To the editor:
A recent article in Blood demonstrated that the Pim kinase inhibitor SGI-1776 has efficacy in acute myeloid leukemia.1 This inhibitor reduced phosphorylation of multiple targets of the Pim kinases, including c-Myc at serine 62 (S62). Because no therapies directly target c-Myc, modulating S62 phosphorylation is a potential approach for reducing c-Myc protein levels in acute myeloid leukemia and other cancers. S62 phosphorylation can regulate c-Myc activity,2,3 and it controls c-Myc levels by altering its stability in both normal and cancer cells.4,–6 As c-Myc S62 phosphorylation is studied further, the reliability of tools used to study it is of utmost importance.
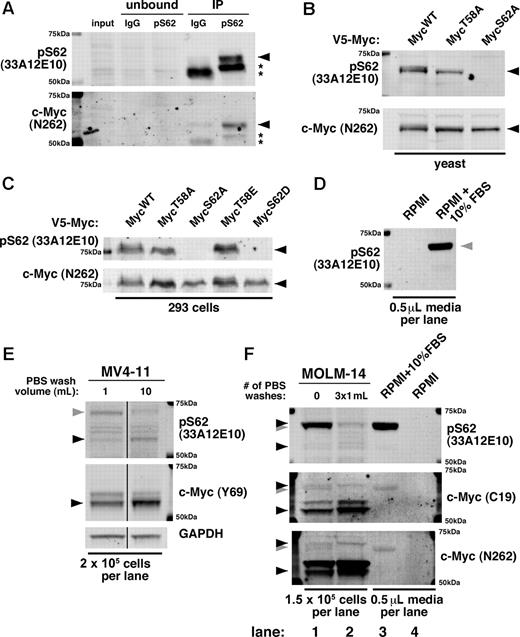
At present, only 1 antibody (33A12E10; Abcam and Bioacademia) is commercially available for targeting phosphorylation at the S62 site alone. Given the importance of c-Myc S62 phosphorylation, we validated the specificity of 33A12E10. When used to immunoprecipitate endogenous c-Myc, the 33A12E10 antibody enriches a band that reacts with both 33A12E10 and the polyclonal c-Myc antibody N262 (Figure 1A black arrow). We further found, by expressing point mutants of c-Myc in yeast and 293 cells, that 33A12E10 recognizes ectopic c-Myc only when S62 is not mutated, and this band overlaps with the band recognized by N262 (Figure 1B-C black arrow). These results demonstrate that 33A12E10 can specifically recognize c-Myc and is dependent on the S62 residue.
The monoclonal pS62 antibody 33A12E10 recognizes c-Myc but also cross-reacts with a serum protein. (A) pS62 (33A12E10) immunoprecipitates c-Myc. Cells (3×107 JY) were lysed in Ab lysis buffer and incubated overnight at 4°C with either mouse IgG or 33A12E10, followed by protein A beads for 1 hour. Bound protein was washed with Ab lysis buffer, boiled in SDS sample buffer, and separated by SDS-PAGE. Immunoblotting was performed with 33A12E10 and N262 antibodies. Asterisks indicate IgG heavy chain. (B-C) Serine 62 is required for recognition of the pS62 (33A12E10) antibody. The indicated V5-tagged mouse c-Myc constructs were expressed in yeast (B) or 293 cells (C), and lysates were separated by SDS-PAGE. Immunoblotting was performed with 33A12E10 and N262 antibodies. (D) 33A12E10 robustly detects a protein present in trace amounts of FBS. RPMI (10 μL) or RPMI + 10% FBS (10 μL) without cells was resuspended in 100 μL of SDS sample buffer and boiled, and 5 μL were separated by SDS-PAGE and immunoblotted with 33A12E10. (E) Washing cells with PBS reduces cross-reactivity of 33A12E10. One million MV4-11 cells were washed with either 1 mL or 10 mL of PBS, then cells were lysed in SDS sample buffer. Lysate from 2 × 105 cells was separated by SDS-PAGE, and immunoblotting was performed with 33A12E10 and Y69 antibodies. Irrelevant intervening lanes were removed. (F) A form of pS62 c-Myc co-migrates with the cross-reacting serum protein. MOLM-14 cells were harvested with and without 3 sequential PBS washes (1 mL each). Cells were counted before final spin (to control for cells lost during washing), and pellets were lysed in SDS sample buffer. Lysate from 1.5 × 105 cells was separated by SDS-PAGE, and immunoblotting was performed with 33A12E10, C19, and N262 antibodies. For all immunoblots, proteins were separated by SDS-PAGE, transferred to Immobilon-FL membrane, and blocked with Aquablock. Blots were incubated with the indicated primary antibodies followed by goat anti–mouse or anti–rabbit secondary antibodies conjugated to either Alexa Fluor 680 or IRDye800 and imaged on a LI-COR Odyssey scanner. The following primary antibodies were used: pS62 c-Myc (33A12E10; 1:500) and Y69 (1:1000; both from Abcam); N262 (1:1000) and C-19 (1:100; both from Santa Cruz Biotechnology); and GAPDH (1:10 000; Ambion). c-Myc is indicated with a black arrowhead; FBS cross-reacting band is indicated with a gray arrowhead.
The monoclonal pS62 antibody 33A12E10 recognizes c-Myc but also cross-reacts with a serum protein. (A) pS62 (33A12E10) immunoprecipitates c-Myc. Cells (3×107 JY) were lysed in Ab lysis buffer and incubated overnight at 4°C with either mouse IgG or 33A12E10, followed by protein A beads for 1 hour. Bound protein was washed with Ab lysis buffer, boiled in SDS sample buffer, and separated by SDS-PAGE. Immunoblotting was performed with 33A12E10 and N262 antibodies. Asterisks indicate IgG heavy chain. (B-C) Serine 62 is required for recognition of the pS62 (33A12E10) antibody. The indicated V5-tagged mouse c-Myc constructs were expressed in yeast (B) or 293 cells (C), and lysates were separated by SDS-PAGE. Immunoblotting was performed with 33A12E10 and N262 antibodies. (D) 33A12E10 robustly detects a protein present in trace amounts of FBS. RPMI (10 μL) or RPMI + 10% FBS (10 μL) without cells was resuspended in 100 μL of SDS sample buffer and boiled, and 5 μL were separated by SDS-PAGE and immunoblotted with 33A12E10. (E) Washing cells with PBS reduces cross-reactivity of 33A12E10. One million MV4-11 cells were washed with either 1 mL or 10 mL of PBS, then cells were lysed in SDS sample buffer. Lysate from 2 × 105 cells was separated by SDS-PAGE, and immunoblotting was performed with 33A12E10 and Y69 antibodies. Irrelevant intervening lanes were removed. (F) A form of pS62 c-Myc co-migrates with the cross-reacting serum protein. MOLM-14 cells were harvested with and without 3 sequential PBS washes (1 mL each). Cells were counted before final spin (to control for cells lost during washing), and pellets were lysed in SDS sample buffer. Lysate from 1.5 × 105 cells was separated by SDS-PAGE, and immunoblotting was performed with 33A12E10, C19, and N262 antibodies. For all immunoblots, proteins were separated by SDS-PAGE, transferred to Immobilon-FL membrane, and blocked with Aquablock. Blots were incubated with the indicated primary antibodies followed by goat anti–mouse or anti–rabbit secondary antibodies conjugated to either Alexa Fluor 680 or IRDye800 and imaged on a LI-COR Odyssey scanner. The following primary antibodies were used: pS62 c-Myc (33A12E10; 1:500) and Y69 (1:1000; both from Abcam); N262 (1:1000) and C-19 (1:100; both from Santa Cruz Biotechnology); and GAPDH (1:10 000; Ambion). c-Myc is indicated with a black arrowhead; FBS cross-reacting band is indicated with a gray arrowhead.
However, during our studies using the 33A12E10 antibody, we found that it strongly cross-reacts with a protein in FBS (Figure 1D gray arrow). This cross-reacting band is very similar in size to c-Myc, and substantial washing of cells with PBS is required to diminish its intensity (Figure 1E gray arrow). On further characterization of this cross-reactivity and the multiple 33A12E10-reactive bands, we found that the predominant lower molecular weight band recognized by c-Myc antibodies C19 and N262 is also recognized by 33A12E10 (Figure 1E bottom black arrow, F lanes 1-2), while the higher molecular weight serum protein is detected robustly by 33A12E10 and to a lesser degree by C19 and N262 (Figure 1F lanes 1,3 gray arrow). Importantly, washing multiple times reveals a persistent band that migrates slightly higher than the cross-reacting serum protein, visible with all 3 antibodies (Figure 1F lane 2 vs lane 1 and lane 3 top black arrow). While most of our previous studies used a validated, custom-generated polyclonal pS62 antibody,4,5,7,–9 we recently examined this higher molecular weight c-Myc in breast cancer cell lines using the 33A12E10 antibody.6 We find that under serum-starved conditions and with ample PBS washing, this band can be manipulated with chemicals that alter c-Myc stability6 and with kinase inhibitors (X.Z., unpublished data, July 2009).
The cross-reactivity of the pS62 c-Myc antibody 33A12E10 with a serum protein is of particular concern when working with leukemia cell lines or other cells grown in suspension. As these cells require harvesting by centrifugation, the volume of PBS used during collection can dramatically affect the results generated with 33A12E10 and can potentially confound the study of this higher molecular weight c-Myc. We caution users to rigorously validate this antibody for cross-reactivity under their experimental conditions.
Authorship
Acknowledgments: The authors thank Mushui Dai and members of the Sears laboratory for helpful discussion.
This work was supported by the Tartar Trust Fellowship (D.C.T), the Oregon Health & Science University Training Program in Molecular Hematology (T32HL007781, D.C.T.), a Leukemia & Lymphoma Scholar Award (R.C.S.); and R01 CA129040 (R.C.S.).
Contribution: D.C.T., J.R.E.-P., and X.Z. performed laboratory work; and D.C.T. and R.C.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosalie C. Sears, Department of Molecular and Medical Genetics, Oregon Health & Science University, 3181 SW Sam Jackson Park Rd, Portland, OR 97239; e-mail: searsr@ohsu.edu.
References
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal