Abstract
True long-term nonprogressors (LTNPs)/elite controllers (ECs) maintain durable control over HIV replication without antiretroviral therapy. Herein we describe 4 unique persons who were distinct from conventional LTNPs/ECs in that they had extraordinarily low HIV burdens and comparatively weak immune responses. As a group, typical LTNPs/ECs have unequivocally reactive HIV-1 Western blots, viral loads below the lower threshold of clinical assays, low levels of persistent viral reservoirs, an over-representation of protective HLA alleles, and robust HIV-specific CD8+ T-cell responses. The 4 unique cases were distinguished from typical LTNPs/ECs based on weakly reactive Western blots, undetectable plasma viremia by a single copy assay, extremely low to undetectable HIV DNA levels, and difficult to isolate replication-competent virus. All 4 had at least one protective HLA allele and CD8+ T-cell responses that were disproportionately high for the low antigen levels but comparatively lower than those of typical LTNPs/ECs. These unique persons exhibit extraordinary suppression over HIV replication, therefore, higher-level control than has been demonstrated in previous studies of LTNPs/ECs. Additional insight into the full spectrum of immune-mediated suppression over HIV replication may enhance our understanding of the associated mechanisms, which should inform the design of efficacious HIV vaccines and immunotherapies.
Introduction
True long-term nonprogressors (LTNPs)/elite controllers (ECs) test positively in standard antibody assays and maintain stable CD4 counts and HIV-1 RNA levels below the lower detection threshold of clinical assays without antiretroviral therapy (ART).1 In cohorts defined by stringent criteria that include HIV RNA measurements, remarkable similarities have been observed among LTNPs/ECs.1 Most have very low, but detectable, levels of plasma HIV RNA2-5 and cell-based DNA.5-11 Particular HLA class I alleles, especially B*57, are significantly overrepresented and are the host genetic factors most consistently associated with this unique phenotype.1,12 Their HIV-specific CD8+ T cells are highly functional12-17 and exhibit robust proliferation, up-regulation of perforin, and cytotoxicity after in vitro stimulation with HIV-infected targets over several days.3,17,18 Defective or attenuated viruses have been demonstrated in some cases,19-21 but host factors appear to be primarily responsible for the durable control observed in the majority of LTNPs/ECs.
Even though stricter case definitions have resulted in the recruitment of more homogeneous cohorts, immune-mediated control still does not occur to the same extent and duration in all LTNPs/ECs.1 At the extreme end of the spectrum is a subset of LTNPs/ECs with persistently high CD4 counts and undetectable plasma viremia in ultrasensitive assays for more than 20 years.2-5,8 Included in this subgroup have been a few rare cases exhibiting atypical features relative to most LTNPs/ECs.5,8,22-24 In the present study, we performed a comprehensive analysis of 4 unique persons who presented with weakly reactive Western blots and have demonstrated clear differences from conventional LTNPs/ECs. All 4 cases had viral burdens, including HIV reservoir sizes, bordering on the limits of detection, virtually undetectable replication-competent virus, and comparatively lower HIV-specific antibody profiles and CD8+ T-cell responses than those of typical LTNPs/ECs. These persons occupy the extreme end of the disease spectrum and, as such, provide evidence that nearly complete suppression of HIV replication is possible in humans and might be an attainable goal for future HIV vaccines or immunotherapies.
Case reports
Case 1
A 37-year-old white male with a history of ulcerative colitis and major depression was diagnosed with HIV infection in 2002. He reported high-risk sexual exposures in the mid 1990s with his male partner who died from AIDS-related non-Hodgkin lymphoma in 2004. Despite a protracted influenza-like illness early in their relationship, case 1 (C1) had numerous negative rapid HIV tests through 2002. He was ART-naive and had never been diagnosed with an opportunistic disease. His CD4 counts ranged from 400 to 600 cells/mm3 and he never had a detectable viral load. C1 self-referred to National Institutes of Health (NIH) and was found to have a weakly reactive Western blot at screening.
Case 2
A 47-year-old white male first tested positive for HIV in 1997 on periodic military screening. His Western blot met minimum criteria for reactivity with bands at gp120, p24, and p18 and weakly positive bands at gp160, p51/55, and gp41. He had had multiple negative HIV tests from 1985 to 1994. Case 2 (C2) denied symptoms consistent with an acute retroviral syndrome or significant HIV risk factors. ART was initiated shortly after diagnosis despite undetectable plasma viremia. He also received IL-2 in 2000 through the ESPRIT clinical trial. His HIV status was questioned given consistently undetectable HIV RNA levels, high CD4 counts, and a stable clinical course. ART was discontinued in 2003. Subsequent testing yielded indeterminate or weakly positive Western blots on multiple occasions. In 2005, idiopathic thrombocytopenic purpura was diagnosed, but no improvement occurred with a brief trial of tenofovir, emtricitabine, and efavirenz.
Since his HIV diagnosis, C2 had remained clinically well with CD4 counts exceeding 850 cells/mm3 and only 2 detectable plasma HIV RNA results (of 64 tests): 61 (branched DNA Version 3, 1999) and 300 copies/mL (ultrasensitive HIV-1, Roche Amplicor Version 1.5, 2005). HIV DNA by qualitative PCR was not detected on 4 occasions (2001-2008). During evaluation for in vitro fertilization, HIV could not be detected in semen. Studies in 2006 revealed a weakly reactive Western blot (weak gp160, p24, p40, and p55 bands). In addition, a Multi-spot HIV-1/2 Rapid test, an HIV-1 RNA test (Roche Amplicor HIV-1 Monitor Assay Version 1.5), and a Procleix Discriminatory HIV-1 test were all nonreactive/negative. He was referred to NIH for further evaluation.
Case 3
A 25-year-old male from Buenos Aires was found to have a weakly reactive HIV-1 Western blot in 2008 on preoperative testing for repair of a sports injury sustained in 2006. Because his HIV test in 2006 had been negative, he was referred for enrollment in the Argentine Acute HIV Cohort. Case 3 (C3) denied symptoms compatible with an acute retroviral syndrome or a history of men who have sex with men or intravenous drug use but had received multiple tattoos and had engaged in unprotected heterosexual intercourse from 2006 to 2008. During brief follow-up, he remained healthy and had stable CD4 counts (650-881 cells/mm3) without ART. Five viral load measurements (branched DNA Version 3) were less than 50 copies/mL and 1 was 74 copies/mL. HIV-1/2 DNA and semen analysis for HIV p24 were not detected. Features differing from persons with acute HIV infection or other LTNPs/ECs prompted referral to NIH for further evaluation.
Case 4
A 56-year-old white female with a history of basal cell carcinoma and nephrolithiasis first tested positive for HIV in 1992. She denied symptoms consistent with an acute antiretroviral syndrome and significant HIV risk factors, except for a few instances of unprotected intercourse. On repeat testing in subsequent years, her HIV test was indeterminate or weakly positive on a few occasions. She was ART-naive with CD4 counts always exceeding 1200 cells/mm3 and consistently undetectable viral loads. Case 4 (C4) was identified to be an LTNP/EC and self-referred to our National Institute of Allergy and Infectious Diseases (NIAID) study. Unlike other LTNP/EC subjects, her screening Western blot was weakly reactive.
Methods
Subjects
Cases, chronically HIV-infected patients and HIV-negative controls, signed NIAID institutional review board–approved protocol informed consent documents in accordance with the Declaration of Helsinki and underwent phlebotomy and leukapheresis. Some persons also underwent endoscopy with biopsies performed under a separate institutional review board-approved NIAID study. HIV infection was determined by HIV-1/2 immunoassay (Abbott Laboratories) and Cambridge Biotech HIV-1 Western blot (Maxim Biomedical). LTNPs/ECs (n = 43), viremic progressors (n = 18), and ART recipients with HIV RNA levels of less than 50 copies/mL (Rx<50; n = 53) have been defined previously.3,17 Prior publications included single copy assay results for 26 LTNPs/ECs and 51 Rx<503,17 ; gut biopsy cell counts for 11 LTNPs/ECs, 12 viremic progressors, 16 Rx<50, and 11 HIV-negative controls25,26 ; and HIV DNA results for 10 LTNPs/ECs.10 HLA class I/II typing was performed by sequence-specific hybridization.27 A single-copy assay done in triplicate with a lower detection threshold of 1 copy/mL and CCR5 Δ32 genotyping on samples derived from patients and appropriate controls were performed as reported previously.3,28,29
HIV-1 DNA PCR
HIV-1 DNA in PBMC-derived (2-7.7 × 106) and gut-associated lymphoid tissue (GALT)–derived (9.2-17.8 × 105) CD4+ T cells selected by magnetic automated cell sorting (AutoMACS; Miltenyi Biotec) was amplified by semiquantitative PCR with primers specific for HIV-1 Gag, Env, and LTR, as described.30 Measurements of total and integrated HIV DNA in PBMCs were performed according to published protocols.10 The frequency of negatively selected, purified colonic (1-1.8 × 106) and ileal (1.2 × 106) CD4+ T cells carrying HIV DNA was determined as previously described.31
Colonoscopy biopsy processing and calculation of T-cell numbers
HIC assay
To determine the frequency of CD4+ T cells carrying replication-competent HIV, high-input quantitative coculture (HIC) assays were conducted in which multiple wells, each containing 107 purified CD4+ T cells (1.9-5.6 × 108 total), were activated in 12-well tissue culture plates as described previously.32
Prolonged (120-day) in vitro culture of stimulated CD4+ T cells and CD4+ T-cell infection with HIVSF162
In experiments assessing the presence of replication-competent HIV, PBMC-derived (5.2-140 × 106) and colonic (2-2.4 × 106) purified CD4+ T cells that had been positively selected with anti-CD4 microbeads (Miltenyi Biotec) were stimulated with “blasting” medium for 3 days as previously described.3,17 These cells were subsequently washed and cultured in medium containing 10% human AB serum (HAB; Gemini Bio-Products), human IL-2 (40 IU/mL, Roche Diagnostics), l-glutamine (4mM; Invitrogen), HEPES (10mM; Invitrogen) and the broad-spectrum antimicrobial formulation primocin (0.1 mg/mL; Amaxa Biosystems) for 120 days. Medium was replaced and/or cultures were split 3 times per week. Samples were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) every 48 to 72 hours, stained with surface and intracellular anti-HIV Gag p24 (KC57 RDI; Beckman Coulter) monoclonal antibodies, and analyzed by flow cytometry, as described previously.3 CD8 depletion was performed if CD8+ cells represented more than 1% of T cells. In experiments assessing susceptibility to HIVSF162 infection, stimulated CD4+ T cells were coincubated with magnetized HIVSF162, as previously described.3 In experiments using unstimulated cells, CD4+ T cells were negatively selected with the CD4+ T-cell Isolation Kit II (Miltenyi Biotec) and subjected to further depletion with anti-CD8 microbeads (Miltenyi Biotec).33,34 Purity exceeding 98% was confirmed by flow cytometry. Cells were directly incubated with HIVSF162 (TCID50/mL 2000) for 4 hours, washed twice, resuspended in 10% HAB medium not containing IL-2 at 5 × 105 cells/mL, and incubated in 24-well plates at 37°C as published recently.35,36 Cells were assessed at 96 hours by flow cytometry for p24 expression and CD8+ T-cell overgrowth.
HIV-specific antibody activity
HIV-specific CD8+ T-cell frequency, proliferation, and cytotoxic capacity
Flow cytometry
Multiparameter flow cytometry was performed according to standard protocols. All staining was performed at 4°C for 30 minutes with the following antibodies (BD Biosciences): FITC-conjugated anti-CD3; PerCP-conjugated anti-CD3 and anti-CD8; APC-conjugated anti-CD4 and anti–IFN-γ; PE-conjugated anti-CD8; and PE Cy7-conjugated anti-CD3. Samples were analyzed on a FACSAria multilaser cytometer (BD Biosciences) with FACSDiva Version 6.1.3 software. Data were analyzed using FlowJo Version 9.4.7 software (TreeStar).
Statistical analysis
Comparisons of independent groups were made by the Wilcoxon 2-sample test. Correlations were determined by the Spearman rank method. The Bonferroni method was used to adjust P values for multiple testing.
Results
Weakly reactive HIV-1 Western blots of the cases contrasted with unequivocally reactive Western blots of typical LTNPs/ECs
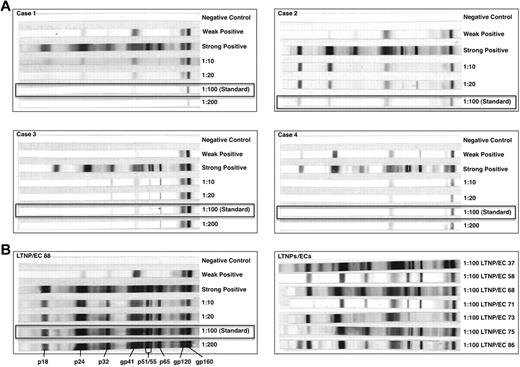
At initial evaluation, the cases had been diagnosed with HIV for a similar duration as viremic progressors (P = .11), but for a shorter period than a well-characterized cohort of typical LTNPs/ECs (P = .04)3,17 and Rx<50 (P = .02; Tables 1 and 2). Despite this relatively brief duration of infection, their HIV-1 Western blots were weakly reactive at standard dilutions in contrast to those of LTNPs/ECs (Figure 1). The Western blot of C3, who was infected for the shortest period, was more reactive than those of the other cases. At higher serum concentrations, additional bands were detected in all cases, confirming the presence of several HIV-specific antibodies (Figure 1).
Patient characteristics
| . | Case 1 . | Case 2 . | Case 3 . | Case 4 . |
|---|---|---|---|---|
| Duration of HIV diagnosis, y | 6 | 12 | 1 | 14 |
| CD4+ T-cell count, cells/mm3 | 650 | 974 | 868 | 1488 |
| HIV-1 RNA levels, copies/mL | < 50 | < 50 | < 50 | < 50 |
| Single-copy assay, copies/mL | < 1 | < 1 | < 1 | < 1 |
| HLA class I B loci | 40, 44 | 15, 57 | 44, 51 | 13, 57 |
| CCR5 genotype | WT/Δ32 | WT/WT | WT/WT | WT/WT |
| Neutralizing antibody activity, ID50* | < 30 | < 30 | < 30 | < 30 |
| Colonic CD4+ T cells, % | NA | 53.2 | 59.6 | 73.7 |
| Colonic CD4+ T cells/g | NA | 4.45 × 106 | 3.35 × 106 | 2.67 × 107 |
| Colonic CD4/CD8 ratio | NA | 1.77 | 1.97 | 3.7 |
| . | Case 1 . | Case 2 . | Case 3 . | Case 4 . |
|---|---|---|---|---|
| Duration of HIV diagnosis, y | 6 | 12 | 1 | 14 |
| CD4+ T-cell count, cells/mm3 | 650 | 974 | 868 | 1488 |
| HIV-1 RNA levels, copies/mL | < 50 | < 50 | < 50 | < 50 |
| Single-copy assay, copies/mL | < 1 | < 1 | < 1 | < 1 |
| HLA class I B loci | 40, 44 | 15, 57 | 44, 51 | 13, 57 |
| CCR5 genotype | WT/Δ32 | WT/WT | WT/WT | WT/WT |
| Neutralizing antibody activity, ID50* | < 30 | < 30 | < 30 | < 30 |
| Colonic CD4+ T cells, % | NA | 53.2 | 59.6 | 73.7 |
| Colonic CD4+ T cells/g | NA | 4.45 × 106 | 3.35 × 106 | 2.67 × 107 |
| Colonic CD4/CD8 ratio | NA | 1.77 | 1.97 | 3.7 |
WT indicates wild type; Δ32, CCR5 delta-32 mutation; ID50, 50% inhibitory dose; and NA, not available.
Examined against a median of 5 viruses.
Comparison of cases with chronic HIV-infected patients
| Measurements . | Cases (n = 4) . | LTNPs/ECs (n = 43) . | Progressors (n = 18) . | Rx<50 (n = 53) . |
|---|---|---|---|---|
| Median duration of HIV diagnosis, y (range) | 9 (1-14) | 20 (5-32)* | 18 (2-25) | 17 (6-27)* |
| Median CD4+ T-cell count, cells/mm3 (range) | 921 (650-1488) | 851 (277-1813) | 343 (142-1040)* | 587 (204-1409)* |
| Median HIV-1 RNA levels, copies/mL (range) | < 50 | < 50 | 62 233 (10 443-160 954)* | < 50 |
| Median single-copy assay, copies/mL (range)† | < 1 | 4 (< 1-48) | NA | < 1 (< 1-53) |
| Prevalence HLA B*57, %‡ | 50 | 63 | 28 | 19 |
| Measurements . | Cases (n = 4) . | LTNPs/ECs (n = 43) . | Progressors (n = 18) . | Rx<50 (n = 53) . |
|---|---|---|---|---|
| Median duration of HIV diagnosis, y (range) | 9 (1-14) | 20 (5-32)* | 18 (2-25) | 17 (6-27)* |
| Median CD4+ T-cell count, cells/mm3 (range) | 921 (650-1488) | 851 (277-1813) | 343 (142-1040)* | 587 (204-1409)* |
| Median HIV-1 RNA levels, copies/mL (range) | < 50 | < 50 | 62 233 (10 443-160 954)* | < 50 |
| Median single-copy assay, copies/mL (range)† | < 1 | 4 (< 1-48) | NA | < 1 (< 1-53) |
| Prevalence HLA B*57, %‡ | 50 | 63 | 28 | 19 |
NA indicates not available.
P < .05, cases versus other patient groups.
Results from 26 LTNPs/ECs and 51 Rx<50 were used in comparison with the cases.
Sample size was too small to determine a significant difference between group frequencies.
HIV-1 Western blots of the cases were weakly reactive in contrast to the unequivocally positive Western blots of typical LTNPs/ECs. (A) HIV-1 Western blots with controls performed with serial, including standard (1:100; boxes), dilutions of patient sera from each of the cases C1 to C4 to diluent. (B) HIV-1 Western blots with controls performed with serial, including standard (1:100; box), dilutions of patient serum to diluent from one representative LTNP/EC (left), and at the standard dilution for a subset of typical LTNPs/ECs (right).
HIV-1 Western blots of the cases were weakly reactive in contrast to the unequivocally positive Western blots of typical LTNPs/ECs. (A) HIV-1 Western blots with controls performed with serial, including standard (1:100; boxes), dilutions of patient sera from each of the cases C1 to C4 to diluent. (B) HIV-1 Western blots with controls performed with serial, including standard (1:100; box), dilutions of patient serum to diluent from one representative LTNP/EC (left), and at the standard dilution for a subset of typical LTNPs/ECs (right).
As expected, CD4 counts of the cases were significantly higher than those of viremic progressors (P = .01) and Rx<50 (P = .02) and most similar to those of LTNPs/ECs (P > .5; Tables 1 and 2). HIV-1 RNA levels were less than 50 copies/mL, as found previously in these cases. Notably, C2 and C4 were HLA B*57+, and all carried at least one allele previously demonstrated to be enriched in LTNPs/ECs, including B*13, 15, 44, and 51 (Tables 1 and 2).1
Total HIV burden, including reservoir size, in the plasma and tissues of the cases is exceptionally low and less than that of typical LTNPs/ECs
HIV RNA levels in the cases were below the lower detection threshold in clinical assays. In ultrasensitive assays with detection limits as low as 1 HIV-1 RNA copy/mL, most LTNPs/ECs have been shown to have detectable plasma viremia.2-5,17 In the present study, all 4 cases had HIV RNA levels of less than 1 copy/mL, which were the same as those found in one-third of conventional LTNPs/ECs with the same assay3,4,17 and were not significantly different overall from those of 26 typical LTNPs/ECs (P = .06) or 51 Rx<50 (P = .25) included herein (Table 2).
HIV DNA was measured in PBMC and GALT samples. From gut biopsies, the percentage of colonic CD4+ T cells and CD4/CD8 ratios of the cases were similar to those of LTNPs/ECs and HIV-negative controls (P > .05 for all comparisons) but were significantly higher than those of viremic progressors (P = .01 for both comparisons) and Rx<50 (P = .01 and P = .02, respectively; Tables 1 and 3). Similar comparisons for absolute numbers of CD4+ T cells per gram of tissue did not reach statistical significance (P > .05 for all comparisons; Table 3). In summary, the cases had preserved gut-associated CD4+ T-cell populations resembling those of LTNPs/ECs and HIV-negative controls.
T-cell composition of gut biopsy samples
| Patient group . | Median colonic CD4+ T-cells, % (range) . | Median colonic CD4+ T cells/g (range) . | Median colonic CD4/CD8 ratio (range) . |
|---|---|---|---|
| Cases (n = 3) | 59.6 (53.2-73.7) | 4.45 × 106 (3.35 × 106-2.67 × 107) | 1.97 (1.77-3.7) |
| HIV-negative (n = 12) | 57. 5 (42.9-70.8) | 3.59 × 106 (4.53 × 105-6.23 × 106) | 2.84 (1.14-4.99) |
| LTNPs/ECs (n = 12) | 54.7 (24.1-69) | 2.74 × 106 (6.32 × 105-1.74 × 107) | 2.23 (0.58-4.46) |
| Progressors (n = 13) | 5.8 (0.1-21.6)* | 5.63 × 105 (1.13 × 104-4.75 × 106) | 0.08 (0.002-0.32)* |
| Rx<50 (n = 16) | 36.6 (24.6-55.8)* | 2.61 × 106 (9.39 × 105-1.33 × 107) | 0.84 (0.47-2.32)* |
| Patient group . | Median colonic CD4+ T-cells, % (range) . | Median colonic CD4+ T cells/g (range) . | Median colonic CD4/CD8 ratio (range) . |
|---|---|---|---|
| Cases (n = 3) | 59.6 (53.2-73.7) | 4.45 × 106 (3.35 × 106-2.67 × 107) | 1.97 (1.77-3.7) |
| HIV-negative (n = 12) | 57. 5 (42.9-70.8) | 3.59 × 106 (4.53 × 105-6.23 × 106) | 2.84 (1.14-4.99) |
| LTNPs/ECs (n = 12) | 54.7 (24.1-69) | 2.74 × 106 (6.32 × 105-1.74 × 107) | 2.23 (0.58-4.46) |
| Progressors (n = 13) | 5.8 (0.1-21.6)* | 5.63 × 105 (1.13 × 104-4.75 × 106) | 0.08 (0.002-0.32)* |
| Rx<50 (n = 16) | 36.6 (24.6-55.8)* | 2.61 × 106 (9.39 × 105-1.33 × 107) | 0.84 (0.47-2.32)* |
P < .05, cases versus other patient groups.
In purified CD4+ T cells from PBMCs and GALT, HIV DNA was not detected by qualitative PCR in a small number of the cases and typical LTNPs/ECs (Table 4). To determine the frequencies of CD4+ T cells carrying total and integrated HIV DNA, quantitative PCR was performed on blinded samples from the same persons (Table 4). Total HIV DNA levels have been shown to be low in typical LTNPs/ECs6,8,11 but were not significantly different from those of Rx<50 when directly compared.9,10 In contrast, significantly lower levels of integrated HIV DNA in the cells of LTNPs/ECs than in those of Rx<50 were recently demonstrated.10 In the present study, the cases had significantly lower levels of both median total (6.56 copies/106 PBMCs) and integrated (0.02 copies/106 PBMCs) HIV DNA than did LTNPs/ECs (323 copies/106 PBMCs, P = .01 and 9.77 copies/106 PBMCs, P = .04, respectively; Table 4). Notably, despite detection in 100% of typical LTNPs/ECs by this repetitive sampling approach, HIV DNA was not detected in C4 after assaying 185 million genomes (Table 4).
HIV proviral DNA in cases and LTNPs/ECs
| Patient groups . | HIV DNA qualitative PCR . | Total HIV DNA* (PBMCs) . | Integrated HIV DNA* (PBMCs) . | Total HIV DNA† . | HIC (PBMCs) . | |||
|---|---|---|---|---|---|---|---|---|
| PBMC . | Colon . | Ileum . | Colon . | Ileum . | ||||
| Cases | ||||||||
| 1 | Undetectable | NA | NA | 6.56 | 0.02 | NA | NA | < 0.0041 |
| 2 | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | 25.2 | 2.47 | 2.8 | NA | < 0.0018 |
| 3 | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | Gag+, Env+, LTR+ | NA | NA | 1.95 | NA | 0.046 |
| 4 | Undetectable | Undetectable | Undetectable | Undetectable | Undetectable | < 2.6 | 42.4 | < 0.0042 |
| Median | 6.56 | 0.02 | 0.0041 | |||||
| LTNPs/ECs | ||||||||
| 4 | Gag+, Env−, LTR+ | Gag+, Env−, LTR− | 3084.48 | 0.42 | NA | NA | 0.0114 | |
| 58 | Gag+, Env−, LTR+ | 506.49 | 18.8 | NA | NA | < 0.0058 | ||
| 71 | Gag−, Env−, LTR+ | Gag−, Env+, LTR+ | 14 498.24 | 3.24 | NA | NA | 0.0236 | |
| 73 | Undetectable | Undetectable | 323.03 | 2.27 | NA | NA | 0.008 | |
| 75 | Gag+, Env+, LTR+ | 235.42 | 51.95 | NA | NA | 0.289 | ||
| 84 | Gag+, Env−, LTR+ | Gag−, Env+, LTR− | 179.80 | 74 | NA | NA | 0.0288 | |
| 86 | Gag+, Env+, LTR+ | 153.33 | 3.71 | NA | NA | 0.0223 | ||
| 88 | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | 61.24 | 16.97 | 1255.46 | NA | 0.026 |
| 96 | Gag+, Env−, LTR+ | Gag+, Env−, LTR+ | 2576.66 | 7.39 | NA | NA | NA | |
| 37 | Gag+, Env+, LTR+ | 201.51 | 9.77 | NA | NA | NA | ||
| 17 | Gag+, Env−, LTR+ | Gag+, Env−, LTR+ | 668.93 | 29.45 | NA | NA | NA | |
| Median | 323‡ | 9.77‡ | NA | NA | 0.023 | |||
| Patient groups . | HIV DNA qualitative PCR . | Total HIV DNA* (PBMCs) . | Integrated HIV DNA* (PBMCs) . | Total HIV DNA† . | HIC (PBMCs) . | |||
|---|---|---|---|---|---|---|---|---|
| PBMC . | Colon . | Ileum . | Colon . | Ileum . | ||||
| Cases | ||||||||
| 1 | Undetectable | NA | NA | 6.56 | 0.02 | NA | NA | < 0.0041 |
| 2 | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | 25.2 | 2.47 | 2.8 | NA | < 0.0018 |
| 3 | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | Gag+, Env+, LTR+ | NA | NA | 1.95 | NA | 0.046 |
| 4 | Undetectable | Undetectable | Undetectable | Undetectable | Undetectable | < 2.6 | 42.4 | < 0.0042 |
| Median | 6.56 | 0.02 | 0.0041 | |||||
| LTNPs/ECs | ||||||||
| 4 | Gag+, Env−, LTR+ | Gag+, Env−, LTR− | 3084.48 | 0.42 | NA | NA | 0.0114 | |
| 58 | Gag+, Env−, LTR+ | 506.49 | 18.8 | NA | NA | < 0.0058 | ||
| 71 | Gag−, Env−, LTR+ | Gag−, Env+, LTR+ | 14 498.24 | 3.24 | NA | NA | 0.0236 | |
| 73 | Undetectable | Undetectable | 323.03 | 2.27 | NA | NA | 0.008 | |
| 75 | Gag+, Env+, LTR+ | 235.42 | 51.95 | NA | NA | 0.289 | ||
| 84 | Gag+, Env−, LTR+ | Gag−, Env+, LTR− | 179.80 | 74 | NA | NA | 0.0288 | |
| 86 | Gag+, Env+, LTR+ | 153.33 | 3.71 | NA | NA | 0.0223 | ||
| 88 | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | Gag−, Env−, LTR+ | 61.24 | 16.97 | 1255.46 | NA | 0.026 |
| 96 | Gag+, Env−, LTR+ | Gag+, Env−, LTR+ | 2576.66 | 7.39 | NA | NA | NA | |
| 37 | Gag+, Env+, LTR+ | 201.51 | 9.77 | NA | NA | NA | ||
| 17 | Gag+, Env−, LTR+ | Gag+, Env−, LTR+ | 668.93 | 29.45 | NA | NA | NA | |
| Median | 323‡ | 9.77‡ | NA | NA | 0.023 | |||
HIC indicates high input quantitative coculture assay, measuring infectious units per 106 (IUPM) CD4+ T cells; NA, not available; Env, envelope; and LTR, long terminal repeat regions of HIV-1.
Copies/106 PBMCs.
Copies/106 CD4+ T cells.
P < .05, cases versus LTNPs/ECs.
In prior studies of Rx<50, the average frequency of CD4+ T cells carrying HIV DNA in GALT was 4887 copies/106 cells, which was 5- to 10-fold higher than that in PBMC.39,40 Similar results were found in a representative B*57+ LTNP/EC 88 in the present study, who had 1255 copies/106 colonic CD4+ T cells, which was 20-fold higher than levels in PBMC. In contrast to both LTNP/EC 88 (Table 4) and Rx<50, very low (C2, C3) or undetectable (C4) HIV DNA levels were observed in colonic CD4+ T cells of the cases who had undergone colonoscopy. In C4, HIV DNA was only detected in terminal ileal samples, but at very low levels (Table 4). Therefore, significantly lower HIV DNA levels in both PBMCs and GALT of the cases support the presence of smaller HIV reservoirs, even in lymphoid tissues than those of Rx<50 and typical LTNPs/ECs.
In a HIC assay used to measure the frequency of cells carrying infectious virus as infectious units per million (IUPM) PBMC-derived CD4+ T cells,31,32 3 of the 4 cases compared with 1 of 8 LTNPs/ECs had no detectable replication-competent virus despite activating 1.9 to 5.6 × 108 purified CD4+ T cells (Table 4). The median frequencies of cells carrying infectious virus tended to be lower in the cases versus LTNPs/ECs (0.0041 vs 0.023 IU/106 CD4+ T cells, respectively), although this difference did not achieve statistical significance with these small sample sizes. Notably, however, the IUPM CD4+ T cells were strongly correlated with integrated HIV DNA levels (R = 0.72, P = .03). In prior work, median frequencies of 0.02 IUPM CD4+ T cells in LTNPs/ECs41 and 0.059 IUPM CD4+ T cells in Rx<5031 were similar to those of typical LTNPs/ECs in the present study, suggesting that the infectious virus burden of the cases may be even lower than those of both typical LTNPs/ECs and Rx<50. In summary, the cases appeared to have smaller HIV reservoirs based on lower HIV DNA levels in PBMCs and GALT and a trend toward lower frequencies of CD4+ T cells carrying replication-competent virus than those of typical LTNPs/ECs. In addition, the HIV reservoir of C4 was virtually undetectable, except for low HIV DNA levels in the terminal ileum.
Replication-competent HIV could not be recovered from PBMC or GALT of the cases despite 120-day culture
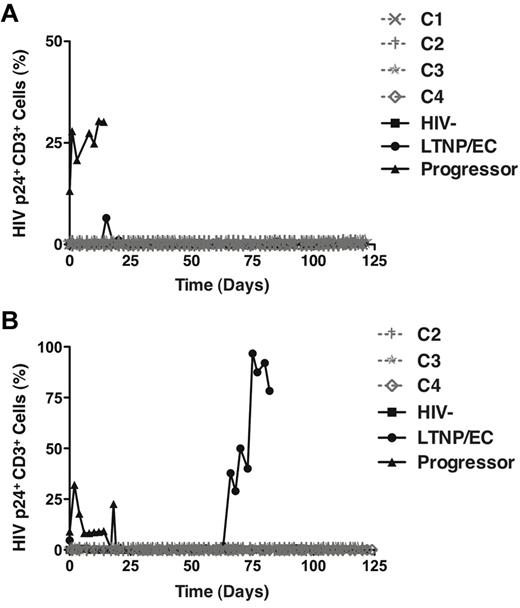
In the HIC assay, cells are cultured for up to 21 days. It was of interest to determine whether replication-competent virus could be recovered with more prolonged culture over a 4-month period and more frequent sampling every 48 to 72 hours. From PBMC-derived CD4+ T cells, significant numbers of p24-expressing cells were only detected in a viremic progressor, but not in the 4 cases or LTNP/EC 88 (Figure 2A). This lack of recovery of infectious virus from PBMC is consistent with previous reports in LTNPs/ECs9,41 and the extremely small reservoir sizes in PBMC of the cases and LTNP/EC 88 (Table 4). Notably, however, with colonic CD4+ T cells, significant numbers of p24-expressing cells from LTNP/EC 88 were detected by day 66, which contrasted with findings in the cases (Figure 2B) and was again consistent with the relative disparities in HIV DNA levels in GALT observed between cases and LTNP/EC 88 (Table 4). In summary, despite prolonged 120-day culture, no replication-competent virus could be isolated from pure populations of up to 1.4 × 108 activated PBMC- or GALT-derived CD4+ T cells of the cases, even though it was ultimately detected at high levels in GALT from a conventional LTNP/EC.
Replication-competent HIV could be recovered after 120-day culture from the activated GALT-derived CD4+ T cells of a typical LTNP/EC, but not from those of the cases. (A-B) Positively selected CD4+ T cells from PBMCs (A; n = 7) or gut biopsy (B; n = 6) samples of the cases (C1, ×; C2, †; C3, ★, C4,◊) and controls were stimulated with medium containing soluble anti-CD3/anti-CD28 monoclonal antibodies and recombinant IL-2 (40 IU/mL) for 3 days and subsequently cultured in IL-2-containing medium for 120 days. Analysis of intracellular HIV p24 expression and overgrowth of CD8+ T cells occurred every 48 to 72 hours. CD8 depletion was performed if CD8+ cells represented more than 1% of the T cells in culture.
Replication-competent HIV could be recovered after 120-day culture from the activated GALT-derived CD4+ T cells of a typical LTNP/EC, but not from those of the cases. (A-B) Positively selected CD4+ T cells from PBMCs (A; n = 7) or gut biopsy (B; n = 6) samples of the cases (C1, ×; C2, †; C3, ★, C4,◊) and controls were stimulated with medium containing soluble anti-CD3/anti-CD28 monoclonal antibodies and recombinant IL-2 (40 IU/mL) for 3 days and subsequently cultured in IL-2-containing medium for 120 days. Analysis of intracellular HIV p24 expression and overgrowth of CD8+ T cells occurred every 48 to 72 hours. CD8 depletion was performed if CD8+ cells represented more than 1% of the T cells in culture.
Viral reservoir sequence analysis could not explain control over virus replication in the cases with detectable cell-associated HIV DNA
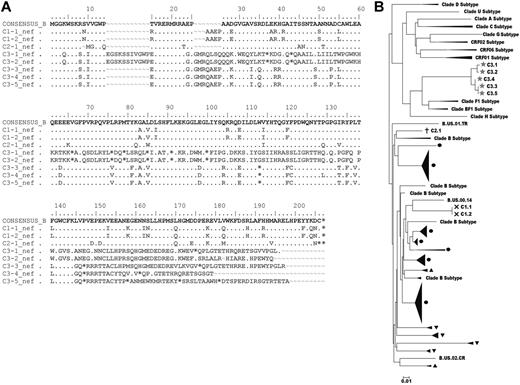
Although it has been demonstrated that the majority of HIV DNA detected in CD4+ T cells is replication-defective42 and plasma virus fitness cannot be reliably determined from analyses of cell-associated provirus sequences,43,44 we attempted to sequence the viral reservoirs (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) in the 3 cases with measurable HIV DNA to search for clues explaining their undetectable levels of infectious virus. Multiple attempts to sequence gag were unsuccessful in all 3 cases despite processing 1 to 1.6 × 107 purified CD4+ T cells, which differed from previous experience with conventional LTNPs/ECs41,44 and again supported the smaller reservoir sizes of these cases relative to LTNPs/ECs. In contrast, 2 independent nef sequences were obtained from C1, which had a 4 amino-acid insertion (between positions 25 and 26) in a region of known length polymorphism (Figure 3A). The only sequence obtained from C2 contained a single amino-acid deletion (position 9), also occurring in a region in which deletions are common (Figure 3A).41 These clade B sequences were distinct for each case and appeared to be functional because they lacked gross nef deletions as have been demonstrated in some LTNPs/ECs,19,23 possessed scattered unique variations that were unlikely to compromise fitness (Figure 3A), and were fairly homologous with isolates obtained from other HIV-infected persons (M.S., personal communication; Figure 3B). In contrast to C1 and C2, 5 nef clones from C3 contained premature stop codons, which could not be attributed primarily to G-to-A hypermutation (Figure 3A). These sequences were most similar to recombinants or clade F isolates from Latin America (Figure 3B), consistent with this patient's residence. Interestingly, although sequences encoding full-length nef were not obtained from C3, he is the only case from whom virus was detected in the HIC assay (Table 4), clearly demonstrating that his cells harbor replication-competent virus. In summary, although full-length proviral sequencing was not possible, the absence of large nef deletions in 2 of the 3 cases and the detection of replication-competent virus in the third case despite premature stop codons suggest that the infecting viruses of the cases were not highly defective and, thus, are unlikely to account for their unique clinical outcomes.
Proviral nef sequence analysis of the cases. (A) nef sequences obtained from C1 to C3 are aligned with the clade B consensus sequence. Dots indicate identity with the consensus sequence; dashes, gaps; and asterisks, stop codons. Sequence variations in C1 and C2 occurred in regions of common length polymorphisms. Premature stop codons occurred in all 5 sequences obtained from C3 and were not due largely to G to A hypermutation. (B) A phylogenetic tree of nef sequences from 3 cases (C1, ×; C2, †; C3, ★) and other chronically infected patients from another study (M.S., personal communication), including conventional LTNPs/ECs (black circles), viremic progressors (solid black upward turning arrow heads), and Rx<50 (solid black downward turning arrow heads) was constructed for each patient by classic, maximum-likelihood and Bayesian methods. Other clade subtype nef sequences are included for comparison.
Proviral nef sequence analysis of the cases. (A) nef sequences obtained from C1 to C3 are aligned with the clade B consensus sequence. Dots indicate identity with the consensus sequence; dashes, gaps; and asterisks, stop codons. Sequence variations in C1 and C2 occurred in regions of common length polymorphisms. Premature stop codons occurred in all 5 sequences obtained from C3 and were not due largely to G to A hypermutation. (B) A phylogenetic tree of nef sequences from 3 cases (C1, ×; C2, †; C3, ★) and other chronically infected patients from another study (M.S., personal communication), including conventional LTNPs/ECs (black circles), viremic progressors (solid black upward turning arrow heads), and Rx<50 (solid black downward turning arrow heads) was constructed for each patient by classic, maximum-likelihood and Bayesian methods. Other clade subtype nef sequences are included for comparison.
CD4+ T cells of the cases were fully receptive to HIV infection, excluding intrinsic resistance to HIV
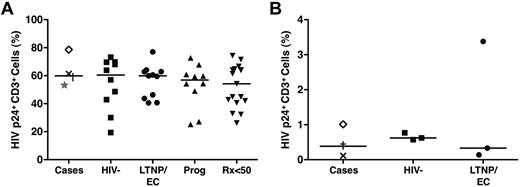
Extremely small HIV-1 burdens in the cases prompted us to explore whether these patients have a partial resistance to HIV-1 infection related to altered expression of HIV coreceptors, such as CCR5. Only C1 was heterozygous for the CCR5 Δ32-bp mutation (Table 1). To further investigate whether these patients had non-Δ32 CCR5 mutations impairing entry of R5-tropic viruses or possessed postentry intrinsic factors that could otherwise restrict HIV replication, we determined the susceptibility of their CD4+ T cells to infection with magnetized HIVSF162.3 Their activated cells were fully receptive to R5-tropic HIV infection based on intracellular p24 staining (median, 59.9%) to similar levels as those of LTNPs/ECs, viremic progressors, Rx<50, and HIV-negative controls (P > .5 for all comparisons; Figure 4A), excluding a significant blockade in co-receptor–mediated entry.
CD4+ T cells of the cases were fully receptive to HIV super-infection, excluding passive intrinsic resistance to HIV. (A) Purified CD4+ T cells derived from the cases (C1, ×; C2, †; C3, ★; C4, ◊), HIV-negative controls (n = 10), typical LTNPs/ECs (n = 11), viremic progressors (n = 10), and Rx<50 (n = 17) were stimulated with medium containing soluble anti-CD3/anti-CD28 monoclonal antibodies and recombinant IL-2 (40 IU/mL) for 3 days, coincubated with magnetized HIVSF162 in the presence of a super-magnet for 15 minutes, and cultured for 36 hours before analysis for intracellular HIV p24 expression. HIV infection is expressed as the percentage of CD3+ lymphocytes expressing p24. Horizontal lines represent median values. Comparisons were made using the Wilcoxon 2-sample test. None of the comparisons was significant. (B) CD4+ T cells from a subset of 3 cases (C1, ×; C2, †; C4, ◊), 3 randomly selected LTNPs/ECs, and 3 HIV-negative controls were negatively selected with magnetic beads and subjected to redepletion of CD8+ cells. Purity was confirmed to exceed 98% by flow cytometry. After 4-hour incubation with HIVSF162 at a TCID50 of 2000, cells were washed twice, resuspended in 10% human AB medium not containing human IL-2 at a concentration of 5 × 105 cells/mL, and incubated in 24-well plates at 37°C for 4 days before assessment by flow cytometry for HIV Gag p24 expression and CD8+ T-cell overgrowth. HIV infection is expressed as the percentage of CD3+ lymphocytes expressing p24.
CD4+ T cells of the cases were fully receptive to HIV super-infection, excluding passive intrinsic resistance to HIV. (A) Purified CD4+ T cells derived from the cases (C1, ×; C2, †; C3, ★; C4, ◊), HIV-negative controls (n = 10), typical LTNPs/ECs (n = 11), viremic progressors (n = 10), and Rx<50 (n = 17) were stimulated with medium containing soluble anti-CD3/anti-CD28 monoclonal antibodies and recombinant IL-2 (40 IU/mL) for 3 days, coincubated with magnetized HIVSF162 in the presence of a super-magnet for 15 minutes, and cultured for 36 hours before analysis for intracellular HIV p24 expression. HIV infection is expressed as the percentage of CD3+ lymphocytes expressing p24. Horizontal lines represent median values. Comparisons were made using the Wilcoxon 2-sample test. None of the comparisons was significant. (B) CD4+ T cells from a subset of 3 cases (C1, ×; C2, †; C4, ◊), 3 randomly selected LTNPs/ECs, and 3 HIV-negative controls were negatively selected with magnetic beads and subjected to redepletion of CD8+ cells. Purity was confirmed to exceed 98% by flow cytometry. After 4-hour incubation with HIVSF162 at a TCID50 of 2000, cells were washed twice, resuspended in 10% human AB medium not containing human IL-2 at a concentration of 5 × 105 cells/mL, and incubated in 24-well plates at 37°C for 4 days before assessment by flow cytometry for HIV Gag p24 expression and CD8+ T-cell overgrowth. HIV infection is expressed as the percentage of CD3+ lymphocytes expressing p24.
These results were consistent with reports demonstrating comparable susceptibility of activated CD4+ T cells from LTNPs/ECs and HIV-negative persons to HIV infection9,11,15,18,33,34 but contrasted with others showing reduced permissiveness to HIV infection in the cells of LTNPs/ECs.11,35 Variable experimental conditions (eg, use of polyclonal stimulation, spinoculation), which could overwhelm intrinsic resistance mechanisms operating in vivo, have been suggested to account for these discrepancies.11,35,36 To examine whether these factors could explain the unique phenotype of the cases, we additionally infected unstimulated CD4+ T cells (purity > 98%) from 3 cases (C1, C2 and C4), 3 typical LTNPs/ECs, and 3 HIV-negative controls according to published methods.34-36 The levels of HIV infection in the cells of the cases (0.12%-1.01%) were similar to those of LTNPs/ECs (0.14%-3.38%) and HIV-negative controls (0.62%-0.77%; Figure 4B), suggesting that the unique phenotype of these cases, including their small HIV reservoirs, could not simply be explained by partial resistance of their CD4+ T cells to HIV infection.
HIV-specific antibody response profiles distinguished cases from other chronically infected patients, including typical LTNPs/ECs
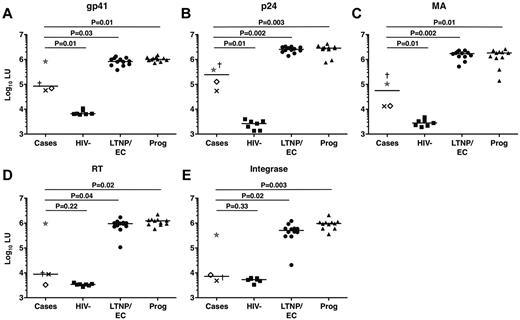
To further examine the low HIV-specific antibody levels in the cases as suggested by their weakly reactive Western blots, HIV-specific antibody responses were analyzed in a LIPS assay that generates highly quantitative antibody response profiles.38 Interestingly, the profiles of the cases differed significantly from those of typical LTNPs/ECs (Figure 5). Indeed, the response profiles of the cases targeting the reverse transcriptase and integrase gene products were most similar to those of HIV-negative controls (P = .22 and P = .33, respectively; Figure 5). C3 tended to have the highest responses among the cases, consistent with his greater Western blot reactivity. A longitudinal analysis of 5 serum samples from C4 spanning a 6-year period revealed no evidence of a significant decline in antibody levels during that interval (data not shown).
HIV-specific antibody response profiles clearly distinguished cases from other chronically infected patients, including typical LTNPs/ECs. (A-E) Antibody response profiles specific for the HIV-1 gene products gp41 (A), p24 (B), matrix (MA) protein (C), reverse transcriptase (RT; D), and integrase (E), as determined by the LIPS assay and reported in log10 luminometer units (LU), are shown for the cases (C1, ×; C2, †; C3, ★; and C4, ◊), HIV-negative controls (n = 7), typical LTNPs/ECs (n = 12), and viremic progressors (Prog; n = 11). Horizontal lines represent median values. Comparisons were made using the Wilcoxon 2-sample test. Only P values referring to comparisons between the responses of the cases and other groups are shown.
HIV-specific antibody response profiles clearly distinguished cases from other chronically infected patients, including typical LTNPs/ECs. (A-E) Antibody response profiles specific for the HIV-1 gene products gp41 (A), p24 (B), matrix (MA) protein (C), reverse transcriptase (RT; D), and integrase (E), as determined by the LIPS assay and reported in log10 luminometer units (LU), are shown for the cases (C1, ×; C2, †; C3, ★; and C4, ◊), HIV-negative controls (n = 7), typical LTNPs/ECs (n = 12), and viremic progressors (Prog; n = 11). Horizontal lines represent median values. Comparisons were made using the Wilcoxon 2-sample test. Only P values referring to comparisons between the responses of the cases and other groups are shown.
HIV-specific antibodies in conventional LTNPs/ECs tend not to exhibit potent or broad neutralizing capacity.37,43,45 Consistent with these observations, none of the cases had significant neutralizing antibody activity (Table 1), suggesting that their unique clinical outcomes were not a result of superior antibody neutralization capacity.
Low-frequency HIV-specific T cells exhibited significant cytotoxic responses after 6-day expansion that were less than those of typical LTNPs/ECs
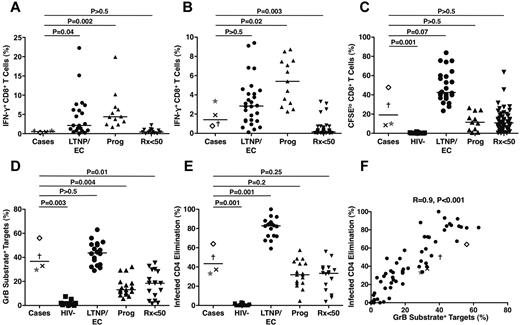
HIV-specific CD8+ T cells targeting peptides spanning the major HIV-1 gene products Nef, Gag, and Pol were detected in all 4 cases at very low frequencies (Figure 6A). Their total responses (median, 0.53%) were significantly lower than those of LTNPs/ECs (2.13%, P = .04) and viremic progressors (4.39%, P = .002) but similar to those of Rx<50 (0.42%, P > .5). Using autologous HIV-infected CD4+ T-cell targets to measure the responses targeting all HIV-1 gene products, the responses of the cases (median, 1.43%) were higher than when stimulated by peptide pools and not significantly different from those of LTNPs/ECs (2.88%, P > .5; Figure 6B). In contrast, the HIV-specific CD8+ T-cell responses of the cases measured by this approach were intermediate between those of viremic progressors (5.4%, P = .02) and Rx<50 (0.14%, P = .003).
Low frequencies of HIV-specific CD8+ T cells in the cases exhibited significant proliferative and cytotoxic responses after 6-day stimulation that were less than those of typical LTNPs/ECs. (A) Frequencies of IFN-γ–secreting CD8+ T cells measured in response to peptide pools spanning the HIV-1 Nef, Gag, and Pol gene products are shown for the cases (C1, ×; C2, †; C3, ★; C4, ◊), typical LTNPs/ECs (n = 21), viremic progressors (Prog; n = 12), and Rx<50 (n = 14). (B) Frequencies of IFN-γ–secreting CD8+ T cells measured in response to autologous HIVSF162-infected CD4+ T-cell targets are shown for the 4 cases, typical LTNPs/ECs (n = 27), viremic progressors (Prog; n = 13), and Rx<50 (n = 50). (C) Proliferation as assessed by CFSE dilution over 6 days in response to autologous HIVSF162-infected CD4+ T-cell targets is shown for the 4 cases, HIV-negative controls (n = 11), typical LTNPs/ECs (n = 24), viremic progressors (Prog; n = 11), and Rx<50 (n = 55). (D-E) Total cytotoxic responses measured by GrB target cell activity (D) or ICE (E) with day 6 CD8+ T cells are shown for the 4 cases, HIV-negative controls (n = 11), typical LTNPs/ECs (n = 18), viremic progressors (Prog; n = 17), and Rx<50 (n = 17). Horizontal lines represent median values. Comparisons were made using the Wilcoxon 2-sample test. Only P values referring to comparisons between the responses of the cases and other groups are shown. (F) Using day 6 CD8+ T cells, GrB target cell activity correlates directly with ICE (n = 67). Statistical analysis was performed using the Spearman correlation.
Low frequencies of HIV-specific CD8+ T cells in the cases exhibited significant proliferative and cytotoxic responses after 6-day stimulation that were less than those of typical LTNPs/ECs. (A) Frequencies of IFN-γ–secreting CD8+ T cells measured in response to peptide pools spanning the HIV-1 Nef, Gag, and Pol gene products are shown for the cases (C1, ×; C2, †; C3, ★; C4, ◊), typical LTNPs/ECs (n = 21), viremic progressors (Prog; n = 12), and Rx<50 (n = 14). (B) Frequencies of IFN-γ–secreting CD8+ T cells measured in response to autologous HIVSF162-infected CD4+ T-cell targets are shown for the 4 cases, typical LTNPs/ECs (n = 27), viremic progressors (Prog; n = 13), and Rx<50 (n = 50). (C) Proliferation as assessed by CFSE dilution over 6 days in response to autologous HIVSF162-infected CD4+ T-cell targets is shown for the 4 cases, HIV-negative controls (n = 11), typical LTNPs/ECs (n = 24), viremic progressors (Prog; n = 11), and Rx<50 (n = 55). (D-E) Total cytotoxic responses measured by GrB target cell activity (D) or ICE (E) with day 6 CD8+ T cells are shown for the 4 cases, HIV-negative controls (n = 11), typical LTNPs/ECs (n = 18), viremic progressors (Prog; n = 17), and Rx<50 (n = 17). Horizontal lines represent median values. Comparisons were made using the Wilcoxon 2-sample test. Only P values referring to comparisons between the responses of the cases and other groups are shown. (F) Using day 6 CD8+ T cells, GrB target cell activity correlates directly with ICE (n = 67). Statistical analysis was performed using the Spearman correlation.
Despite the relatively low frequencies of HIV-specific CD8+ T cells determined in a 6-hour assay, the capacity of these cells to proliferate over 6 days was reasonably high (median, 19.12%) and significantly different from HIV-negative controls (0.25%, P = .001; Figure 6C). It approached the level observed in traditional LTNPs/ECs (42.45%, P = .07) but also overlapped with results obtained with cells from viremic progressors (11.43%, P > .5) and Rx<50 (10.9%, P > .5; Figure 6C).
Perhaps the most robust measurements distinguishing the responses of LTNPs/ECs from those of noncontrollers are ones of HIV-specific CD8+ T-cell cytotoxic capacity in response to HIV-infected CD4+ T-cell targets after 6-day stimulation.3 In the cases, the capacity of HIV-specific CD8+ T cells to efficiently deliver active granzyme B (GrB) to HIV-infected targets was fairly high (median, 36.7%) and most similar to that of LTNPs/ECs (43.7%, P > .5; Figure 6D). As a second measure of killing performed in the same sample, infected CD4+ T-cell elimination mediated by the HIV-specific CD8+ T cells of the cases was investigated (median, 43.45%) and found to be most similar to that of chronic progressors, either viremic (31.96%, P = .2) or Rx<50 (33.4%, P = .25), but significantly less than that of typical LTNPs/ECs (82.8%, P = .001; Figure 6E). These 2 measurements of killing were strongly correlated when all patients were analyzed (Figure 6F), as shown previously.3,17,46 In summary, the HIV-specific CD8+ T-cell killing responses of the cases as a group were disproportionately high for the extremely low HIV RNA levels but were relatively low compared with those of typical LTNPs/ECs.
Discussion
Initial suspicion that these 4 healthy persons with stable CD4 counts and HIV RNA levels of less than 50 copies/mL were different from classic LTNPs/ECs arose from findings of weakly reactive, nonevolving HIV-1 Western blots (all 4 cases) and undetectable HIV DNA by qualitative PCR (C2 and C3). We confirmed that they are distinct from conventional LTNPs/ECs based on very low to undetectable viral burdens, including lower HIV DNA levels and more difficult to isolate replication-competent virus in PBMCs and GALT, atypical HIV-specific antibody response profiles, and HIV-specific CD8+ T-cell responses that were disproportionately high for the level of viremia but lower than those of typical LTNPs/ECs. Despite these differences, the cases shared some features with typical LTNPs/ECs, including intact gut-associated CD4+ T-cell populations, CD4+ T cells that were fully susceptible to HIV infection, and lack of potent neutralizing antibody activity. In addition, they carried at least one protective HLA class I allele known to be enriched in LTNP/EC cohorts,1 suggesting the mechanisms leading to this unusual phenotype might be the same ones causing durable control over HIV infection in conventional LTNPs/ECs.
Based on both direct measurements and as suggested indirectly by reduced frequencies of HIV-specific immune responses, total HIV burdens were barely detectable in these cases and were lower than those of typical LTNPs/ECs. In addition to HIV RNA levels of less than 1 copy/mL in all 4 cases, these persons had extremely low to undetectable total and integrated HIV DNA levels in PBMCs, which were significantly less than those of conventional LTNPs/ECs. These low to undetectable HIV DNA levels were predictive of the negligible recovery of replication-competent virus from PBMCs in both the HIC assay and by flow cytometric p24 detection during 120-day culture. The infectious viral burdens of the cases, as measured in the HIC assay, directly correlated with the levels of integrated HIV DNA and appeared to be lower than those of most LTNPs/ECs (found herein and described previously) and Rx<50.31,41,47
In addition to low HIV DNA levels and low infectious viral burdens in PBMCs, very low HIV DNA levels in colonic CD4+ T cells of the cases were also demonstrated, which contrasted with prior investigations in Rx<50 revealing 5- to 10-fold higher HIV DNA levels in GALT compared with PBMCs.39,40 Low HIV DNA levels in the GALT of the cases also differed from the results of the representative LTNP/EC 88 in whom HIV DNA levels were 20-fold higher in the colon than in PBMCs and in whose colonic CD4+ T cell replication-competent virus was eventually detected. Despite limited data on HIV DNA levels in mucosal tissues of LTNPs/ECs, recent reports of higher SIV RNA and DNA levels in GALT versus PBMCs of LTNP/EC Chinese rhesus macaques48 and significantly higher T-cell responses in the rectal mucosa than in PBMCs of LTNP/EC human subjects16 provide evidence that GALT- versus PBMC-derived CD4+ T cells of LTNPs/ECs may support greater virus replication as in Rx<50. Therefore, the very low to undetectable levels of HIV DNA and difficult to isolate replication-competent virus observed in both GALT and PBMCs of the cases differed from those of most LTNPs/ECs and Rx<50. These observations suggest that the cases have exceedingly small, and virtually undetectable, total viral burdens and, as such, might experience the least amount of ongoing virus replication compared with other patients with low plasma viremia.
Low-frequency HIV-specific immune responses observed among the cases likely reflect infrequent stimulation in vivo because of persistently low antigen levels. At low sera dilution, additional reactivity was detected on their HIV-1 Western blots, suggesting broader reactivity than initially suspected and confirming unequivocal HIV infection. Atypical HIV-specific antibody profiles in the highly quantitative LIPS assay corroborated the Western blot findings and suggested that there had been adequate priming of HIV-specific B-cell responses. Indeterminate or weakly reactive Western blots have been reported in other rare cases. Australian patient C135, who is HLA B*57+ and part of the Sydney Blood Bank Cohort, had repeatedly indeterminate Western blots exhibiting only faint gp160 reactivity despite evidence of HIV infection for 18 years.22 In a case from San Francisco, weakly reactive, but stable Western blots were observed over time.5 These cases, like ours, also had consistently undetectable HIV RNA levels by ultrasensitive assays, small HIV reservoirs and comparatively weak T-cell responses.5,22-24 Low antibody levels in these cases were also reminiscent of the gradual decline in Western blot reactivity reported in the “Berlin patient” who may have been cured of HIV infection after CCR5 Δ32/Δ32 stem cell transplantation.49 Longer follow-up and additional studies might provide further insight into the source and mechanisms responsible for maintenance of these unusual antibody profiles.
The low frequencies of HIV-specific CD8+ T cells of the 4 cases were also consistent with infrequent antigen re-encounter in vivo. After 6-day stimulation, however, the capacity of their HIV-specific CD8+ T cells to proliferate and lyse HIV-infected CD4+ T-cell targets was surprisingly high, although the overall magnitude tended to be lower than that of typical LTNPs/ECs. These results bear some similarities to French “weak responder” LTNPs/ECs who exhibited less frequent viral blips, lower starting frequencies of IFN-γ-secreting CD8+ T cells, and weaker virus suppressive capacity than “high responder” LTNPs/ECs.8 In our study, it is notable that the 2 cases (C2 and C4) with the highest CD8+ T-cell proliferative and cytotoxic responses were both HLA B*57+. This is consistent with prior investigations of chronically infected patients and HIV-uninfected recipients of a replication incompetent adenovirus serotype 5/HIV vaccine, which suggested the series of events starting with proliferation and culminating in cytotoxicity are preferentially preserved in persons with protective HLA alleles.3,17,18,46,50
Despite an exhaustive evaluation, we cannot definitively conclude what mechanisms are responsible for the unique phenotype of the cases and, for that matter, whether the same ones are operating in all 4 or to the same extent. Some possibilities include aborted infection occurring after acquisition of attenuated viruses, robust immune responses, or both. Although gross nef deletions were not observed in 2 cases and replication-competent virus was detected in the third despite numerous premature truncations, viral factors cannot be completely excluded because autologous viruses from these cases were not isolated and fully characterized. Very early viral suppression as occurred in the cases does not exclude an immune-mediated mechanism because spontaneous viral control appears to be established shortly after infection in conventional LTNPs/ECs.51,52 CD8+ T-cell-responses have recently been shown to mediate profound early control of pathogenic SIVmac239 infection after mucosal challenge in macaques immunized with SIV vaccines that include rhesus cytomegalovirus vectors.53 Notably, these animals shared several features with the human cases in the current study, including waning HIV-specific antibodies, low SIV DNA levels, and lack of recoverable replication-competent SIV that differed from conventional LTNPs/ECs.53 Therefore, it is plausible that virus-specific CD8+ T-cell responses could mediate higher-level control than has been observed in conventional LTNPs/ECs and do so shortly after infection. In the present study, favorable HLA haplotypes and reasonably strong HIV-specific CD8+ T-cell responses of the cases suggest that CD8+ T cells play a major role. However, it is also possible that these robust immune responses are not the cause of control, but rather have resulted from preserved immunity in patients with viral replication that had been limited during acute infection by the immune response or because of attenuated viruses. Further studies and longer follow-up of these cases may shed additional light on the mechanisms of control in each of them.
Investigations of LTNPs/ECs have been conducted to identify the mechanisms responsible for durable control of HIV replication. In this study, we describe 4 unique patients who are distinct from most LTNPs/ECs in that they possess the lowest antigen levels studied to date with disproportionately high immune responses. As such, they may represent the best examples of naturally occurring “functional cure” and prove that greater control over HIV replication is possible than was originally suspected from earlier investigations in LTNPs/ECs. Further study of these persons may provide additional insights into the mechanisms associated with nearly complete suppression of HIV replication and inform the development of efficacious HIV vaccines and immunotherapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the study participants for their generous donations of time and study specimens and Julia Metcalf, Dr H. Clifford Lane, Dr Philip Murphy, and Dr Joel N. Blankson for thoughtful discussions related to this project.
This work was supported in part by the Intramural Research Programs of NIAID and National Institute of Dental and Craniofacial Research (NIDCR) and (NIAID contract HHSN261200800001E; V.N., R.L.D., and S.J.).
National Institutes of Health
Authorship
Contribution: D.M., S.A.J., B.A.P., V.N., M.S., R.L.D., P.D.B., N.A.D.-R., E.H.G., J.H.G., J.N.H., W.L.T., U.O., T.-W.C., and S.A.M. performed research; D.M., V.N., M.S., R.L.D., P.D.B., N.A.D.-R., E.H.G., J.H.G., J.N.H., W.L.T., C.W.H., U.O., T.-W.C., and S.A.M. analyzed and interpreted data; N.A.C., C.L.C., C.A.R., O.S., S.A.P., R.J.O., and S.A.M. coordinated inclusion of patients and collected clinical data from patients; C.W.H., J.A.K., I.S., O.S., S.A.P., R.J.O., and M.C. discussed results; D.M., P.D.B., E.H.G., U.O., T.-W.C., and S.A.M. designed the research; and D.M. and S.A.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen A. Migueles, HIV-Specific Immunity Section, Laboratory of Immunoregulation, NIAID, NIH, 10 Center Dr, Bldg 10, Rm 11B-07, Bethesda, MD 20892; e-mail: smigueles@niaid.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal