Abstract
The Na+/H+ exchanger regulatory factor-2 (NHERF-2) is an integral component of almost all endothelial cells (ECs), yet its endothelial function is not known. Here, we found that NHERF-2, is a key regulator of endothelial homeostasis because NHERF-2–silenced ECs proliferate at a much higher rate even in the absence of mitogens such as VEGF compared with control ECs. We further show that the hyperproliferation phenotype of NHERF-2–silenced EC is because of an accelerated cell cycle that is probably caused by a combination of the following factors: increased cytoplasmic calcium, increased expression of c-Myc, increased expression of cyclin D1, and reduced expression of p27. Using an experimental mouse model of human hemangioma, we found that the endothelial neoplasms derived from NHERF-2–silenced cells were much larger in volume than those derived from control cells. Thus, NHERF-2 is a negative regulator of endothelial proliferation and may have important roles in endothelial homeostasis and vascular modeling.
Introduction
The maintenance of endothelial homeostasis is critical in preventing uncontrolled angiogenesis, permeability, thrombosis, and inflammation.1 In this context, the VEGF is a critical regulator of both physiologic and pathologic angiogenesis.2 Originally identified on the basis of its permeability-inducing ability,3 it is now recognized as a potent inducer of endothelial proliferation, migration, and survival. The effects of VEGF and its family members are mediated by structurally related receptors termed VEGFR-1, VEGFR-2, and more recently neuropilin 1 (NRP-1). Among these receptors VEGFR-2 has emerged as the predominant mediator of endothelial proliferation and migration.4 In contrast VEGFR-1 is thought to mediate inhibitory and/or decoy effects in endothelial cells (ECs).5 NRP-1, by contrast, was first found to act as a coreceptor enhancing VEGF binding to VEGFR-2.6 However, we previously reported that in HUVECs, NRP-1 mediates ligand-induced migration but not proliferation.7
The Na+/H+ exchanger regulatory factors, NHERF-1 and NHERF-2, are 2 structurally related protein adapters that contain tandem PDZ domains.8,9 They are primarily expressed in the brush border membrane of the proximal tubule, small intestine, and colon and regulate protein kinase A–mediated inhibition of the sodium-hydrogen exchanger (NHE-3).10,11 In the recently developed NHERF-2 knockout mice Ca2+ or cGMP-mediated inhibition of NHE-3 is abolished, resulting in higher basal fluid absorption rates in the ileum.12 NHE-3 comprises a family of NHEs that extrude H+ (equivalents) generated metabolically in exchange for extracellular Na+ by an antiport mechanism.13 Activation of NHE is a universal response to mitogenic stimulation14 and has a permissive effect in promoting cell proliferation.15
NHERF-2 is a human scaffold protein that connects plasma membrane proteins with members of the ezrin/moesin/radixin family and thereby helps to link them with the actin cytoskeleton and regulates their surface expression.16 NHERF-2 interacts with various G protein–coupled receptors, including parathyroid hormone 1 receptor, lysophosphatidic acid receptor 2, purinergic receptor, and metabotropic glutamate receptor 5, and can enhance their phospholipase C β (PLCβ)–mediated signaling.16 In epithelial cells NHERF-2 has been shown to interact with the PDGFR, N-cadherin/β catenin (N-Cad/Cat) complex, and to regulate lamellopodia formation and cell migration.17 Recent studies have reported endothelial-specific expression of NHERF-218 ; however, its function in the endothelium remains unknown.
We report here that NHERF-2 is a critical regulator of endothelial homeostasis because NHERF-2–silenced cells continued to proliferate even in the absence of growth factor such as VEGF. This hyperproliferation phenotype in NHERF-2–silenced ECs is because of an accelerated cell cycle that is probably caused by a combination of the following factors: increased basal cytoplasmic calcium, increased expression of c-Myc, increased expression of cyclin D1, and reduced expression of p27. Using an experimental mouse model of human hemangioma, we found that the endothelial neoplasms derived from NHERF-2–silenced cells were much larger in volume than those derived from control cells.
Methods
Reagents
VEGF was obtained from R&D Systems. The Abs to VEGFR-2, PLCβ3, phosphoVEGFR-2 (951), VE-Cadherin, and βCat were purchased from Santa Cruz Biotechnology; NHERF-1, phosphoVEGFR-2 (1059), phosphoVEGFR-2 (1175), pRb, Cyclin A, Cyclin B1, and Cyclin D1 were obtained from Cell Signaling Technology. The Abs to NHERF-2 and β-actin were from Sigma-Aldrich. Small interfering RNA (siRNA) for scrambled control and NHERF-2 was from QIAGEN. NHERF-2 pCMV-6 expression plasmid was from Origene Technologies. Human sinusoidal EC (HSEC) RNA was a kind gift from Dr Vijay Shah (Mayo Clinic). HAEND cells were kindly provided by Dr V. Vetvicka, University of Louisville. Polyoma middle T-transformed murine brain endothelial (bEnd.3) cells were obtained from ATCC and cultured in DMEM (4500 mg glucose/L; Sigma-Aldrich) supplemented with 10% FBS, l-glutamine (14 mL/L), and antibiotic/antimycotic (14 mL/L; Sigma-Aldrich).
Intracellular Ca2+ release
HUVECs were transfected with control-scrambled or NHERF-2 siRNA with the use of Oligofectamine reagent in OPTI-MEM medium. After transfection, at 4 hours, the cells were serum-starved overnight, loaded with Fura2-AM and then stimulated with 10 ng/mL VEGF165. Intracellular Ca2+ concentrations were measured with the DeltaScan illumination system (Photon Technology International) with the use of Felix 32 Version 1.1 software as described before.19 Experiments were repeated ≥ 3 times. Statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
Real-time PCR
Total RNA was isolated from cells with the use of TRIzol reagent (Invitrogen). RNA was first retrotranscribed with TaqMan Reverse Transcription Kit (Applied Biosystems), and then real-time PCR was performed with a TaqMan SYBR Green Master Mix (Applied Biosystems). The primers for human and murine NHERF-2, NHE-1, NHE-2, NHE-3, and β-actin were from SA Biosciences. The comparative Ct method was used to calculate the relative abundance of mRNA compared with that of β-actin expression.20 The experiment was performed in triplicate, and significance was determined with 2-sided Student t test, and P < .05 was considered significant.
Immunoprecipitation and Western blot analysis
HUVECs were transfected with control-scrambled or NHERF-2 siRNA with the use of Oligofectamine reagent in OPTI-MEM medium. After transfection, at 4 hours, the cells were starved overnight and stimulated with VEGF 10 ng/mL to collect lysates for Western blot analysis or immunoprecipitation. Cell lysates were prepared in RIPA buffer supplemented with protease inhibitor cocktail. The lysates were collected after centrifugation at 14 000g for 10 minutes at 4°C and separated by SDS-PAGE.21 Cell lysates (500 mg) were immunoprecipitated with 1 μg of Ab and of protein A/G-conjugated agarose beads and immunoblotted with Abs as described in the figures. When immunoprecipitation with NHERF-2 was performed, the 1-hour immunoprecipitation kit from Genscript was used to visualize the NHERF-2 band. This kit masks the IgG heavy chain and allows visualization of proteins ∼ 55 kDa. Western blot analyses were repeated ≥ 4 times.
Immunofluorescence
HUVECs (2 × 104) were seeded on collagen-coated coverslips in 6-well plates. Forty-eight hours later the cells were serum starved overnight and treated with or without VEGF (10 ng/mL). Coverslips were washed in PBS, fixed in 4% paraformaldehyde, and permeabilized with 0.05% Triton X-100 at room temperature. Coverslips were washed in PBS, blocked in 10% goat serum, and stained with NHERF-2 and βCat Abs (1:200) in 1% goat serum for 2 hours. Coverslips were washed in PBS and incubated for 1 hour in respective secondary Alexa Flour Ab at a dilution of 1:200 followed by postfixing in 4% paraformaldehyde and mounting in Vectashield containing DAPI (4′-6′-diamidine-2-phenylindole; Vector Labs). Confocal microscopy was performed with a Zeiss LSM 510 confocal laser scan microscope with C-Apochromat 63×/NA1.2 water-immersion lens. Absence of signal crossover was established with the use of single-labeled samples.
Proliferation assay
HUVECs were transfected with control-scrambled or NHERF-2 siRNA with the use of Oligofectamine reagent in OPTI-MEM medium. After transfection, at 36 hours cells were trypsinized and seeded (2 × 104) in 24-well plates, cultured for 24 hours in EGM, were serum-starved (0.1% serum) overnight, and then treated with VEGF165 (10 ng/mL). After culture for 20 hours, 1 μCi (0.037 Bq) of [3H]-thymidine was added to each well, and 4 hours later cells were washed with chilled PBS, fixed with 100% cold methanol, and collected for the measurement of trichloroacetic acid–precipitable radioactivity.21 Experiments were repeated ≥ 3 times each in triplicate. Proliferation for HAEND cells was performed exactly as in HUVECs, except the cells were maintained in RPMI 1640 supplemented with 10% FBS and 1% penicillin/streptomycin.
Cell-cycle analysis
DNA content was measured after staining cells with propidium iodide (PI). HUVECs were transfected with control-scrambled or NHERF-2 siRNA with the use of Oligofectamine reagent in OPTI-MEM medium. After transfection, at 36 hours, cells were starved overnight and treated with or without VEGF 10 ng/mL and collected 20 hours after treatment. The cells were trypsinized, washed in PBS, and fixed in 95% ethanol for 1 hour. Cells were rehydrated, washed in PBS, and treated with RNase A (1 mg/mL) followed by staining with PI (100 mg/mL).21 Flow cytometric quantification of DNA was performed with a FACScan (BD Biosciences), and data were analyzed with Modfit LT Version 3.1 software. Experiments were repeated ≥ 3 times. Statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
Boyden chamber migration assay
Serum-starved HUVECs (siRNA-transfected) were detached from tissue culture plates with the use of 4 mL of collagenase solution (0.2 mg/mL collagenase, 0.2 mg/mL soybean trypsin inhibitor, 1 mg/mL BSA, and 2mM EDTA in PBS). Then cells were seeded as 1 × 105/well in 500 μL of endothelial basal medium with 0.1% FBS into the transwells coated with Vitrogen (30 mg/mL), and the transwells were inserted into a 24-well plate containing 750 μL of the same medium. The cells were incubated at 37°C for 45 minutes to allow them to attach. Next, VEGF was added at a concentration of 10 ng/mL, and an additional 4-hour incubation was performed.21 The migrated cells were stained with 0.1% crystal violet and counted with the use of Gel Count (Oxford Optronics). All experiments were repeated ≥ 3 times. Statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
ChIP assay
HUVECs transfected with control-scrambled or NHERF-2 siRNA were synchronized and treated with or without VEGF 10 ng/mL for 12 hours. Then, ChIP assay was performed with the ChIP assay kit from Upstate Biotechnology. Briefly, 6 × 106 cells were used for each assay. Protein-DNA cross-linking was performed by the addition of 1% formaldehyde directly to the cell cultures, followed by incubation at 37°C for 10 minutes. After the cells were thoroughly washed with ice-cold PBS, the cells were scraped off and harvested. Cells were lysed with 200 μL of SDS lysis buffer (1% SDS, 10mM EDTA, 50mM Tris, pH 8.1) supplemented with protease inhibitors (1mM phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, and 1 μg/mL pepstatin A). Sonication was then performed on ice with the use of a sonicator (Lab-Line Ultra Tip; Lab-Line Instrument) preset for 10-second pulses with 10-second intervals. Ten repeated sonication cycles (as previously standardized by us) were applied to achieve chromatin fragmentation in the 200- to 1000-bp range. Fragmented chromatin was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 2mM EDTA, 16.7mM Tris, pH 8.1, and 150mM NaCl). Diluted chromatin fragments were precleared by incubation with protein A agarose beads under constant rotation for 2 hours at 4°C. For immunoprecipitations, an Ab specific for βCat or the respective IgG control was used for an overnight incubation with constant rotation at 4°C. The protein-DNA-Ab complex was pulled down by protein A agarose-salmon sperm DNA beads. After thorough and sequential washings with low-salt, high-salt, and LiCl-containing buffers, the resulting immune complex was eluted with 1% SDS and 0.1M NaHCO3. Formaldehyde cross-links were reversed by adding 5M NaCl and heating at 65°C for 4 hours. DNA fragments were then recovered by ethanol precipitation after proteinase K digestion and phenol-chloroform extraction. PCR was performed with the cyclin D1 promoter-specific primers amplifying the region between forward, 5′-GAAACTTGCACAGGGGTTGTGTG-3′, and reverse, 5′-GCGACTGCATCTTCTTTCAT- TTTCA-3′, as described before.22
In vivo tumorigenesis studies
Lentiviral supernatant fluids were prepared by transfecting 293T cells with NHERF-2 shRNA or scrambled control shRNA (2 μg; Sigma-Aldrich) along with gag-pol (1.5 μg) and vesicular stomatitis virus glycoprotein (0.5 μg) with the use of Effectene (QIAGEN) reagent. bEnd.3 cells were then infected overnight with 2 mL of supernatant fluid and 4 mL respective medium. To select stable clones, cells were treated with 2 μg/mL puromycin for 7 days. Next, bEnd.3 NHERF-2 shRNA or control shRNA containing cells were injected (1 × 106) subcutaneously into 5 nude mice each. Mice were monitored, tumors were measured 14 days after injection, mice were killed, and tumors were fixed in formalin. Tumor volumes were calculated with the use of the standard formula ½ (length × width2).23 Animal studies were approved by the Mayo Clinic Institutional Animal Care and Use Committee.
Immunohistochemistry
Slides were processed for H&E staining, or Ag retrieval was performed on 5-μM paraffin sections in citrate buffer, followed by deparaffinization and staining according to the INC select kit from Chemicon with the use of Ki67 Ab from Sigma-Aldrich at a dilution of 1:100. Images were acquired at a magnification of ×40 with the use of AxioPlanII (Carl Zeiss Inc). The nuclei of the single layer of cells lining the vascular lumen were quantitated for Ki67 positivity, and SD among 10 was determined.
Statistical analysis
All values are expressed as means ± SD. Statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
Results
Knockdown of NHERF-2 increases endothelial proliferation
NHERF-2 was one of a set of 58 gene transcripts identified to be specifically expressed in ECs,24 but its function in ECs has not been defined. Therefore, we first determined that NHERF-2 but not NHERF-1 was robustly expressed in HUVECs by Western blot analysis (Figure 1A). Next, 4 different siRNAs against NHERF-2 were tested, and all 4 showed significant down-regulation of the NHERF-2 protein 48 hours after transfection (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To determine the functional effect of NHERF-2 silencing in ECs, we first transfected HUVECs with control-scrambled or NHERF-2 siRNA with the use of oligofectamine for 4 hours. The cells were starved overnight and stimulated with 10 ng/mL VEGF for 24 hours followed by [3H]-tritiated thymidine incorporation assay. A significant increase in baseline proliferation without VEGF was observed in the NHERF-2 knockdown cells; this was further exacerbated on the addition of VEGF compared with the control cells (Figure 1B). Similar results were obtained in MTT assays (data not shown). We also confirmed the increased proliferation phenotype on NHERF-2 silencing in another EC line (HAEND) originally isolated from patients with human liver angiosarcoma. This cell line has been extensively characterized and has shown positivity for VWF, Ulex europeus agglutinin-1, and uptake of Dil-Ac-LDL.25,26 Compared with the primary HSECs, HAEND cells expressed 5-fold higher NHERF-2 mRNA as determined by RT-PCR (Figure 1C left). We next silenced NHERF-2 in HAEND cells and determined proliferation with the use of the [3H]-thymidine incorporation assay. Efficient knockdown of NHERF-2 was confirmed by RT-PCR (Figure 1C middle), and, similar to HUVECs, an ∼ 2-fold increase in proliferation occurred on knockdown (Figure 1C right). However, in the Boyden-chamber migration assay, knockdown of NHERF-2 did not significantly affect migration in response to VEGF (10 ng/mL) in HUVECs (supplemental Figure 2). It should be noted that the migration assay was performed after 4 hours of VEGF stimulation; hence, any contribution from increased proliferation in NHERF-2–silenced cells was not expected. This uncontrolled proliferation suggests that NHERF-2 is a key regulator of endothelial homeostasis.
Knockdown of NHERF-2 increases endothelial proliferation. (A) Only NHERF-2 was expressed in HUVECs. The breast cancer cell line MCF-7 expressed both NHERF-1 and NHERF-2. (B) HUVECs were transfected with control-scrambled or NHERF-2 siRNA for 48 hours. The cells were starved overnight and stimulated with 10 ng/mL VEGF for 24 hours, followed by [3H]-titrated thymidine incorporation assay represented here in fold with respect to the control-scrambled unstimulated cells as in C-V. (C) Approximately 5-fold higher expression of NHERF-2 in HAEND cells compared with HSECs (left). Efficient knockdown of NHERF-2 in HAEND cells as determined by real-time PCR (middle). HAEND cells were transfected with control-scrambled or NHERF-2 siRNA for 48 hours, followed by [3H]-titrated thymidine incorporation assay (right). Statistical significance was determined with 2-sided Student t test, *P < .05 was considered significant.
Knockdown of NHERF-2 increases endothelial proliferation. (A) Only NHERF-2 was expressed in HUVECs. The breast cancer cell line MCF-7 expressed both NHERF-1 and NHERF-2. (B) HUVECs were transfected with control-scrambled or NHERF-2 siRNA for 48 hours. The cells were starved overnight and stimulated with 10 ng/mL VEGF for 24 hours, followed by [3H]-titrated thymidine incorporation assay represented here in fold with respect to the control-scrambled unstimulated cells as in C-V. (C) Approximately 5-fold higher expression of NHERF-2 in HAEND cells compared with HSECs (left). Efficient knockdown of NHERF-2 in HAEND cells as determined by real-time PCR (middle). HAEND cells were transfected with control-scrambled or NHERF-2 siRNA for 48 hours, followed by [3H]-titrated thymidine incorporation assay (right). Statistical significance was determined with 2-sided Student t test, *P < .05 was considered significant.
Previously reported functions of NHERF-2 include protein kinase A–mediated inhibition of the NHE-3, thus regulating cellular pH and proliferation. Therefore, with the use of real-time PCR we next determined which of the NHEs were expressed in HUVECs. Interestingly, the mRNA expression pattern was NHE-1 > NHE-3 > NHE-2 (supplemental Figure 1B). Compared for fold expression with respect to NHE-2 (least expression), mRNA expression of NHE-3 was 2.7-fold and that of NHE-1 was 77.5-fold higher (Figure 1C). However, siRNA-mediated knockdown of NHE-1 in NHERF-2–silenced cells did not attenuate the increased proliferation phenotype observed in NHERF-2–silenced HUVECs (data not shown), thus suggesting NHE-1 function to be dispensable. Because VEGF by acting through VEGFR-2 induces proliferation of HUVECs, we next determined the effect of NHERF-2 knockdown on VEGF-mediated signaling pathways.
Influence of NHERF-2 on VEGF signaling pathways
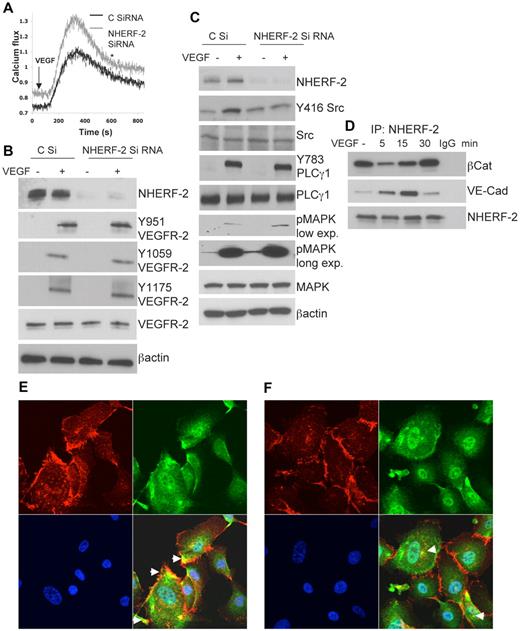
VEGF by acting through VEGFR-2 induces intracellular Ca2+ release in HUVECs, and this is important for proliferation.5 Therefore, we first examined whether calcium homeostasis was affected in HUVECs with the use of the ratiometric Fura2-AM dye. Baseline fluorescence was consistently higher in the NHERF-2–silenced cells than in the control cells (Figure 2A). Furthermore VEGF (10 ng/mL) induced Ca2+ release was also consistently higher in NHERF-2–silenced cells than in the control cells (Figure 2A). We next examined the level of tyrosine phosphorylation of specific residues on VEGFR-2 in VEGF-\stimulated NHERF-2–silenced HUVECs. Knockdown of NHERF-2 did not affect tyrosine phosphorylation of any of the VEGFR-2 residues tested that have been previously implicated in endothelial proliferation and migration (Figure 2B). Possible changes in localization of VEGFR-2, surface versus internal, was also examined, and no differences were observed between NHERF-2–silenced and control HUVECs (data not shown). Downstream from the receptors, other signal-plexes implicated in the VEGF pathway include Src, PLCγ1, and p42/44 MAPK. With VEGF stimulation, control cells showed Y416 autophosphorylation at the active site of Src; however, this was reduced to baseline in NHERF-2–silenced cells (Figure 2C). This is in accordance with our previous observation that in HUVECs inhibition of Src results in augmented DNA synthesis.27 On VEGF stimulation PLCγ1 was robustly phosphorylated at Y783, and no difference was observed between NHERF-2–silenced and control cells (Figure 2C). Because phosphorylation of PLCγ1 is tightly linked to tyrosine phosphorylation of VEGFR-2,28 no change in NHERF-2–silenced cells suggests preservation of the VEGFR-2 signaling pathway. In NHERF-2 knockdown cells, p42/44 MAPK phosphorylation was mildly but consistently increased in the presence or absence of VEGF (Figure 2C). Taken together these data indicate that in NHERF-2–silenced ECs VEGF-induced signaling starting with tyrosine phosphorylation of residues on VEGFR-2, PLCγ1 phosphorylation followed by MAPK phosphorylation is not significantly affected; only Src phosphorylation at the active site (Y416) was disrupted. Previous studies have reported that VE-Cad remains in a complex with Src and Csk (C-terminal Src kinase), a negative regulator of Src activation. Therefore, we next examined whether NHERF-2 interacts with the VE-Cad/βCat complex in ECs.
Effect of NHERF-2 silencing on VEGF-mediated signaling. (A) Serum-starved NHERF-2 or control siRNA-transfected HUVECs were loaded with Fura2-AM and stimulated with VEGF at 10 ng/mL. (B) HUVECs transfected with control-scrambled or NHERF-2 siRNA for 48 hours were serum-starved and stimulated with 10 ng/mL VEGF for 5 minutes, and Western blot analysis was performed to detect tyrosine phosphorylation of the VEGFR-2 residues, Y951, Y1059, and Y1175 or (C) Y416 Src, Y783 PLCγ1, or p42/44 MAPK. Experiments were repeated 3 times, and Western blot analyses were performed ≥ 4 times for each marker. (D) Serum-starved HUVECs were stimulated with or without VEGF at 10 ng/mL. Lysates were immunoprecipitated with Ab against NHERF-2 and immunoblotted with Ab for VE-Cad and βCat. To detect total NHERF-2 pull-down, the 1-hour immunoprecipitation kit from Genscript was used. This kit masks the IgG heavy chain and allows visualization of proteins ∼ 55 kDa. (E) HUVECs without VEGF stimulation were double stained with Abs against βCat (red) and NHERF-2 (green). (F) HUVECs with VEGF (10 ng/mL) stimulation were double stained with Abs against βCat (red) and NHERF-2 (green). Nuclei appear blue because of DAPI staining. Arrows indicate colocalization (yellow). Original magnification, ×63.
Effect of NHERF-2 silencing on VEGF-mediated signaling. (A) Serum-starved NHERF-2 or control siRNA-transfected HUVECs were loaded with Fura2-AM and stimulated with VEGF at 10 ng/mL. (B) HUVECs transfected with control-scrambled or NHERF-2 siRNA for 48 hours were serum-starved and stimulated with 10 ng/mL VEGF for 5 minutes, and Western blot analysis was performed to detect tyrosine phosphorylation of the VEGFR-2 residues, Y951, Y1059, and Y1175 or (C) Y416 Src, Y783 PLCγ1, or p42/44 MAPK. Experiments were repeated 3 times, and Western blot analyses were performed ≥ 4 times for each marker. (D) Serum-starved HUVECs were stimulated with or without VEGF at 10 ng/mL. Lysates were immunoprecipitated with Ab against NHERF-2 and immunoblotted with Ab for VE-Cad and βCat. To detect total NHERF-2 pull-down, the 1-hour immunoprecipitation kit from Genscript was used. This kit masks the IgG heavy chain and allows visualization of proteins ∼ 55 kDa. (E) HUVECs without VEGF stimulation were double stained with Abs against βCat (red) and NHERF-2 (green). (F) HUVECs with VEGF (10 ng/mL) stimulation were double stained with Abs against βCat (red) and NHERF-2 (green). Nuclei appear blue because of DAPI staining. Arrows indicate colocalization (yellow). Original magnification, ×63.
In epithelial cells, NHERF-2 has been shown to interact with the N-Cad/βCat complex.17 In ECs, the VE-Cad/βCat complex has been shown to be important in regulating proliferation and in maintaining intercellular junctional integrity.29 To determine whether NHERF-2 interacted with these junctional proteins, we next performed coimmunoprecipitation of NHERF-2 followed by Western blot analysis for VE-Cad and βCat. Significant constitutive association of NHERF-2 with βCat was observed, and this decreased at 5-15 minutes after VEGF treatment but was restored at 30 minutes (Figure 2D). Interaction of NHERF-2 with VE-Cad gradually increased from 0 to 15 minutes and returned to baseline levels at 30 minutes (Figure 2D). In corroboration, βCat (red) colocalized with NHERF-2 (green) at the cell periphery in HUVECs without VEGF stimulation (Figure 2E). Some colocalization could also be seen in the cytoplasm. On VEGF stimulation, however, βCat staining became disrupted at the cell periphery; thus, colocalization with NHERF-2 if any was mostly visible in the cytoplasm (Figure 2F). These data raise the possibility that in NHERF-2–silenced ECs the VE-Cad/βCat complex might be disturbed. VEGF stimulation releases Csk from VE-Cad by recruiting protein phosphatase SHP2, leading to an increase in Src activation.30 We find significant VEGF-induced association between VE-Cad and NHERF-2, leading to the possibility that this association is required for loss of Csk and activation of Src. Therefore, the increased proliferation phenotype observed in NHERF-2–silenced cells in the presence of VEGF can be explained by the decrease in Src activation, but that in the absence of growth factor stimulation remains unexplained. Therefore, we next proceeded to determine whether knockdown of NHERF-2 affected the cell cycle in ECs.
NHERF-2 regulates cell cycle in ECs
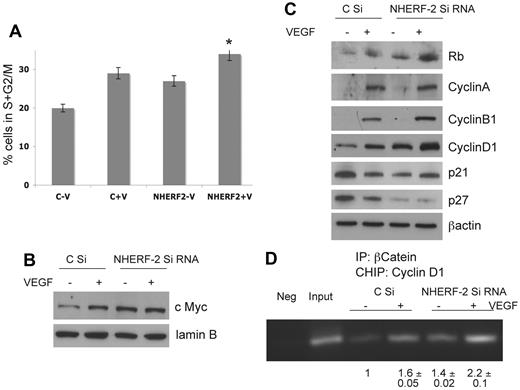
Cell-cycle progression from G0/G1 to S phase is characterized by a series of transcriptional events that involves the expression of key cell cycle–related genes. According to previous reports, passage 3 HUVECs have a doubling time of ∼ 19 hours.31 Hence, HUVECs transfected with control-scrambled or NHERF-2 siRNA were synchronized and treated with or without VEGF 10 ng/mL for 20 hours and then subjected to PI staining for cell-cycle determination or nuclear proteins, and whole-cell lysate were prepared. Percentage of cells in S+G2/M increased significantly on VEGF treatment as expected. However, significant increases in the percentage of cells in S+G2/M in NHERF-2 knockdown cells treated without VEGF was also observed (Figure 3A). Western blot analysis with the use of the nuclear lysate showed an increase in c-Myc levels on VEGF stimulation in control ECs; c-Myc levels elevated further on NHERF-2 knockdown (Figure 3B; supplemental Figure 3A). Lamin B was used as a loading control for nuclear lysates (Figure 3B). In the whole-cell lysates, VEGF treatment induced expression of pRb, cyclin A, cyclin B1, and cyclin D1. Importantly, the expression of pRb and cyclin D1 were significantly up-regulated in NHERF-2 knockdown cells even without VEGF (Figure 3C; supplemental Figure 3B). Expression of cyclin A and p21 remained unchanged in NHERF-2 knockdown cells compared with the control cells, whereas that of p27, a cdk inhibitor, was completely down-regulated in NHERF-2 knockdown cells (Figure 3C; supplemental Figure 3C). c-Myc is a well-recognized activator of cell proliferation and cell cycle. c-Myc represses the p27 promoter32 and induces expression of cyclin D1, which sequester and inhibit p27.33 c-Myc also induces expression of Cullin 134 and cyclin-dependent kinase subunit 1B,35 which are involved in p27 proteolysis. All of these data suggest tight regulation of the EC cycle by NHERF-2.
NHERF-2 regulates cell cycle in ECs. HUVECs transfected with control-scrambled or NHERF-2 siRNA were synchronized and treated with or without VEGF 10 ng/mL for 20 hours and then subjected to (A) PI staining for cell-cycle determination, percentage of S+G2/M phase population was calculated from 30 000 cells of each group (n = 3); or (B) nuclear lysates were collected and subjected to Western blot analysis with the use of c-Myc Ab and lamin B as control (n = 3); or (C) whole-cell lysates collected and subjected to Western blot analysis with Abs against pRb, Cyclin A, Cyclin B1, Cyclin D1, p21, p27 (N = 3). (D) HUVECs transfected with control-scrambled or NHERF-2 siRNA were synchronized and treated with or without VEGF 10 ng/mL for 12 hours and then cross-linked chromatin-protein complexes were isolated with βCat Ab as well as control immunoglobulin (Neg). After reverse cross-linking, DNA fragments were isolated, and the cyclin D1 promoter was amplified by PCR with the use of promoter-specific primers and run on a 2% agarose gel. The Input sample represents 5% of total chromatin DNA. Representative gel is shown with band intensities normalized with respect to control siRNA-transfected cells without VEGF (n = 3). When applicable, statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
NHERF-2 regulates cell cycle in ECs. HUVECs transfected with control-scrambled or NHERF-2 siRNA were synchronized and treated with or without VEGF 10 ng/mL for 20 hours and then subjected to (A) PI staining for cell-cycle determination, percentage of S+G2/M phase population was calculated from 30 000 cells of each group (n = 3); or (B) nuclear lysates were collected and subjected to Western blot analysis with the use of c-Myc Ab and lamin B as control (n = 3); or (C) whole-cell lysates collected and subjected to Western blot analysis with Abs against pRb, Cyclin A, Cyclin B1, Cyclin D1, p21, p27 (N = 3). (D) HUVECs transfected with control-scrambled or NHERF-2 siRNA were synchronized and treated with or without VEGF 10 ng/mL for 12 hours and then cross-linked chromatin-protein complexes were isolated with βCat Ab as well as control immunoglobulin (Neg). After reverse cross-linking, DNA fragments were isolated, and the cyclin D1 promoter was amplified by PCR with the use of promoter-specific primers and run on a 2% agarose gel. The Input sample represents 5% of total chromatin DNA. Representative gel is shown with band intensities normalized with respect to control siRNA-transfected cells without VEGF (n = 3). When applicable, statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
Cyclin D1 is an important regulator of cellular proliferation that helps to initiate transition from the late G1 to the S phase of the cell cycle.22 That βCat regulates the expression of cyclin D1 has been reported in colon cancer cells.36,37 Therefore, we next determined whether βCat was able to activate the cyclin D1 promoter activity in NHERF-2 knockdown ECs. HUVECs transfected with control-scrambled or NHERF-2 siRNA were synchronized and treated with or without VEGF 10 ng/mL for 12 hours and then subjected to CHIP assay with the use of βCat Ab as well as control immunoglobulin (IgG). Next, PCR was performed with primers specific for the cyclin D1 promoter. Significant increases in the cyclin D1 promoter amplicon were observed after treatment of control cells with VEGF or in NHERF-2 knockdown cells without VEGF (Figure 3D). The band intensity of the amplified product was highest in the NHERF-2 knockdown cells treated with VEGF (Figure 3D). In addition, increased nuclear βCat was noted in NHERF-2–silenced cells compared with control cells after VEGF treatment (supplemental Figure 3D). These data entirely corroborate our previous observations and elucidate a plausible mechanism whereby lack of NHERF-2 increases available nuclear βCat that binds the cyclin D1 promoter, thereby increasing cyclin D1 protein levels.
Overexpression of NHERF-2 arrests cell proliferation
To complement our knockdown studies, we next transfected HUVECs with the empty vector (EV) or NHERF-2 plasmid for 48 hours with the use of nucleofection. Next, equal numbers of trypsinized cells were plated in triplicate in 6-well plates and counted with trypan blue after 24 hours or lysed for Western blot analysis. A significant decrease in proliferation was observed in the NHERF-2–overexpressing cells compared with the EV control (Figure 4A). Simultaneously significant increase in p27 and decrease in c-Myc was observed in the NHERF-2–overexpressing cells compared with the EV control (Figure 4B). The data thus far suggested that NHERF-2 acts as a negative regulator of proliferation in ECs, and we next tested this hypothesis in vivo.
Overexpression of NHERF-2 arrests cell proliferation. HUVECs were transfected with 2 μg of EV or NHERF-2 plasmid with the use of nucleofection for 48 hours. Next, equal number of trypsinized cells were plated in triplicate in 6-well plates and after 24 hours were either (A) counted with trypan blue dye or (B) lysed and subjected to Western blot analysis with the use of Abs against c Myc, NHERF-2, p27, and β actin. Statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
Overexpression of NHERF-2 arrests cell proliferation. HUVECs were transfected with 2 μg of EV or NHERF-2 plasmid with the use of nucleofection for 48 hours. Next, equal number of trypsinized cells were plated in triplicate in 6-well plates and after 24 hours were either (A) counted with trypan blue dye or (B) lysed and subjected to Western blot analysis with the use of Abs against c Myc, NHERF-2, p27, and β actin. Statistical significance was determined with 2-sided Student t test, and a value of P < .05 was considered significant.
NHERF-2 knockdown increases tumor growth in vivo
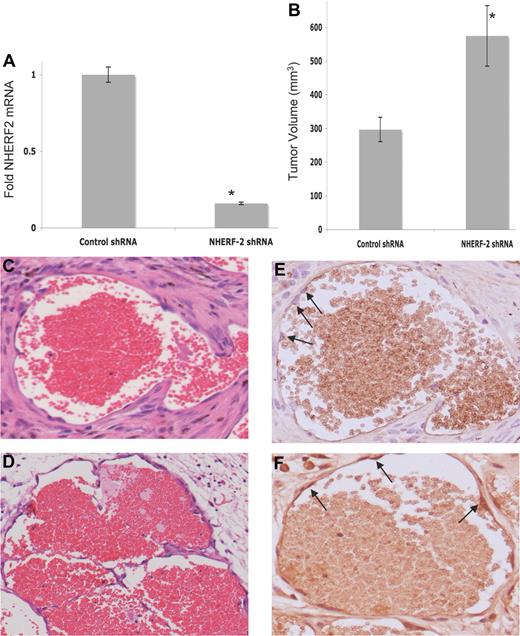
We wanted to examine the role of NHERF-2 silencing in ECs in an in vivo setting. Therefore, with the use of an experimental model of human hemangioma, in which the bEnd.3 cells are grafted subcutaneously into nude mice,38 we compared hemangioma growth between NHERF-2–silenced and control cells. Hemangiomas are EC-derived neoplasms that histologically consist of clusters of ECs surrounding vascular lumens of various diameters.39 Control-scrambled and NHERF-2 shRNA bearing bEnd.3 cells were selected with puromycin after infection with respective lentiviruses. Efficient knockdown of NHERF-2 in cells was confirmed by real-time PCR before injecting subcutaneously (Figure 5A). Two weeks after injection, tumor volumes from the NHERF-2–silenced cells were ∼ 50% greater than those derived from the control cells (Figure 5B). Morphologic and histologic analyses with the use of H&E staining found typical cavernous hemangiomas (ECs lining vascular lumen containing red blood cells) in both endothelioma-derived hemangiomas (Figure 5C-D). Immunohistochemical staining with Ki67 found positive nuclei in ∼ 20% ± 5% of the cells lining the vascular lumen derived from the NHERF-2–silenced cells (Figure 5F) versus none from the control cell–derived hemangioma (Figure 5E). These data substantiate our in vitro findings that NHERF-2 acts as a negative regulator of endothelial proliferation.
Effect of silencing NHERF-2 on in vivo tumorigenesis. (A) Efficient knockdown of NHERF-2 in bEnd.3 cells by lentiviral shRNA as determined by real-time PCR. (B) Average tumor volume (in mm3) in control shRNA- versus NHERF-2 shRNA-bearing groups. Ten mice were injected with 1 × 106 cells, and animals were killed on day 14. Statistical significance was determined with 2-sided Student t test, and *P < .05 was considered significant. (C) Histologic analysis by H&E staining found typical cavernous hemangiomas in control-shRNA and (D) NHERF-2 shRNA–bearing groups. Original magnification, ×20. (E) Ki67 staining of the control shRNA–bearing hemangioma. Arrows indicate lack of Ki67 (brown) staining; instead hematoxylin counter-stained blue nuclei are visible. (F) Ki67 staining of the NHERF-2 shRNA–bearing hemangioma. Arrows indicate positive Ki67 (brown) staining. Original magnification, ×40.
Effect of silencing NHERF-2 on in vivo tumorigenesis. (A) Efficient knockdown of NHERF-2 in bEnd.3 cells by lentiviral shRNA as determined by real-time PCR. (B) Average tumor volume (in mm3) in control shRNA- versus NHERF-2 shRNA-bearing groups. Ten mice were injected with 1 × 106 cells, and animals were killed on day 14. Statistical significance was determined with 2-sided Student t test, and *P < .05 was considered significant. (C) Histologic analysis by H&E staining found typical cavernous hemangiomas in control-shRNA and (D) NHERF-2 shRNA–bearing groups. Original magnification, ×20. (E) Ki67 staining of the control shRNA–bearing hemangioma. Arrows indicate lack of Ki67 (brown) staining; instead hematoxylin counter-stained blue nuclei are visible. (F) Ki67 staining of the NHERF-2 shRNA–bearing hemangioma. Arrows indicate positive Ki67 (brown) staining. Original magnification, ×40.
Discussion
ECs in the adult mammal are among the least proliferative cell types, with ∼ 1 in 10 000 cells entering the cell cycle at any given time.40 This quiescence is rapidly reversed in response to growth factors during pathologic neovascularization, particularly during tumorigenesis. The robust proliferative switch of the quiescent endothelium is a complex process that is governed by a network of checks and balances. In this context, we report here for the first time a functional role for NHERF-2 in ECs.
We found that NHERF-2 regulates endothelial homeostasis, both basal and growth factor induced, by preventing uncontrolled proliferation. Importantly, we found that ECs expressed only NHERF-2 and not NHERF-1, therefore providing an ideal system to study the role of NHERF-2 in these cells. Silencing NHERF-2 increased basal and VEGF-induced intracellular Ca2+ in HUVECs. In neuronal and epithelial cells, NHERF-2 coprecipitates with and activates the plasma membrane Ca2+-ATPase (PMCA) efflux pumps, including PMCA 1b, the isoform expressed in ECs.41-43 It is possible then that loss of interaction with NHERF-2 blocks activation of PMCA, thereby facilitating retention of cytosolic Ca2+(Figure 6).
Schematic illustration of a hypothetical model of interactions of NHERF-2 in the ECs. In NHERF-2–silenced cells, loss of interaction with NHERF-2 inhibits PMCA localization and activation and increases free βCat. Thus, PMCA cannot pump calcium out, resulting in increased cytosolic Ca2+ that can induce c-Myc, which can then repress the p27 promoter and induce expression of cyclin D1. Cyclin D1 also sequesters and inhibits p27. Furthermore, c-Myc also induces expression of Cullin 1 and cyclin-dependent kinase subunit 1B, which are involved in p27 proteolysis. Free βCat also increases expression of cyclin D1 by binding to its promoter, all of which result in increased proliferation in NHERF-2–silenced cells.
Schematic illustration of a hypothetical model of interactions of NHERF-2 in the ECs. In NHERF-2–silenced cells, loss of interaction with NHERF-2 inhibits PMCA localization and activation and increases free βCat. Thus, PMCA cannot pump calcium out, resulting in increased cytosolic Ca2+ that can induce c-Myc, which can then repress the p27 promoter and induce expression of cyclin D1. Cyclin D1 also sequesters and inhibits p27. Furthermore, c-Myc also induces expression of Cullin 1 and cyclin-dependent kinase subunit 1B, which are involved in p27 proteolysis. Free βCat also increases expression of cyclin D1 by binding to its promoter, all of which result in increased proliferation in NHERF-2–silenced cells.
In NHERF-2–silenced cells the VEGF signaling pathway was preserved, starting with tyrosine phosphorylation of VEGFR-2, followed by tyrosine phosphorylation of PLCγ1 and activation of pMAPK; however Src phosphorylation at the active site Y416 was abolished. This is in accordance with our observation that in HUVECs inhibition of Src results in augmented DNA synthesis.27 Previous studies have reported that VE-Cad remains in a complex with Src and Csk, a negative regulator of Src activation. VEGF stimulation releases Csk from VE-Cad by recruiting protein phosphatase SHP2, leading to an increase in Src activation.30 We find significant VEGF-induced association between VE-Cad and NHERF-2, leading to the possibility that this association is required for loss of Csk and activation of Src.
Our most significant finding is that loss of NHERF-2 increases endothelial proliferation and accelerates cell cycle even in the absence of growth factors. This could be because of a combination of a number of effects of NHERF-2 knockdown that tie in to each other, beginning with increased cytoplasmic calcium, increased expression of c-Myc, increased expression of cyclin D1, and reduced expression of p27. Ionophore-induced increases in cytosolic calcium induce c-Myc expression.44 Hence, increases in intracellular calcium could be a reason for c-Myc induction in NHERF-2–silenced cells (Figure 6). The consequences of c-Myc induction are multiple; it is a well-recognized activator of cell proliferation and cell cycle. c-Myc represses the p27 promoter32 and induces expression of cyclin D1, which sequester and inhibit p27.33 c-Myc also induces expression of Cullin 134 and cyclin-dependent kinase subunit 1B,35 which are involved in p27 proteolysis. This explains our observations on p27 inhibition and cyclin D1 induction in NHERF-2–silenced cells. In corroboration, proliferation was inhibited with simultaneous up-regulation of p27 and down-regulation of c-Myc in NHERF-2–overexpressing cells. In addition, with the use of phage display, NHERF-2 was identified as a binding partner for βCat.17 Two separate studies have shown that in colon cancer accumulated βCat activates transcription of cyclin D1.36,37 We observed significant constitutive association of NHERF-2 with βCat, and in NHERF-2 knockdown HUVECs there was increased βCat binding to the cyclin D1 promoter compared with control cells treated with or without VEGF. Therefore, it is possible that in knockdown cells in which NHERF-2 cannot sequester βCat, it is available to activate the cyclin D1 promoter, thereby stimulating proliferation (Figure 6). Finally, with the use of the mouse model of human hemangioma, we found that NHERF-2 acted as a negative regulator of proliferation in vivo as well. Previously, reactive oxygen species generated by NADPH oxidases (Nox) have been shown to control hemangioma growth.45,46 Nox can be activated by intracellular calcium.47 Hence, it is possible that in NHERF-2 knockdown endothelioma cells increased calcium activates Nox, resulting in increased reactive oxygen species, leading to a larger hemangioma than with control cells.
Overall, our results show an important role for NHERF-2 in maintaining endothelial proliferation and thereby homeostasis.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (grants CA78383, HL072178, and HL70567) and by a grant from the American Cancer Society (D.M.).
National Institutes of Health
Authorship
Contribution: R.B. conceived and directed all of the experiments; E.W. performed majority of Western blot analyses; S.K.D. performed ChIP assays; P.K.V. performed immunohistochemistry; G.E. helped with confocal experiments; Y.S.P. performed calcium determinations; and D.M. provided help with writing, conception, and funding for the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Debabrata Mukhopadhyay, Department Biochemistry and Molecular Biology, Guggenheim 1321C, Mayo Foundation, 200 First St SW, Rochester, MN 55905; e-mail: mukhopadhyay.debabrata@mayo.edu.

![Figure 1. Knockdown of NHERF-2 increases endothelial proliferation. (A) Only NHERF-2 was expressed in HUVECs. The breast cancer cell line MCF-7 expressed both NHERF-1 and NHERF-2. (B) HUVECs were transfected with control-scrambled or NHERF-2 siRNA for 48 hours. The cells were starved overnight and stimulated with 10 ng/mL VEGF for 24 hours, followed by [3H]-titrated thymidine incorporation assay represented here in fold with respect to the control-scrambled unstimulated cells as in C-V. (C) Approximately 5-fold higher expression of NHERF-2 in HAEND cells compared with HSECs (left). Efficient knockdown of NHERF-2 in HAEND cells as determined by real-time PCR (middle). HAEND cells were transfected with control-scrambled or NHERF-2 siRNA for 48 hours, followed by [3H]-titrated thymidine incorporation assay (right). Statistical significance was determined with 2-sided Student t test, *P < .05 was considered significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/20/10.1182_blood-2011-11-392563/4/m_zh89991288870001.jpeg?Expires=1769107754&Signature=RxsWy-VfuGmlD7s69D4UF-bVRdLaIfwmQMtX~tkNDkAJi4-mPlQs73MzggeoneEieruGWGeKoG1uM2UDeVqHnAqreVUfPtP9GzdiqXzildQ48DpYajm189dbERXWBVg~QzjHdmk3yVTFN6il2bAHXzfTwRd8vofBSmms3BNEWHzyIKSk8Cb4Ha5~IjEbnw2B-Rdl3VXcTojwUZrF9wLpZpETHVmOiBH2R-oWVGf5Kep-8k3xSsccg~uitgzwekEA2bd2VlEV34Vo1F7zo0r8d61dMH5cDlK~7kmuclN44epU3a3~FgKFtgNQdb0rKuNEKUk-smq~eDTuUYkhZ~djNQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal