrafted with ns-shRNA cells (Figure 2D). Therefore, Shp2 plays a role in maintaining in vitro autonomous proliferation and clonogenicity of leukemic cells, as well as in in vivo tumorigenicity.
To assess further the role of Shp2 in cell growth, we investigated whether Shp2 depletion would affect cell death and/or the cell cycle. We observed that the reduced proliferation of leukemic cells transduced with Shp2-shRNA was associated with an increased cell death as evaluated by trypan blue exclusion assay (data not shown). To determine whether cells died by apoptosis, the percentage of cells expressing the active form of caspase 324 was assessed by flow cytometric analysis. The number of apoptotic cells was increased more than 2-fold for both 606HS2 and 931HS2 cells when Shp2 was knocked down (Figure 2E), whereas no significant change in cell-cycle distribution could be detected (supplemental Figure 3). These data suggest that Shp2 extinction impairs proliferation by increasing apoptosis.
The Shp2/Mek/Erk pathway induces autonomous survival of leukemic cells
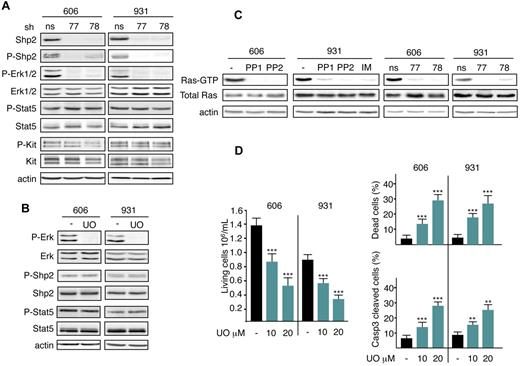
Other studies have reported that Shp2 triggers Erk activation.19 This led us to investigate whether Erk1/2 and Shp2 activation belong to the same signaling pathway downstream of mutant Kit. The phosphorylation level of Erk1/2 in HS2 cells was evaluated in comparison with the Shp2 level. Erk1/2 was not phosphorylated when Shp2 was knocked down (Figure 3A). Conversely, when Erk1/2 signaling was inactivated by treatment with the Mek inhibitor UO126, Shp2 phosphorylation was not modified (Figure 3B). These data demonstrate that Erk1/2 activation occurs downstream of Shp2.
Shp2 mediates Mek/Erk1/2 pathway activation for survival downstream of the Kit mutant in leukemic cells. (A) Whole-cell extracts from 606HS2 and 931HS2 cells transduced with Shp2-shRNA77 or Shp2-shRNA78 or control ns-shRNA were analyzed by immunoblotting with the indicated Abs 72 hours after infection. (B) Lysates from 606HS2 and 931HS2 cells treated or not (−; DMSO) for 4 hours with UO126 (UO; 20μM) were analyzed by immunoblotting with the indicated Abs. In panels A and B, data are from 1 representative experiment of 5. (C) The amount of activated Ras (Ras-GTP) was measured by Ras-GTP pull-down assays using the Ras-GTP–binding domain of Raf1 and lysates from 606HS2 and 931HS2 cells treated with Kit inhibitors or not (−) and lysates from 606HS2 and 931HS2 cells transducing Shp2-shRNA77 or Shp2-shRNA78 or control ns-shRNA 72 hours after infection. Anti-Ras immunoblotting of the corresponding lysates was performed to control the amount of total Ras in each sample. Anti–β-actin Ab was used as a loading control. Data are from 1 representative experiment of 3. (D) Cells (606HS2 and 931HS2) were plated at 1 × 105 cells/mL and cultured for 48 hours in the absence (−; DMSO) or in the presence of the indicated concentrations of UO126. The number of living cells, the percentage of dead cells evaluated by trypan blue exclusion, and the percentage of apoptotic cells evaluated by flow cytometry using anti-active caspase 3 Abs were determined. Data are means ± SEM (n = 3). Statistical differences from the value of the control (−) are indicated as follows: **P < .01; ***P < .001 (by Student t test).
Shp2 mediates Mek/Erk1/2 pathway activation for survival downstream of the Kit mutant in leukemic cells. (A) Whole-cell extracts from 606HS2 and 931HS2 cells transduced with Shp2-shRNA77 or Shp2-shRNA78 or control ns-shRNA were analyzed by immunoblotting with the indicated Abs 72 hours after infection. (B) Lysates from 606HS2 and 931HS2 cells treated or not (−; DMSO) for 4 hours with UO126 (UO; 20μM) were analyzed by immunoblotting with the indicated Abs. In panels A and B, data are from 1 representative experiment of 5. (C) The amount of activated Ras (Ras-GTP) was measured by Ras-GTP pull-down assays using the Ras-GTP–binding domain of Raf1 and lysates from 606HS2 and 931HS2 cells treated with Kit inhibitors or not (−) and lysates from 606HS2 and 931HS2 cells transducing Shp2-shRNA77 or Shp2-shRNA78 or control ns-shRNA 72 hours after infection. Anti-Ras immunoblotting of the corresponding lysates was performed to control the amount of total Ras in each sample. Anti–β-actin Ab was used as a loading control. Data are from 1 representative experiment of 3. (D) Cells (606HS2 and 931HS2) were plated at 1 × 105 cells/mL and cultured for 48 hours in the absence (−; DMSO) or in the presence of the indicated concentrations of UO126. The number of living cells, the percentage of dead cells evaluated by trypan blue exclusion, and the percentage of apoptotic cells evaluated by flow cytometry using anti-active caspase 3 Abs were determined. Data are means ± SEM (n = 3). Statistical differences from the value of the control (−) are indicated as follows: **P < .01; ***P < .001 (by Student t test).
To investigate whether activation of Mek/Erk1/2 by Shp2 occurred through activated Ras GTPases, as was observed previously in response to growth factors,25 we investigated whether the active forms of Ras proteins were detected in HS2 cells. This was evaluated by the amount of GTP-bound Ras found in Ras pull-down assays. Figure 3C shows that Ras GTPases were activated in HS2 cells. After treatment of 606HS2 and 931HS2 cells with Kit inhibitors or after Shp2 knockdown, no GTP-bound Ras could be detected, whereas the level of total Ras was unchanged, indicating that mutant Kit and Shp2 contribute to Ras activation. These data identify Shp2/Ras/Mek/Erk1/2 as being a biochemical pathway activated downstream of the Kit mutant.
It was predictable that Erk1/2 would exert an antiapoptotic function in leukemic cells. Treatment with UO126 inhibited the growth of 606HS2 and 931HS2 cells (an approximately 35% decrease with 10μM and a 60% decrease with 20μM UO126; Figure 3D). This effect was associated with an increase in apoptotic cell number according to UO126 dose, whereas the cell-cycle progression was not affected (data not shown). Therefore, Erk1/2 activation prevents apoptosis, as was also shown above for Shp2. These data identify Shp2/Ras/Mek/Erk1/2 as being a biochemical pathway preventing apoptosis in leukemic cells.
Depletion of Stat5 causes cell-cycle arrest in G1
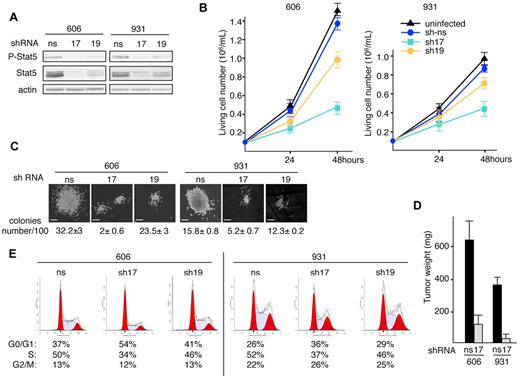
Two lentiviruses encoding independent shRNAs for Stat5 (sh17 and sh19) were generated. Compared with control ns-shRNA, the expression of Stat5-shRNA17 or sh19 in 606HS2 and 931HS2 cells reduced the Stat5 protein level by 90% or 50%, respectively (Figure 4A). Proliferation assays over 48 hours (Figure 4B) showed that Stat5 extinction by Stat5-shRNA17 reduced the number of 606HS2 or 931HS2 cells by 65% or 55%, respectively, compared with control cells. Consistent with its weaker effects on Stat5 extinction, Stat5-shRNA19 reduced cell numbers by 30% or 20% for 606HS2 or 931HS2 cells, respectively. These reduced proliferative capacities were also observed in clonogenic assays. The cloning efficiency of 606HS2 and 931HS2 cells was reduced by 94% and 67%, respectively, after transduction with Stat5-shRNA17, and by 27% and 22%, respectively after transduction with Stat5-shRNA19. Moreover, the size of the colonies derived from both cell lines was drastically reduced compared with control cells (Figure 4C). We also investigated whether extinction of Stat5 expression would affect tumor development in vivo. 606HS2 and 931HS2 cells transduced with Stat5-shRNA17 were engrafted subcutaneously into nude mice and all recipients developed tumors within 3 weeks. However, tumor weights were significantly reduced (by 75%-90%) compared with tumors generated by control cells (Figure 4D). Therefore, tumor development was strongly restrained when Stat5 was depleted in leukemic cells.
Knockdown of Stat5 reduces proliferation and alters cell cycle of leukemic cells. (A) Cells (606HS2 and 931HS2) were infected with a lentiviral vector transducing Stat5-shRNA17 or Shp2-shRNA19 or control ns-shRNA and sorted for green fluorescent protein expression. Experiments were performed 72 hours after infection. Whole-cell extracts were immunoblotted with anti–P-Stat5 and anti-Stat5 Abs and anti–β-actin Ab as a loading control. (B) Cells were plated at 1 × 105 cells/mL. Viable cells were evaluated by trypan blue exclusion at 24 and 48 hours. Data are means ± SEM (n = 4, performed in duplicate). (C) Representative colonies grown into methylcellulose for 6 days. Bars correspond to 80 μm. The average number of colonies per 100 seeded cells ± SEM is indicated below (n = 3, performed in duplicate). (D) Subcutaneous tumors in nude mice engrafted with 606HS2 and 931HS2 cells transducing Stat5-shRNA17 or control ns-shRNA were taken 3 weeks after injection and weighed. Bar represents the mean weight ± SEM of 8 tumors. (E) Representative cell-cycle distribution of green fluorescent protein–sorted cells plated at 1 × 105 cells/mL for 24 hours. The percentages of living cells in the G0/G1, S, and G2/M phases of the cell cycle are indicated below. The values are from 1 of 3 independent experiments performed at 48 hours. Similar results were obtained when experiments were performed at 24 hours (not shown).
Knockdown of Stat5 reduces proliferation and alters cell cycle of leukemic cells. (A) Cells (606HS2 and 931HS2) were infected with a lentiviral vector transducing Stat5-shRNA17 or Shp2-shRNA19 or control ns-shRNA and sorted for green fluorescent protein expression. Experiments were performed 72 hours after infection. Whole-cell extracts were immunoblotted with anti–P-Stat5 and anti-Stat5 Abs and anti–β-actin Ab as a loading control. (B) Cells were plated at 1 × 105 cells/mL. Viable cells were evaluated by trypan blue exclusion at 24 and 48 hours. Data are means ± SEM (n = 4, performed in duplicate). (C) Representative colonies grown into methylcellulose for 6 days. Bars correspond to 80 μm. The average number of colonies per 100 seeded cells ± SEM is indicated below (n = 3, performed in duplicate). (D) Subcutaneous tumors in nude mice engrafted with 606HS2 and 931HS2 cells transducing Stat5-shRNA17 or control ns-shRNA were taken 3 weeks after injection and weighed. Bar represents the mean weight ± SEM of 8 tumors. (E) Representative cell-cycle distribution of green fluorescent protein–sorted cells plated at 1 × 105 cells/mL for 24 hours. The percentages of living cells in the G0/G1, S, and G2/M phases of the cell cycle are indicated below. The values are from 1 of 3 independent experiments performed at 48 hours. Similar results were obtained when experiments were performed at 24 hours (not shown).
To determine the defect in cellular proliferation induced by Stat5 extinction, cell viability and the cell cycle were investigated. The reduced cell-growth rate after extinction of Stat5 was not associated with apoptosis (supplemental Figure 4A). Flow cytometric analysis of the cell-cycle distribution (Figure 4E) showed that Stat5 extinction by Stat5-shRNA17 led to an increase of cells in the G0/G1 phase (33% for 606HS2 and 28.3% for 931HS2 cells) and a reduction of cells in the S phase (30.8% in 606HS2 cells and 22.3% in 931HS2 cells; P < .001 by Student t test). According to the partial extinction of Stat5, Stat5-shRNA19 had moderate effects on the cell cycle (+9.8% or +9.2% of cells in G0/G1 phase and −11.9% or −11% of cells in the S phase in 606HS2 or 931HS2 cells compared with control cells). These data suggest that Stat5 contributes to the autonomous proliferation of leukemic cells by inducing cells to exit from the G1 phase of the cell cycle, but with no effect on survival.
The PI3K/AKT pathway promotes an accumulation of cells in the S phase of the cell cycle
The PI3K/Akt pathway is required for extensive proliferation of HS2 leukemic cells.9,18 Indeed, p85 was found in the phosphoproteome. Its expression was not changed by Kit inhibitors (Figure 1B) and, like Stat5, it interacted constitutively with Kit in the coimmunoprecipitation assay (Figure 1D). Therefore, we investigated the role of the PI3K/Akt pathway in the leukemic phenotype. Cell survival and cell-cycle progression of 606HS2 and 931HS2 cells treated with the PI3K inhibitor LY294002 were analyzed. Treatment caused a 35%-60% inhibition in proliferation depending on LY294002 dose (5 or 10μM), with no detectable cell death shown by trypan blue assay or caspase 3 cleavage (Figure 5A). Conversely, flow cytometric analysis revealed a significant increase of cells in the G0/G1 phase (30.5% or 49.9% in 606HS2 or 931HS2 cells, respectively) concomitantly with a decrease in cells in the S phase (17.1% or 24.7% in 606HS2 or 931HS2 cells, respectively; P < .001 by Student t test; Figure 5B).
PI3K/AKT pathway induces an accumulation of cells in the S phase of cell cycle and cooperates with Stat5 to promote cell growth. (A) Cells (606HS2 and 931HS2) were plated at 1 × 105 cells/mL and cultured for 48 hours in the absence (−; ETOH) or in the presence of indicated concentrations of LY294002 (LY) or NVP-BEZ235 (BEZ). The number of living cells, the percentage of dead cells (trypan blue exclusion), and the percentage of apoptotic cells (by flow cytometry using anti-active caspase 3 Abs) were determined. Bar is the mean ± SEM (n = 3). Statistical differences from the value of the living cells control (−) are indicated as follows: ***P < .001 (by Student t test). Mortality after LY294002 or NVP-BEZ235 treatment was not significantly different (P > .05 by Student t test). (B) Representative cell-cycle distribution of HS2 cells treated or not (−; ETOH) with LY294002 (10μM) or NVP-BEZ235 (120nM) for 24 hours. The percentages of living cells in the G0/G1, S, and G2/M phases of the cell cycle are indicated below. The values are from 1 of 3 independent experiments performed at 24 and 48 hours that gave similar results. (C) Whole-cell extracts from 606HS2 and 931HS2 cells transducing Stat5-sh-RNA17 or Stat5-shRNA19 or control ns-shRNA were analyzed by immunoblotting with the indicated Abs. (D) Lysates from 606HS2 and 931HS2 cells treated or not (−; ETOH) with LY294002 (10μM) or NVP-BEZ235 (120nM) for 4 hours were analyzed by immunoblotting with the indicated Abs. (E) Cells (606HS2 and 931HS2) transducing Stat5-shRNA-17 or control ns-shRNA were plated 72 hours after infection at 1 × 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of LY294002 (10μM). The number of viable cells was scored at 24 and 48 hours. Data are means ± SEM (n = 3). Statistical differences from the value of living cells control (sh-ns + V) are P < .001. Statistical differences between the value of living cells (sh-ns + LY) and (sh17 + LY) and between the value of living cells (sh17 + V) and (sh17 + LY) were P < .001 (by Student t test).
PI3K/AKT pathway induces an accumulation of cells in the S phase of cell cycle and cooperates with Stat5 to promote cell growth. (A) Cells (606HS2 and 931HS2) were plated at 1 × 105 cells/mL and cultured for 48 hours in the absence (−; ETOH) or in the presence of indicated concentrations of LY294002 (LY) or NVP-BEZ235 (BEZ). The number of living cells, the percentage of dead cells (trypan blue exclusion), and the percentage of apoptotic cells (by flow cytometry using anti-active caspase 3 Abs) were determined. Bar is the mean ± SEM (n = 3). Statistical differences from the value of the living cells control (−) are indicated as follows: ***P < .001 (by Student t test). Mortality after LY294002 or NVP-BEZ235 treatment was not significantly different (P > .05 by Student t test). (B) Representative cell-cycle distribution of HS2 cells treated or not (−; ETOH) with LY294002 (10μM) or NVP-BEZ235 (120nM) for 24 hours. The percentages of living cells in the G0/G1, S, and G2/M phases of the cell cycle are indicated below. The values are from 1 of 3 independent experiments performed at 24 and 48 hours that gave similar results. (C) Whole-cell extracts from 606HS2 and 931HS2 cells transducing Stat5-sh-RNA17 or Stat5-shRNA19 or control ns-shRNA were analyzed by immunoblotting with the indicated Abs. (D) Lysates from 606HS2 and 931HS2 cells treated or not (−; ETOH) with LY294002 (10μM) or NVP-BEZ235 (120nM) for 4 hours were analyzed by immunoblotting with the indicated Abs. (E) Cells (606HS2 and 931HS2) transducing Stat5-shRNA-17 or control ns-shRNA were plated 72 hours after infection at 1 × 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of LY294002 (10μM). The number of viable cells was scored at 24 and 48 hours. Data are means ± SEM (n = 3). Statistical differences from the value of living cells control (sh-ns + V) are P < .001. Statistical differences between the value of living cells (sh-ns + LY) and (sh17 + LY) and between the value of living cells (sh17 + V) and (sh17 + LY) were P < .001 (by Student t test).
We also used another PI3K inhibitor, NVP-BEZ235, which is currently undergoing phase 1/2 clinical trials in cancer patients26 (www.clinicaltrials.gov identifier NCT00620594). NVP-BEZ235 inhibited the proliferation of HS2 cells (35% decrease with 60nM and 60% decrease with 120nM; Figure 5A). This effect was associated with cell -cycle modifications (+34% or +24% of cells in the G0/G1 phase and −24% or −20% of cells in the S phase in 606HS2 or 931HS2 cells compared with control cells (P < .001 by Student t test) without cell death (Figure 5A-B). Increasing the NVP-BEZ235 concentration never led to complete inhibition of cell proliferation, even at higher doses (data not shown), as was observed with LY294002 treatment.18 These results emphasize the role of the PI3K/Akt pathway in promoting exit from the G1 phase of cell cycle.
Because the cell-cycle defects associated with Stat5 depletion and PI3K/Akt inhibition were similar, we speculated that Stat5 might be a mediator of the PI3K/Akt pathway. However, Stat5 depletion did not affect Akt phosphorylation (Figure 5C). Reciprocally, no change in Stat5 phosphorylation level could be observed in HS2 cells treated with LY294002 or NVP-BEZ235 (Figure 5D). In view of this absence of signaling interference, we could not detect an association of p85 with Stat5 in coimmunoprecipitation assays (data not shown).
Because no cross-talk between the PI3K/Akt and Stat5 pathways could be observed, we investigated whether the depletion of Stat5 and the inhibition of PI3K/Akt might have additive effects in inhibiting cell proliferation. Treatment of Stat5-shRNA17-cells with LY294002 resulted in an 80% inhibition of proliferation, which was superior to that induced by either Stat5 depletion alone (approximately 60%) or LY294002 treatment alone (approximately 55%; Figure 5E) and a slight reduction of cells in the S phase (supplemental Figure 4B), without significant cell death (supplemental Figure 4C). This result indicates that PI3K/Akt and Stat5 cooperate to stimulate HS2-cell proliferation.
Combined inhibition of cell cycle and survival is required to eradicate leukemic cell growth
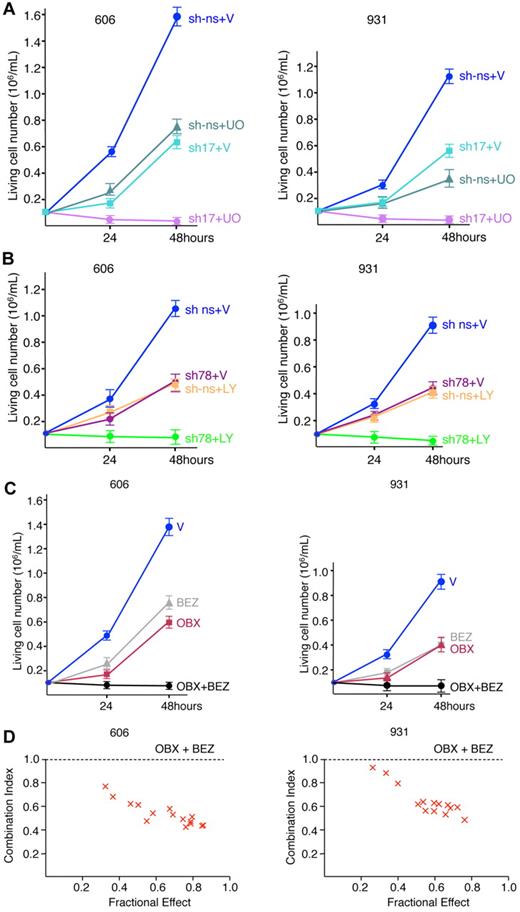
Our data thus far had provided evidence that Stat5 contributes with PI3K/Akt to cell-cycle progression with no effect on survival, whereas Shp2 contributes to cell survival with no effect on the cell cycle, strongly suggesting that 2 distinct and independent signaling pathways are involved in the maintenance of leukemic cells. Indeed, when only one of these pathways (depletion of Shp2 or Stat5 or inhibition of PI3K or Erk1/2) or 2 pathways on the same biologic process (depletion of Stat5 and inhibition of PI3K) was modulated, proliferation was reduced but cell growth was never completely abolished. We then combined Stat5 depletion by Stat5-shRNA17 with pharmacologic inhibition of the Erk1/2 pathway with UO126, which resulted in a total loss of cell proliferation (Figure 6A). In parallel, proliferation assays were performed with cells depleted in Shp2 by Shp2-shRNA78 in combination with inhibition of the PI3K/Akt pathway by LY294002 (Figure 6B) and, again, the association of Shp2 and PI3K/Akt inhibition completely inhibited cellular proliferation.
Combined inhibition of cell cycle and survival eradicate leukemic cell growth. (A) Cells (606HS2 and 931HS2) transducing Stat5-shRNA17 or control ns-shRNA were plated 72 hours after infection at 1 × 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of UO126 (20μM). The number of viable cells was scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing sh-ns + V with sh-ns + UO or sh17 + V and comparing sh-ns + UO or sh17 + V with sh17 + UO (by Student t test). (B) Cells (606HS2 and 931HS2) transducing Shp2-shRNA78 or control ns-shRNA were plated 72 hours after infection at 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of LY294002 (10μM). Viable cells were scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing sh-ns + V with sh-ns + LY or sh78 + V and comparing sh-ns + LY or sh78 + V with sh78 + LY (by Student t test). (C) Cells (606HS2 and 931HS2) were plated at 1 × 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of NVP-BEZ235 (100nM) and/or Obatoclax (OBX; 100nM). Viable cells were scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing control cells with cells grown in the presence of NVP-BEZ235 and/or OBX (by Student t test). (D) The effects of BEZ and OBX on cell proliferation were analyzed according to the Chou-Talalay method using CalcuSyn software.30 Cells (606HS2 and 931HS2) were treated with suboptimal concentrations of BEZ combined with suboptimal concentrations of OBX. The combination index values were determined from these dose-response curves for each combination. Combination index values above 1.1 indicate antagonistic, 0.9-1.1 additive, 0.7-0.9 moderately synergistic, and 0.3-0.7 synergistic effects.
Combined inhibition of cell cycle and survival eradicate leukemic cell growth. (A) Cells (606HS2 and 931HS2) transducing Stat5-shRNA17 or control ns-shRNA were plated 72 hours after infection at 1 × 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of UO126 (20μM). The number of viable cells was scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing sh-ns + V with sh-ns + UO or sh17 + V and comparing sh-ns + UO or sh17 + V with sh17 + UO (by Student t test). (B) Cells (606HS2 and 931HS2) transducing Shp2-shRNA78 or control ns-shRNA were plated 72 hours after infection at 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of LY294002 (10μM). Viable cells were scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing sh-ns + V with sh-ns + LY or sh78 + V and comparing sh-ns + LY or sh78 + V with sh78 + LY (by Student t test). (C) Cells (606HS2 and 931HS2) were plated at 1 × 105 cells/mL and cultured in the absence (V, vehicle) or in the presence of NVP-BEZ235 (100nM) and/or Obatoclax (OBX; 100nM). Viable cells were scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing control cells with cells grown in the presence of NVP-BEZ235 and/or OBX (by Student t test). (D) The effects of BEZ and OBX on cell proliferation were analyzed according to the Chou-Talalay method using CalcuSyn software.30 Cells (606HS2 and 931HS2) were treated with suboptimal concentrations of BEZ combined with suboptimal concentrations of OBX. The combination index values were determined from these dose-response curves for each combination. Combination index values above 1.1 indicate antagonistic, 0.9-1.1 additive, 0.7-0.9 moderately synergistic, and 0.3-0.7 synergistic effects.
To validate our data with inhibitors currently in clinical trials, we used NVP-BEZ235, a PI3K inhibitor, and Obatoclax, which is known to antagonize all Bcl2 prosurvival members and to be potent against a variety of solid and hematologic malignancies.27-29 We determined that Obatoclax inhibited proliferation (35% decrease with 60nM and 60% decrease with 120nM) and induced apoptosis in HS2 leukemic cells without cell-cycle modification (supplemental Figure 5), but failed to completely abolish proliferation even when doses were increased (data not shown). When Obatoclax and NVP-BEZ235 were used in combination, each at a concentration resulting in a 50% reduction in cell proliferation (IC50), cell proliferation was totally suppressed (Figure 6C). Therefore, the combined inhibition of a signaling pathway controlling the cell cycle and a signaling pathway controlling survival is a powerful strategy to eradicate leukemic growth.
Finally, we analyzed whether these 2 drugs had additive or synergistic effects on proliferation. The presence of synergy was examined using the Chou-Talalay method.30 This method determines whether the combination of the 2 drugs produces an effect that exceeds that predicted by the simple addition of the individual drugs. Figure 6D shows that Obatoclax and NVP-BEZ235 are synergistic inhibitors of leukemic proliferation.
Combined inhibition of cell cycle and survival eradicate the proliferation of human Kit-driven leukemia
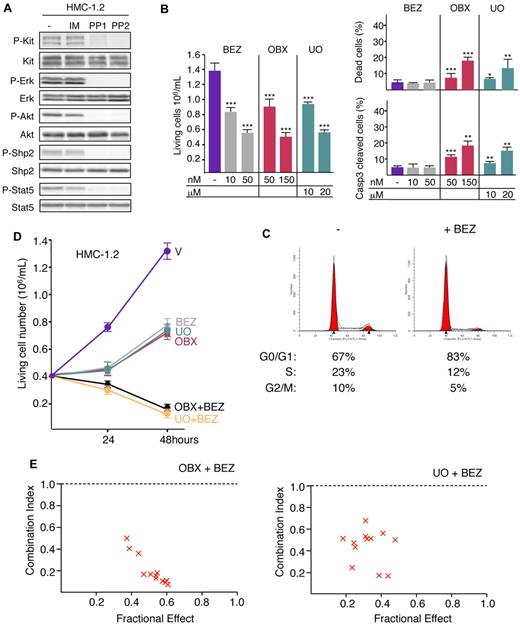
To determine whether the synergistic effect mediated by inhibition of both the cell-cycle and survival pathways was effective in human cells, experiments were performed with the HMC-1.2 cell line derived from an AML patient with mast cell leukemia.11,12 HMC-1.2 cells express mutant KitD816V and are resistant to IM,11 but treatment with PP1 or PP2 inhibits their proliferation and induces apoptosis in a dose-dependent manner (supplemental Figure 6). Accordingly, the constitutive phosphorylation of Kit and activation of Shp2, Erk1/2, Akt, and Stat5 were sensitive to PP1 and PP2 but not to IM (Figure 7A). HMC-1.2 cells were treated with NVP-BEZ235 or UO126. Both compounds inhibited cell proliferation in a dose-dependent manner (Figure 7B). NVP-BEZ235 induced dephosphorylation of Akt without modifications on other signaling pathways studied previously (supplemental Figure 7). At IC50, NVP-BEZ235 treatment modified the cell cycle (+32% of cells in the G0/G1 phase and −50% of cells in the S phase compared with control cells; P < .001 by Student t test; Figure 7C) but had no effect on cell death (Figure 7B). At IC50, UO126 treatment induced apoptosis (Figure 7B) and inhibited Erk1/2 activation without changing other signaling pathways (supplemental Figure 7). Therefore, mutant KitD816 in humans, like mutant KitD814 in mice, promotes the survival of leukemic cells through activation of the Erk1/2 pathway and controls the G1/S transition through activation of the PI3K pathway. Treatment of HMC-1.2 cells with Obatoclax inhibited proliferation in a dose-dependent manner by inducing apoptosis (Figure 7B) without cell-cycle modifications (supplemental Figure 8). HMC-1.2 cells were treated with the combination of NVP-BEZ235 with either Obatoclax or UO126. When NVP-BEZ235 and Obatoclax or UO126 were used in combination, each at a concentration resulting in a 50% reduction in cell proliferation (IC50), cell proliferation was totally suppressed whatever the combination of compounds tested (Figure 7D).
Combining NVP-BEZ235 with Obatoclax or UO126 eradicates proliferation of human HMC-1.2 leukemia cell line. (A) Whole-cell lysates from HMC-1.2 cells treated or not (−; DMSO) for 4 hours with IM (10μM), PP1 (10μM), or PP2 (10μM) were analyzed by immunoblotting with the indicated Abs. Data are from 1 representative experiment of 3. (B) HMC-1.2 cells were plated at 4 × 105 cells/mL and cultured in the absence (−; DMSO) or in the presence of indicated concentrations of NVP-BEZ235, Obatoclax (OBX), or UO126. Number of living cells, percentage of dead cells (trypan blue exclusion), and percentage of apoptotic cells (flow cytometry using anti-active caspase 3 Abs) were determined at 48 hours. Bar is mean ± SEM (n = 3). Statistical differences from the value of living cells control (−) are indicated as follows: ***P < .001; **P < .01; *P < .05 (by Student t test). Mortality after NVP-BEZ235 treatment was not significantly different (P > .05 by Student t test). (C) Representative cell-cycle distribution of HMC-1.2 cells treated or not (−; DMSO) with NVP-BEZ235 (50nM) for 48 hours. The percentages of living cells in the G0/G1, S, and G2/M phases of the cell cycle are indicated below. Data are from 1 of 3 independent experiments. (D) HMC-1.2 cells were plated at 4 × 105 cells/mL and cultured in the absence (V: DMSO) or in the presence of NVP-BEZ235 (25nM), Obatoclax (150nM), or UO126 (15μM) and the combination of NVP-BEZ235 (25nM) with Obatoclax (150nM) or NVP-BEZ235 (25nM) with UO126 (15μM). Viable cells were scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing control cells with cells grown in the presence of NVP-BEZ235, Obatoclax, and/or UO126 (by Student t test). (E) The effects of BEZ and OBX or UO126 on cell proliferation were analyzed according to the Chou-Talalay method using CalcuSyn software.30 HMC-1.2 cells were treated with suboptimal concentrations of BEZ combined with suboptimal concentrations of OBX or UO126. The combination index values were determined from these dose-response curves for each combination. Combination index values of 0.9-1.1 indicate additive, 0.7-0.9 moderately synergistic, 0.3-0.7 synergistic, and less than 0.3 strongly synergistic effects.
Combining NVP-BEZ235 with Obatoclax or UO126 eradicates proliferation of human HMC-1.2 leukemia cell line. (A) Whole-cell lysates from HMC-1.2 cells treated or not (−; DMSO) for 4 hours with IM (10μM), PP1 (10μM), or PP2 (10μM) were analyzed by immunoblotting with the indicated Abs. Data are from 1 representative experiment of 3. (B) HMC-1.2 cells were plated at 4 × 105 cells/mL and cultured in the absence (−; DMSO) or in the presence of indicated concentrations of NVP-BEZ235, Obatoclax (OBX), or UO126. Number of living cells, percentage of dead cells (trypan blue exclusion), and percentage of apoptotic cells (flow cytometry using anti-active caspase 3 Abs) were determined at 48 hours. Bar is mean ± SEM (n = 3). Statistical differences from the value of living cells control (−) are indicated as follows: ***P < .001; **P < .01; *P < .05 (by Student t test). Mortality after NVP-BEZ235 treatment was not significantly different (P > .05 by Student t test). (C) Representative cell-cycle distribution of HMC-1.2 cells treated or not (−; DMSO) with NVP-BEZ235 (50nM) for 48 hours. The percentages of living cells in the G0/G1, S, and G2/M phases of the cell cycle are indicated below. Data are from 1 of 3 independent experiments. (D) HMC-1.2 cells were plated at 4 × 105 cells/mL and cultured in the absence (V: DMSO) or in the presence of NVP-BEZ235 (25nM), Obatoclax (150nM), or UO126 (15μM) and the combination of NVP-BEZ235 (25nM) with Obatoclax (150nM) or NVP-BEZ235 (25nM) with UO126 (15μM). Viable cells were scored at 24 and 48 hours. Data are means ± SEM (n = 3). P < .001 comparing control cells with cells grown in the presence of NVP-BEZ235, Obatoclax, and/or UO126 (by Student t test). (E) The effects of BEZ and OBX or UO126 on cell proliferation were analyzed according to the Chou-Talalay method using CalcuSyn software.30 HMC-1.2 cells were treated with suboptimal concentrations of BEZ combined with suboptimal concentrations of OBX or UO126. The combination index values were determined from these dose-response curves for each combination. Combination index values of 0.9-1.1 indicate additive, 0.7-0.9 moderately synergistic, 0.3-0.7 synergistic, and less than 0.3 strongly synergistic effects.
The Chou-Talalay method30 was again applied to the determination of synergism (Figure 7E). The combinations of NVP-BEZ235 with UO126 or NVP-BEZ235 with Obatoclax exhibited strong synergistic effects for inhibiting the proliferation of HMC-1.2 cells.
Our findings are consistent with the principle that the combined inhibition of survival and the cell cycle is a powerful strategy to eradicate leukemic growth.
Discussion
Persistent kinase activity mediated by Kit mutations is associated with several hematopoietic and nonhematopoietic tumors. The identification of selective inhibitors of Kit has provided therapeutic approaches, but relapses to treatment occur. Characterizing the pathways that link mutant Kit to cell survival and proliferation may facilitate the development of new targeted therapies. The present study was designed to identify the biochemical networks downstream of mutant Kit in leukemia using the model of leukemic progression in spi-1/PU.1-TG mice. Starting from a phosphoproteomic screen, we showed that Kit-driven leukemogenesis depends on a linear Kit/Shp2/Ras/Mek/Erk pathway that prevents apoptosis, and that both the Kit/Stat5 and PI3K/Akt pathways cooperate to drive the G1/S transition during the cell cycle. The combined inhibition of survival and proliferation induces full eradication of leukemic cell growth, a way to counteract the resistance to Kit inhibitors.
We have applied a global profiling of PY peptides in HS2 cells. The phosphoproteome analysis identified a limited number of PY peptides. Our proteomic findings cannot be considered as a fully representative molecular signature of Kit signaling in leukemic cells because the phosphoproteome technology introduces a variety of pitfalls, such as incompatibility between trypsin treatment and LC-MS/MS analysis or low signal levels for a given peptide. However, this strategy allowed the identification of 2 signaling effectors absolutely required for the oncogenic function of the Kit mutant whatever the Kit mutation (KitD814Y or KitD818Y). We validated that Shp2 and Stat5 were phosphorylated in HS2 leukemic cells and that persistent phosphorylation depends on mutant Kit activity. Their physical interaction with Kit suggests that Shp2 and Stat5 are substrates of Kit kinase, but a role for a cytoplasmic tyrosine kinase (apart from Jak and Src kinases) in mediating their phosphorylation remains possible.
In the present study, we have shown for the first time that Shp2 is a target of mutant Kit kinase, leading to the persistent activation of Shp2 in leukemic cells. In a cellular context in which Kit is wild-type, as in preleukemic HS1 cells, Shp2 phosphorylation was inducible by SCF, as would be expected from the documented role of Shp2 in mediating cytokine signaling.19 The persistence of Shp2 phosphorylation is evocative of leukemogenic mechanisms implicating somatic Shp2 mutations in humans.31,32 Mutations in PTPN11, the gene encoding Shp2, generate proteins that most frequently exhibit an increased and persistent phosphatase activity33 and have been causally linked to juvenile myelomonocytic leukemia and acute leukemias.31,32 We have shown herein that Shp2 belongs to the Kit/Shp2/Ras/Mek/Erk1/2 signaling pathway, making it the main factor in the resistance to apoptosis in leukemic cells. How Shp2 controls persistent Erk activation is not known. The phosphatase activity of Shp2 has been reported to down-regulate the activity of wild-type Kit,34 but Shp2 depletion did not change the phosphorylation of the Kit mutant in leukemic cells. Therefore, the loss of a negative feedback control on Kit activity might account for the persistent activation of Shp2. Ras GTPases are constitutively activated downstream of mutant Kit/Shp2 in leukemic cells and Shp2 might dephosphorylate a Ras negative regulator, as has been reported in EGF and HGF signaling.19
Our data herein demonstrate that the permanent phosphorylation of Stat5 depends on Kit kinase activity. Inappropriate activation of Stats is associated with various types of cancer.35 In particular, in chronic myeloid leukemia and AML, Stat5 is activated by oncogenic tyrosine kinases such as BCR-ABL, TEL-JAK2, and the FLT3 mutant.36 Targeting of Stat5 by Kit is specific to the mutant Kit in leukemic cells as opposed to the wild-type Kit in preleukemic cells, raising the question of the underlying mechanism. Mutated forms of FLT3 in human AML induce the persistent activation of Stat5 when they are anchored in the endoplasmic reticulum.37,38 Recent data in human neoplastic mast cells showed that the KitD816V mutant induces permanent activation of Stat539 and promotes signaling from the endoplasmic reticulum.40,41 Nevertheless, we did not find any difference between mutant Kit localization in leukemic cells and wild-type Kit localization in preleukemic cells (data not shown).
In the present study, we have shown that Stat5 favors the G1/S transition during cell-cycle progression, whereas Shp2 promotes cell survival. In addition, the PI3K/Akt pathway also favors cell-cycle progression and the effects of Stat5 and PI3K/Akt on the cell cycle are additive. Accordingly, PI3K/Akt is not an effector of Stat5 and vice versa. Moreover, there is no evidence for cross-talk between the Kit/Shp2 and Kit/Stat5 pathways. Therefore, a striking feature arising from these data are the absence of cross-talk between the signaling pathways we studied. It is generally accepted that RTK signaling networks are connected internally. For example, the Erk and PI3K/Akt pathways are linked with effectors such as Ras that regulate both pathways,42,43 and Shp2 can modulate the activation of the PI3K/Akt pathway44,45 and Stat factors.46,47 One explanation may be that the experiments reported herein were performed with doses of chemical inhibitors near IC50 and in a context of monoallelic expression of Kit, with HS2 cells being heterozygotic for Kit mutation. It follows that the strength and persistence of signal are conditioned by transcription of the mutated allele without the confounding influences associated with ectopic expression of signaling molecules.
Our findings highlight that a simultaneous inhibition of both proliferation and survival is necessary to abolish the growth of leukemic cells. This prompted us to use a therapeutic combination of inhibitors targeting biochemical effectors of these 2 biologic processes. In agreement with this notion, we have shown that the combined use of NVP-BEZ235 and Obatoclax is an effective antileukemic strategy. The combination of NVP-BEZ235 with Obatoclax greatly improves their respective efficiency in inhibiting the proliferation of murine and human leukemic cells. This appears of particular interest because very few drug combinations have been reported to exert synergistic antiproliferative effects in Kit-dependent hematopoietic neoplasms,48-50 and because reduced doses of drugs contribute to limit adverse effects and risk of drug resistance.
In conclusion, the results of the present study show that phosphoproteome profiling of individual tumors provides a molecular rationale for the identification of dominant signaling pathways governing malignant cell behavior, which might be “druggable” in therapy. With the prospect of individualized therapies, such a systematic characterization of signaling networks according to the oncogenic stimulus and cell lineage will provide key insights into new therapy options.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Z. Maciorowski of the cytometry department at Institut Curie for assistance with flow cytometry and cell-cycle analysis; F. Wendling for valuable comments on the manuscript; and Dr P. Dubreuil for providing IM.
This study was supported by Inserm and Institut Curie, Association pour la Recherche sur le Cancer (grant 5078), Fondation de France (grant 2618), and the Association Christelle Bouillot. D.B. is a postdoctoral researcher supported by Ligue Nationale Contre le Cancer and Association pour la Recherche sur le Cancer.
Authorship
Contribution: D.B. performed proteomic profiling, proliferation assays, lentiviral infections, cell-cycle analysis, tumorigenic assays, and immunoblotting; I.G. and O.K. performed the coimmunoprecipitation assays; E.L. constructed the lentiviral vectors for shStat5; N.D. performed the molecular cloning; B.L. and F.G. performed the proteomic profiling analysis; D.L. and P.M. provided expertise in proteomic profiling; I.D.-F. provided expertise in Stat5 function; C.G. provided expertise in the cell cycle; and F.M.-G. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Françoise Moreau-Gachelin, Inserm U830, Institut Curie, 26 rue d'Ulm, 75248 Paris, France cedex 05; e-mail: framoreau@curie.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal