Abstract
Mouse CD8α+ dendritic cells (DCs) in lymphoid organs and CD103+ CD11b− DCs in nonlymphoid tissues share phenotypic and functional similarities, as well as a unique shared developmental dependence on the transcription factor Batf3. Human DCs resembling mouse CD8α+ DCs in phenotype and function have been identified in human blood, spleen, and tonsil. However, it is not clear whether such cells are also present in human nonlymphoid organs, and their equivalence to mouse CD8α+ DC has recently been questioned. Furthermore, the identification of “CD8α+ DC-like” cells across different tissues and species remains problematic because of the lack of a unique marker that can be used to unambiguously define lineage members. Here we show that mouse CD8α+ DCs and CD103+ CD11b− DCs can be defined by shared high expression of DNGR-1 (CLEC9A). We further show that DNGR-1 uniquely marks a CD11b− human DC population present in both lymphoid and nonlymphoid tissues of humans and humanized mice. Finally, we demonstrate that knockdown of Batf3 selectively prevents the development of DNGR-1+ human DCs in vitro. Thus, high expression of DNGR-1 specifically and universally identifies a unique DC subset in mouse and humans. Evolutionarily conserved Batf3 dependence justifies classification of DNGR-1hi DCs as a distinct DC lineage.
Introduction
Dendritic cells (DCs) comprise a heterogeneous group of leukocytes with functions in both innate and adaptive immunity. In mouse, rat, and human, multiple subtypes of DCs can be defined in lymphoid and nonlymphoid tissues and in blood on the basis of differential expression of selected markers, as well as quantitative differences in antigen processing ability and in production of specific cytokines after stimulation.1-9 However, the relationship between different DC subsets in different tissues and the equivalence of DC subtypes across species has been difficult to establish and remains controversial. Furthermore, the increased refinement of flow cytometric approaches, allowing cross-correlation of an ever-expanding number of surface markers, has led to a marked increase in the number of putative DC subsets that can be defined phenotypically. For that reason, recent efforts have been directed toward supplementing phenotypic characterization with ontogenetic criteria, as well as using comparative analysis of global gene expression patterns, to define related DC subtypes across tissues and species.6,10
The use of ontogenetic relationships has been successful in formally establishing the relatedness of mouse CD8α+ DCs from lymphoid tissues and CD103+ CD11b− DCs from nonlymphoid tissues. CD8α+ DCs were originally identified as a discrete population of DCs that expressed low levels of CD11b and SIRPα, high levels of CD8α, CD24, DEC-205, and DNGR-1 (CLEC9A), and variable levels of CD207 (langerin) and CD103.11 CD8α-expressing DCs have not generally been found outside mouse lymphoid tissues. However, mouse nonlymphoid tissues include a population of CD11b− SIRPα− DCs that expresses CD103, CD24, and CD207. These cells are additionally found in lymph nodes, where they are thought to have immigrated from nonlymphoid tissues via the afferent lymph.11 The phenotypic resemblance as well as strong dependence on Flt3L for their development led to the proposition that CD8α+ DCs resident in lymphoid tissues and CD103+ CD11b− DCs from nonlymphoid tissues and lymph nodes are indeed closely related.12 Consistent with that notion, both types of DCs have a similar transcriptome13 and uniquely require the transcription factors IRF8,12,14 Id2,12,15 and Batf316,17 for their development. The dependence on Batf3 is particularly remarkable as that transcription factor is expressed by other DC subtypes yet appears dispensable for their differentiation. Indeed, Batf3-deficient mice, particularly on a 129S6/SvEv background, lack CD8α+ DCs from lymphoid tissues and CD103+ CD11b− DCs from nonlymphoid tissues and lymph nodes while retaining other DC types.16-18 Thus, CD103+ CD11b− DCs and CD8α+ can be argued to constitute an ontogenetically distinct lineage of Batf3-dependent “CD8α+-like DCs.”
In addition to their common ontogenetic origin, CD8α+-like DCs also share a superior ability to cross-present exogenous antigens to CD8+ T cells and play a key role in priming of CTL responses against some viruses and tumors.11,16,17 However, identification of CD8α+-like DCs in mice remains problematic and requires judicious use of flow cytometric markers. For example, CD103 levels on these cells can vary depending on the cytokine environment,19-21 and loss of CD103 staining can be misinterpreted as absence of the cells in question.18,22 In addition, CD103 is highly expressed by a prominent population of CD11b+ DCs in gut and mesenteric lymph nodes (MLNs) that are Batf3-independent and unrelated to the CD8α+-like DC lineage.16 Thus, there is at present a dearth of cell surface markers shared between CD8α+ DCs and CD103+ CD11b− DCs that can be used to unambiguously identify the family of CD8α+-like DCs across mouse tissues.
The identification of CD8α+-like DCs in humans has been similarly problematic. We and others have recently described a discrete population of DCs in human spleen, blood, and tonsil that can variably be defined as DNGR-1+, BDCA3+, or XCR-1+ and that resemble mouse lymphoid tissue CD8α+ DCs in expression of selected markers and functional properties.23-26 On the basis of such correlative analyses, it has been proposed that these cells constitute the long-sought human equivalents of CD8α+ DCs.27 However, this notion has been put in doubt recently by the observation that some of the supposedly characteristic functions of these cells are shared by other human DC types. For example, a comprehensive comparison of blood and spleen human DCs suggests that BDCA3+ DCs do not display a higher cross-presentation capability than other human DC subsets, such as BDCA1+ DCs.28 Moreover, the ontogenetic equivalence of BDCA3+ DCs and mouse CD8α+ DCs has been questioned by the recent report that patients harboring an autosomal dominant mutation in IRF8 display a loss of BDCA-1+ DCs but not BDCA3+ DCs in peripheral blood.29 Finally, there is at present no characterized equivalent of CD103+ CD11b− DCs in human nonlymphoid tissues. As such, the existence of a Batf3-dependent lineage of CD8α+-like DCs in human lymphoid and nonlymphoid tissues remains unproven.
To address these issues, we compared mouse and human DCs across lymphoid and nonlymphoid tissues and tested their Batf3 dependence, as well as their ability to be systematically identified using a limited set of markers. Here we show that high expression of DNGR-1 and low expression of CD11b universally and unambiguously mark mouse and human CD8α+-like DCs in different tissues. We further show that the development of the human cells is Batf3-dependent, as in the mouse. Thus, a Batf3-dependent family of CD8α+-like DCs is found across species and can be identified uniquely and consistently by high DNGR-1 expression.
Methods
Human tissue
Umbilical cord blood (CB) was collected from mothers attending the Royal London Hospital, London, United Kingdom, after informed consent in accordance with the Declaration of Helsinki through a protocol approved by the East London and City Research Ethics Committee. Mononuclear cells obtained by Ficoll density centrifugation and ammonium chloride red cell lysis were depleted of lineage marker-positive cells using the StemStep system (StemCell Technologies) to generate lineage-negative (Lin−) hematopoietic stem cells (HSCs)/hematopoietic progenitor cells (HPCs). Healthy terminal ileum and MLNs were obtained from patients undergoing cystectomy with neo-bladder reconstruction (n = 11). All tissues were obtained with informed patient consent and in accordance with local ethical approval from the Regional Ethics committee in Lund, Sweden.
Mice and humanized mice
Animal protocols were approved by the London Research Institute Ethics Committee and United Kingdom Home Office (Animal Scientific Procedures Act 1986). C57BL/6 (B6) and Batf3−/− (C57BL/6 background18 ) and NOD/SCID/IL-2Rγ-null (NSG) mice were bred at the London Research Institute. NSG mice 8 to 12 weeks of age were sublethally irradiated (3.75 Gy) up to 24 hours before intravenous injection of 100 000 human Lin− CB cells. Mice were analyzed 7 to 8 weeks after reconstitution. For lentivirus (LV) infections, the Lin− cells were prestimulated for 4 to 6 hours with 150 ng/mL SCF, 150 ng/mL Flt3L, 20 ng/mL G-CSF, 10 ng/mL IL-6, and 20 ng/mL TPO, and infected overnight at a multiplicity of infection of 30 before transplantation.
LV production
Synthetic oligonucleotides corresponding to the targeting sequences were cloned into the CS-RfA-EG lentivector (a kind gift from Dr A. Miyawaki, Riken Tsukuba Institute). The targeting sequences were as follows: sh-CTRL, ACGCTGAGTACTTCGAAAT (targeting luciferase) or GCTCCAAGGTGTACGTGAA (targeting red fluorescent protein); sh1-Batf3, GCTCAGAGAAGTCGGAAGA; sh2-Batf3: GCACTGAAGGAGCACGAGA. LV were produced as previously described.30
In vitro–differentiated DCs
Human Lin− CB cells were differentiated into DCs using a 2-step protocol, as described previously,26 except that round-bottom well plates were used during the differentiation culture. For LV infection, 2.5 × 105 amplified cells were plated and cultured as for the differentiation step. After 1 day, medium was removed and a small volume of LV was added (final multiplicity of infection of 30) and cells were incubated for 2 hours before adding back medium and continuing the differentiation culture. Human BMDCs were generated by culturing total bone marrow from humanized mice at 5 × 105 cells/mL in the same conditions as the differentiation culture used for the CB cells.
Flow cytometry and cell sorting
Anti–human antibodies, anti–HLA-DR (L243), anti–BDCA-3/CD141 (AD5-14H12), anti-CD11c (B-ly6), anti-CD14 (MϕP9), anti-CD16 (3G8), anti-CD19 (SJ25C1), anti-CD20 (L27), and anti-CD56 (B159) were purchased from BD Biosciences. Anti-CD3 (OKT3), anti-CD103 (B-Ly7), anti-CD11b (ICRF44), and isotype-matched control antibodies were purchased from eBioscience. Anti–mouse antibodies, anti-CD11c (HL3), anti-CD45.2 (104), anti-CD103 (M290), and anti-CD11b (M1/70) antibodies were all purchased from BD Biosciences. Anti-CD207 antibody (929F3) was from Dendritics, and the anti-CD45 (30-F11) was from BioLegend. Anti-CD16/CD32 (93), anti–MHC class II (M5/114), and anti-CD8α antibodies were purchased from eBioscience. The anti–human DNGR-1 (8F9) and anti–mouse DNGR-1 (1F6) were described previously.31 Antibodies were tested for staining against appropriate negative and positive controls.
Human and mouse cells were preincubated on ice with mouse serum (Jackson ImmunoResearch Laboratories) plus purified IgG2a (BD Biosciences), or anti–mouse CD16/CD32 (93; eBiosciences), respectively, to block Fc receptors, and then stained with appropriate antibody combinations. Analysis was performed on an LSRII (BD Biosciences) or FACSAria (BD Biosciences) flow cytometer. Doublets and dead cells were excluded by a combination of scatter gating and 4,6-diamidino-2-phenylindole exclusion (for unfixed cells) or a LIVE/DEAD Fixable Violet Dead-Cell Stain Kit (Life Technologies; for fixed cells; used for some analyses of tissues from humanized mice). Analysis was performed using FlowJo Version 9.4.7 software (TreeStar). For cell sorting (FACSAria), cells from humanized mice were stained and live Lin− (CD3/14/16/19/20/56) HLA-DR+ cells negative for mouse CD45 were sorted into the indicated subsets.
Fluorescence microscopy
Endogenous peroxidase activity of acetone-fixed human ileal cryosections (7 μm) was blocked for 10 minutes with 0.5% H202. Sections were then incubated with donkey serum (10%) in PBS-Tween 20 (0.05%; Sigma-Aldrich) and avidin-biotin blocking kit (Vector Laboratories). Tissue was then stained with mouse anti–human CD11c (10 μg/mL, clone 3.9, eBioscience) for 45 minutes followed by DyLight-649 labeled donkey anti–mouse antibody (Jackson ImmunoResearch Laboratories). After washing and blocking with 10% mouse serum for 30 minutes, sections were stained with biotinylated mouse anti–human DNGR-1 (3 μg/mL) and FITC-conjugated mouse anti–human HLA-DR (1.25 μg/mL, LN3, eBioscience) in the presence of 10% mouse serum for 45 minutes. Slides were visualized using a biotinyl-tyramide signal amplification kit (PerkinElmer Life) with Alexa-555–conjugated streptavidin and Alexa-488–conjugated rabbit anti-FITC (Invitrogen). Images were acquired with an Axiovert 200M microscope equipped with a 20× 0.8 Plan-Apochromat air objective and Hamamatsu Ocra-ER camera (Carl Zeiss MicroImaging and Volocity Version 5.2.1 software; Improvision; PerkinElmer Life).
DC isolation
Mouse and human DCs were isolated from lymphoid and nonlymphoid organs as previously described.12,32 In brief, the organs were first cut in small pieces and incubated with a mixture of collagenase (type II for the spleen and lymph nodes or type IV for kidney and lung; Worthington Biochemical) and of DNase (Sigma-Aldrich). After red blood cell lysis (Sigma-Aldrich), total single cell suspensions were stained for analysis.
RNA isolation and quantitative RT-PCR
RNA isolation, cDNA preparation, and quantitative RT-PCR were as described previously.26 Levels of mRNA for the specific gene being measured were divided by those for GAPDH (normalized expression).
Statistical analyses
All statistical analyses were performed using Prism Version 5 software (GraphPad Software).
Results
Mouse lymph nodes contain migratory DNGR-1+ CD103+ DCs
Like other secondary lymphoid tissues, skin-draining lymph nodes contain resident DCs with high expression of CD11c and intermediate expression of MHC class II (Figure 1A top left panel, gate I). Phenotypic subdivision of resident DCs into CD8α+ and CD11b+ DCs (Figure 1A top middle panel, gate A and B) showed that only the CD8α+ subset stained for DNGR-1, as reported for spleen DCs31,33 (Figure 1A top right panel, gates A and B). Unlike spleen, lymph nodes also contain an additional population of migratory DC (mig-DC) that can be distinguished from resident DCs by their lower expression of CD11c and higher expression of MHC class II (Figure 1A top left panel, gate II). Mig-DC include the tissue-derived CD103+ CD11b− and CD103− CD11b+ subsets (Figure 1A bottom middle panel, gates C and D). As shown in Figure 1A, CD103+ CD11b− mig-DCs also expressed high levels of DNGR-1 (bottom right panel, gate C). In contrast, DNGR-1 expression was completely absent from CD103− CD11b+ DCs (Figure 1A bottom right panel, gate D). When CD207 was additionally analyzed, DNGR-1+ mig-DCs were clearly CD207+ (Figure 1B), even though not all CD207+ DCs expressed DNGR-1: a population of CD207+ CD103− CD11b+ DCs, probably corresponding to migratory Langerhans cells,34 did not express DNGR-1 (Figure 1B blue gate), consistent with the fact that anti–DNGR-1 does not stain Langerhans cells in epidermis (D.S. and C.R.e.S., unpublished observations, April 2007). When the 3 markers (CD103, CD207, and DNGR-1) were analyzed simultaneously, there was a perfect correlation between expression of CD103 and DNGR-1 and all double-positive cells were CD11b− (Figure 1B red gate). Batf3-independent CD103+ CD11b+ mig-DCs in MLN did not express DNGR-1 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In sum, in mouse lymph nodes, high expression of DNGR-1 is found exclusively on resident CD8α+ DCs and on migratory CD103+ CD11b− DCs, which together compose the Batf3-dependent CD8α+-like DC lineage.
Mouse lymph nodes contain resident and migratory DNGR-1+ CD8α+-like DCs. (A) Flow cytometric analysis of cutaneous draining lymph node cell suspensions from B6 mouse. Live cells were analyzed for the expression of CD11c versus MHCII (top right panel). Two populations were defined (gate I, CD11chigh MHCIIinter tissue-resident DCs; gate II, CD11cinter to high MHCIIhigh migratory DCs) and analyzed for the expression of, respectively, CD8 versus CD11b (for gate I), or CD103 versus CD11b (for gate II, top left panels). Four populations were defined (gate A, CD8+, CD11b−; gate B, CD8−, CD11b+; gate C, CD103+, CD11b−; and gate D, CD103−, CD11b+) and analyzed for the expression of DNGR-1 (red) or for an isotype control antibody (blue). (B) CD11cinter to high MHCIIhigh migratory DCs were analyzed for the expression of CD207 versus DNGR-1 (top left panel). Two populations were defined (blue gate, CD207+, DNGR-1−; and red gate, CD207+, DNGR-1+) and analyzed for the expression of CD103 versus CD11b. Migratory DCs were analyzed also for the expression of CD103 versus DNGR-1 (bottom left panel). Two populations were defined (blue gate, CD103−, DNGR-1−; and red gate, CD103+, DNGR-1+) and analyzed for the expression of CD207 and CD11b. Numbers indicate percentage of cells in each of the indicated gates. Arrows indicate gating strategy. (A-B) Data are representative of 2 independent experiments.
Mouse lymph nodes contain resident and migratory DNGR-1+ CD8α+-like DCs. (A) Flow cytometric analysis of cutaneous draining lymph node cell suspensions from B6 mouse. Live cells were analyzed for the expression of CD11c versus MHCII (top right panel). Two populations were defined (gate I, CD11chigh MHCIIinter tissue-resident DCs; gate II, CD11cinter to high MHCIIhigh migratory DCs) and analyzed for the expression of, respectively, CD8 versus CD11b (for gate I), or CD103 versus CD11b (for gate II, top left panels). Four populations were defined (gate A, CD8+, CD11b−; gate B, CD8−, CD11b+; gate C, CD103+, CD11b−; and gate D, CD103−, CD11b+) and analyzed for the expression of DNGR-1 (red) or for an isotype control antibody (blue). (B) CD11cinter to high MHCIIhigh migratory DCs were analyzed for the expression of CD207 versus DNGR-1 (top left panel). Two populations were defined (blue gate, CD207+, DNGR-1−; and red gate, CD207+, DNGR-1+) and analyzed for the expression of CD103 versus CD11b. Migratory DCs were analyzed also for the expression of CD103 versus DNGR-1 (bottom left panel). Two populations were defined (blue gate, CD103−, DNGR-1−; and red gate, CD103+, DNGR-1+) and analyzed for the expression of CD207 and CD11b. Numbers indicate percentage of cells in each of the indicated gates. Arrows indicate gating strategy. (A-B) Data are representative of 2 independent experiments.
Batf3-dependent DNGR-1+ CD103+ CD11b− DCs are found in mouse nonlymphoid tissues
To investigate DNGR-1 expression by mouse DCs in peripheral tissues, we analyzed mouse kidney (Figure 2A) and lung (Figure 2B). Kidney DCs were defined as CD45+ leukocytes expressing high levels of CD11c and MHC class II (Figure 2A top left) and were subdivided into CD103+ CD11b− DCs (Figure 2A top middle panel, gate A) and CD103− CD11b+ DCs (Figure 2A top middle panel, gate B).12 Consistent with the data obtained in lymph nodes, only the CD103+ CD11b− kidney DCs expressed DNGR-1 (Figure 2A top panels). Similarly, lung DCs (defined as CD45+ CD11c+ MHCII+ cells bearing low levels of autofluorescence [autolow]) only showed DNGR-1 staining of the CD103+ CD11b− DC subset, as recently reported35 (Figure 2B top panels). No staining was detected in DCs from kidneys or lungs of Batf3−/− mice, in agreement with the absence of CD103+ CD11b− DCs in those animals (Figure 2A-B bottom panels). Thus, DNGR-1 specifically marks Batf3-dependent CD103+ CD11b− mouse DCs in nonlymphoid tissues.
DNGR-1+ CD103+ CD11b− DCs are found in mouse nonlymphoid tissues. (A) CD11c+ MHCII+ CD45+ live kidney cells from Batf3-sufficient mice (B6; top left panel) or Batf3−/− mice (bottom left panel) were analyzed as in Figure 1 for the expression of CD103 versus CD11b. For the Batf3+/+ mice, 2 populations were defined: gate A, CD103+, CD11b−; and gate B, CD103−, CD11b+. For the Batf3−/− mice, only 1 DC gate was defined encompassing gates A and B. Cells within these gates were analyzed for staining with anti–DNGR-1 (black) or an isotype-matched control antibody (grey). (B) CD11c+ MHCII+ CD45+ autolow live lung cells were analyzed as described for the kidney. Numbers indicate percentage of cells in each of the indicated gates. Arrows indicate gating strategy. (A-B) Data are representative of 2 independent experiments.
DNGR-1+ CD103+ CD11b− DCs are found in mouse nonlymphoid tissues. (A) CD11c+ MHCII+ CD45+ live kidney cells from Batf3-sufficient mice (B6; top left panel) or Batf3−/− mice (bottom left panel) were analyzed as in Figure 1 for the expression of CD103 versus CD11b. For the Batf3+/+ mice, 2 populations were defined: gate A, CD103+, CD11b−; and gate B, CD103−, CD11b+. For the Batf3−/− mice, only 1 DC gate was defined encompassing gates A and B. Cells within these gates were analyzed for staining with anti–DNGR-1 (black) or an isotype-matched control antibody (grey). (B) CD11c+ MHCII+ CD45+ autolow live lung cells were analyzed as described for the kidney. Numbers indicate percentage of cells in each of the indicated gates. Arrows indicate gating strategy. (A-B) Data are representative of 2 independent experiments.
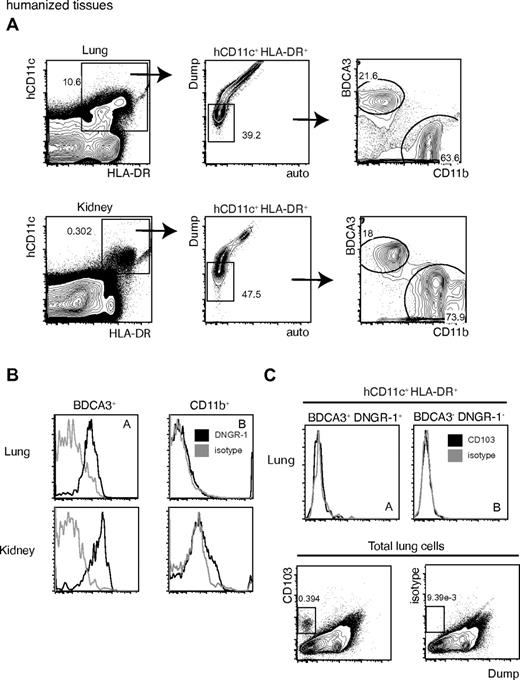
Human DNGR-1+ BDCA3+ DCs are found in nonlymphoid tissues of humanized mice
We have previously reported that DNGR-1 marks a human DC population in spleens of humans and in spleens of humanized mice that resembles murine spleen CD8α+ DCs in phenotype and function.26 Given the results in the preceding section showing that DNGR-1 additionally marks nonlymphoid tissue Batf3-dependent mouse DCs, we analyzed several nonlymphoid organs from humanized mice for the presence of human DNGR-1+ DCs. Dual expression of human CD11c (hCD11c) and HLA-DR was used to identify human DC in single cell suspensions from lungs (Figure 3A top panels) and kidneys (Figure 3A bottom panels) of mice transplanted with human CB HSCs/HPCs (Figure 3A left panels). To refine our analysis, we used one empty channel to exclude autofluorescent cells and another channel (called “dump”) to exclude any cells that stained with human lymphoid cell-lineage markers (CD3/CD19/CD56), or that expressed mouse CD45 or that failed to exclude a cell viability fluorescent dye (Figure 3A middle panels). Using this approach, we could find 2 prominent subsets of human DCs in both tissues that were clearly defined by mutually exclusive expression of BDCA3 and CD11b (Figure 3A right panels). Notably, DNGR-1 staining was found in the BDCA3+ DC subset but not in the CD11b+ DCs (Figure 3B), reminiscent of the situation in the mouse where DNGR-1 expression is similarly restricted to the CD11b− subset (see preceding section). Interestingly, CD103 was not detected on either DNGR-1+ BDCA3+ or DNGR-1− BDCA3− DCs (Figure 3C top panel) despite the fact that the anti-CD103 mAb could clearly mark other human cell types in the same preparation (Figure 3C bottom panel). In sum, the nonlymphoid tissues of humanized mice contain 2 discrete populations of human DCs that can be defined as DNGR-1+ BDCA3+ CD11b− or DNGR-1− BDCA3− CD11b+ DCs, the former of which probably represents the human equivalent of mouse CD103+ CD11b− DCs.
DNGR-1+ BDCA3+ DCs are found in nonlymphoid tissues of humanized mice. (A) hCD11c+ HLA-DR+ lung (top left panel) or kidney (bottom left panel) cells were analyzed as shown for the expression of BDCA3 versus CD11b (right panel), after having gated out Dump+ auto+ cells (middle panels). Two populations were defined: gate I, BDCA3+, CD11b−; and gate II, BDCA3−, CD11b+. The Dump channel contains the following stainings grouped together: a Lin cocktail (anti–human CD3/CD19/CD56), an anti–mouse CD45, and a viability marker. (B) Lung (top panels) and kidney (bottom panel) BDCA3+ (left panels) or CD11b+ (right panels) cells, as defined in panel A by gates I and II, were analyzed for the expression of DNGR-1 (black) or for an isotype control Ab (grey). (C) hCD11c+ HLA-DR+ BDCA3+ lung cells, as gated in panel A, expressing DNGR-1+ (top left) or not (top right) were analyzed for CD103 expression (black) or for an isotype control antibody (grey). As an internal control, total lung cells were also analyzed for these markers versus the Dump channel (bottom). Numbers indicate percentage of cells in each of the indicated gates. Arrows show gating strategy. (A-B) Data are representative of at least 3 independent experiments.
DNGR-1+ BDCA3+ DCs are found in nonlymphoid tissues of humanized mice. (A) hCD11c+ HLA-DR+ lung (top left panel) or kidney (bottom left panel) cells were analyzed as shown for the expression of BDCA3 versus CD11b (right panel), after having gated out Dump+ auto+ cells (middle panels). Two populations were defined: gate I, BDCA3+, CD11b−; and gate II, BDCA3−, CD11b+. The Dump channel contains the following stainings grouped together: a Lin cocktail (anti–human CD3/CD19/CD56), an anti–mouse CD45, and a viability marker. (B) Lung (top panels) and kidney (bottom panel) BDCA3+ (left panels) or CD11b+ (right panels) cells, as defined in panel A by gates I and II, were analyzed for the expression of DNGR-1 (black) or for an isotype control Ab (grey). (C) hCD11c+ HLA-DR+ BDCA3+ lung cells, as gated in panel A, expressing DNGR-1+ (top left) or not (top right) were analyzed for CD103 expression (black) or for an isotype control antibody (grey). As an internal control, total lung cells were also analyzed for these markers versus the Dump channel (bottom). Numbers indicate percentage of cells in each of the indicated gates. Arrows show gating strategy. (A-B) Data are representative of at least 3 independent experiments.
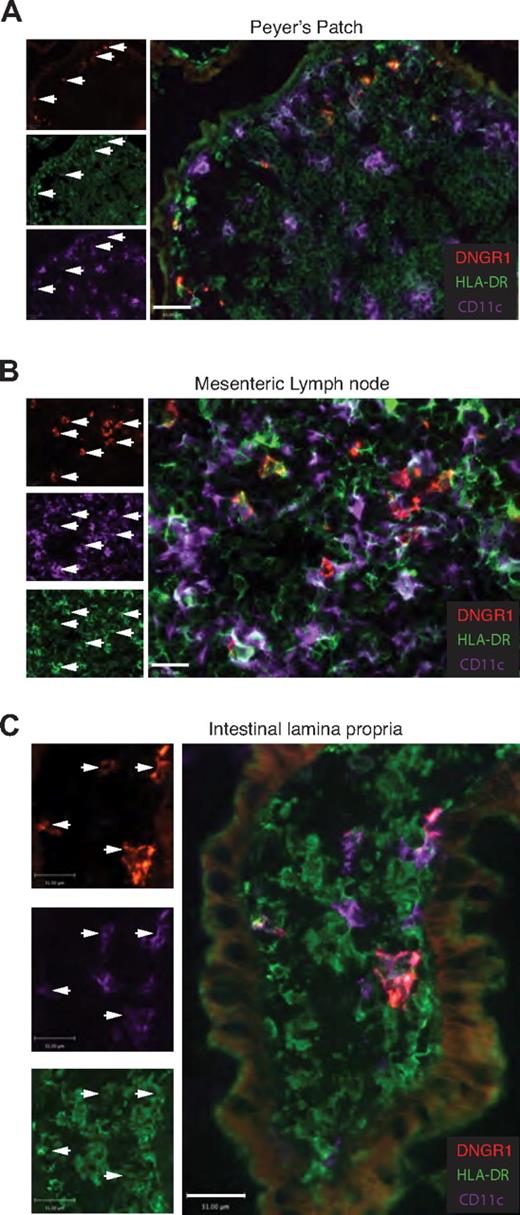
Human DNGR-1+ DCs are found in human small intestine lamina propria and in gut-associated lymphoid tissue
To extend these results to human tissues, we analyzed samples of human small intestine, gut-associated lymphoid tissue, and MLNs. Because of scarcity of tissue, analysis was restricted to staining of frozen tissue sections, which permitted cross-correlation of a more limited set of markers compared with flow cytometry. Nevertheless, as shown in Figure 4A-B, this approach allowed identification of discrete DNGR-1+ cells in the subepithelial dome of human Peyer patch and the T-cell areas of MLNs (Figure 4A-B). DNGR-1+ cells coexpressed HLA-DR and CD11c but, as expected, only a fraction of CD11c+ HLA-DR+ DCs labeled with anti–DNGR-1 (Figure 4A-B arrows). Notably, DNGR-1+ CD11c+ HLA-DR+ DCs were also readily observed within the small intestinal lamina propria (Figure 4C). Therefore, DNGR-1+ DCs are found in both human lymphoid and nonlymphoid tissues.
DNGR-1+ DCs are found in human small intestinal lamina propria, MLN, and Peyer patches. Three-color immunofluorescence staining for DNGR1, CD11c, and HLA-DR in cryosections of human intestinal tissue. The arrowheads indicate DNGR-1+ CD11c+ HLA-DR+ cells in (A) Peyer patches (n = 3), (B) MLNs (n = 3), and (C) intestinal lamina propria (n = 6). (A) Scale bar represents 63 μm. (B-C) Scale bars represent 31 μm
DNGR-1+ DCs are found in human small intestinal lamina propria, MLN, and Peyer patches. Three-color immunofluorescence staining for DNGR1, CD11c, and HLA-DR in cryosections of human intestinal tissue. The arrowheads indicate DNGR-1+ CD11c+ HLA-DR+ cells in (A) Peyer patches (n = 3), (B) MLNs (n = 3), and (C) intestinal lamina propria (n = 6). (A) Scale bar represents 63 μm. (B-C) Scale bars represent 31 μm
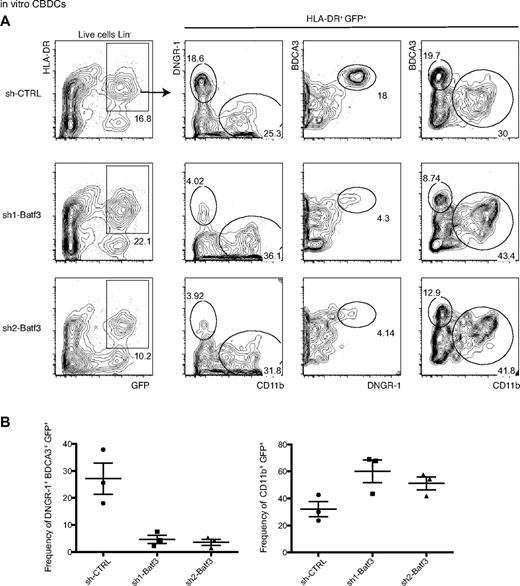
In vitro generated DNGR-1+ BDCA3+ CB-derived DCs are Batf3-dependent
On the basis of the results presented thus far, DNGR-1 would appear to act as a marker for the CD8α+-like DC family across species and across tissues. This family is currently best defined in the mouse on the basis of Batf3 dependence, but this has not been established in the human. The ability to grow DNGR-1+ BDCA3+ DCs from CB HSCs/HPCs in a 2-step culture (CB-derived DCs [CBDCs])26 allowed us to investigate their ontogenetic equivalence with the putative mouse counterparts. First, we screened 10 Batf3-silencing shRNA LV vector constructs bearing a puromycin selection marker and found 2 that appeared to selectively impair the development of puromycin-resistant DNGR-1+ DCs (data not shown). We subcloned these 2 Batf3-silencing hairpins, as well as 2 scrambled sequence controls (CTRL), into another lentiviral vector, in which the puromycin resistance locus is replaced with a gene encoding GFP, which acts as a marker of LV-transduced cells and their progeny. We produced shRNA LVs, used them to infect CBDC differentiation cultures, and monitored the development of the DNGR-1+ BDCA3+ subset. The control viruses (sh-CTRL) were used to optimize the transduction and culture protocols. We found that LV infection at 24 hours after initiation of the differentiation culture reliably allowed generation of GFP+ DCs, identified as live cells that lacked lineage-specific markers, and expressed both GFP and HLA-DR (Figure 5A left column). Among these DCs we could identify a discrete DNGR-1+ BDCA3+ CD11b− subset (Figure 5A top panel; only 1 sh-CTRL depicted). Notably, when cultures infected with control versus Batf3-silencing shRNA LV were compared, we found a significant and reproducible decrease in the frequency of DNGR-1+ BDCA3+ CD11b− DCs among the GFP+ DCs in cultures infected with the latter (Figure 5A middle and bottom panels, sh1-Batf3 and sh2-Batf3; B left panel). Other uncharacterized DNGR-1− BDCA3− CD11b+ DCs that developed in the cultures were not impaired by Batf3 silencing; consequently, their frequency was increased compared with the controls (Figure 5A-B right panel). As an internal control, all cultures contained DNGR-1+ BDCA3+ DCs in the GFP− fraction (data not shown). In addition, LV-mediated Batf3 knockdown did not affect the growth of M-CSF– or IL-34–derived human macrophage-like cells (data not shown). In conclusion, shRNA-mediated knockdown of Batf3 selectively impairs the generation of DNGR-1+ BDCA3+ DCs in vitro, establishing that these human cells are ontogenetically similar to their putative mouse counterparts.
In vitro generated DNGR-1+ BDCA3+ CBDCs are Batf3-dependent. CBDCs were generated and infected with different sh-RNA LVs as described in “LV production.” (A) Lin− (CD3/14/16/19/20/56) live cells were analyzed for the expression of HLA-DR and GFP. HLR-DR+ GFP+ cells were analyzed for the expression DNGR-1 versus CD11b, BDCA3 versus DNGR-1, and BDCA3 versus CD11b. (B) Frequency of DNGR-1+ BDCA3+ GFP+ (left panel) and of CD11b+ GFP+ cells (right panel). (A-B) Data are representative of multiple CBDC cultures with 2 independent pools of CB-derived HSCs/HPCs.
In vitro generated DNGR-1+ BDCA3+ CBDCs are Batf3-dependent. CBDCs were generated and infected with different sh-RNA LVs as described in “LV production.” (A) Lin− (CD3/14/16/19/20/56) live cells were analyzed for the expression of HLA-DR and GFP. HLR-DR+ GFP+ cells were analyzed for the expression DNGR-1 versus CD11b, BDCA3 versus DNGR-1, and BDCA3 versus CD11b. (B) Frequency of DNGR-1+ BDCA3+ GFP+ (left panel) and of CD11b+ GFP+ cells (right panel). (A-B) Data are representative of multiple CBDC cultures with 2 independent pools of CB-derived HSCs/HPCs.
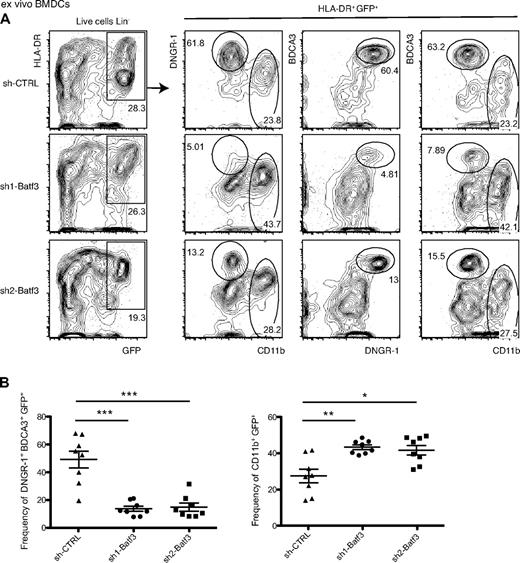
Batf3 silencing is not sufficient to impair DNGR-1+ BDCA3+ DC development in humanized mice
To assess the Batf3 dependence of human DNGR-1+ BDCA3+ DC in vivo, we generated humanized mice with human HSCs/HPCs that had been transduced with the different shRNA LV. To assess the stability of transduction in vivo, we isolated bone marrow from these mice at 7 to 8 weeks after transplantation and used it as a source of progenitors for in vitro DC differentiation (pseudo-“bone marrow–derived DCs” [BMDCs]). We found that this approach allowed generation of a large fraction of GFP+ DCs from all mice, indicating that transduced human HSCs/HPCs had successfully engrafted and retained DC-generating potential. Notably, there was a marked decrease in the frequency of DNGR-1+ BDCA3+ DCs among GFP+ cells grown from bone marrow of mice that received human HSCs/HPCs transduced with either of the 2 shRNA LV targeting Batf3 displayed compared with the shRNA CTRL LV (Figure 6). These results indicate that Batf3 silencing can be maintained in vivo by transduction of human HSCs/HPCs with shRNA LV and further confirm that Batf3 is required to grow DNGR-1+ BDCA3+ DC from human progenitor cells, in this case obtained from the bone marrow of humanized mice.
Ex vivo generated DNGR-1+ BDCA3+ from humanized mice are Batf3-dependent. (A) BMDCs were generated as described in “In vitro–differentiated DCs” from bone marrow of humanized mice and analyzed as in Figure 5. (B) Frequency of DNGR-1+ BDCA3+ GFP+ (left panel) and of CD11b+ GFP+ cells (right panel). Numbers indicate percentage of cells in each of the indicated gates. (A) One representative experiment of 2 is shown, with 4 mice for each sh-RNA. (B) Data are mean ± SEM of 2 independent experiments. *P < .05 (Mann-Whitney test). **P < .01 (Mann-Whitney test). ***P < .001 (Mann-Whitney test). sh-CTRL indicates control.
Ex vivo generated DNGR-1+ BDCA3+ from humanized mice are Batf3-dependent. (A) BMDCs were generated as described in “In vitro–differentiated DCs” from bone marrow of humanized mice and analyzed as in Figure 5. (B) Frequency of DNGR-1+ BDCA3+ GFP+ (left panel) and of CD11b+ GFP+ cells (right panel). Numbers indicate percentage of cells in each of the indicated gates. (A) One representative experiment of 2 is shown, with 4 mice for each sh-RNA. (B) Data are mean ± SEM of 2 independent experiments. *P < .05 (Mann-Whitney test). **P < .01 (Mann-Whitney test). ***P < .001 (Mann-Whitney test). sh-CTRL indicates control.
Given these results, we fully expected to see a decrease in the frequency of DNGR-1+ BDCA3+ DCs among the GFP+ DCs in spleen, lung, and kidney of humanized mice transplanted with Batf3-silenced human HSCs/HPCs. Surprisingly, this was not the case (Figure 7). Although there was a slight trend toward reduction (Figure 7A), pooled analyses of individual animals revealed no statistically significant difference in the overall frequency of GFP+ DNGR-1+ BDCA3+ DCs between mice reconstituted with HSCs/HPCs transduced with sh1-Batf3 LV versus sh-CTRL LV (Figure 7B; and data not shown). To ensure that this was not because of in vivo selection of cells resistant to Batf3 knockdown, we sorted human DCs from the spleens of these animals and confirmed that there was a significant reduction of Batf3 mRNA in the GFP+ (Figure 7C) but not the GFP− (data not shown) fraction. Furthermore, we confirmed that the knockdown was functional by showing that the bone marrow of these mice still failed to generate GFP+ DNGR-1+ BDCA3+ DCs on ex vivo culture (Figure 6). We conclude that Batf3 knockdown prevents development of DNGR-1+ BDCA3+ in vitro but that this can be bypassed in our humanized mouse model.
Batf3 silencing is not sufficient to impair DNGR-1+ BDCA3+ DC development in humanized mice. (A) CD11c+ HLA-DR+ live cells (same as in Figure 3) were analyzed for the expression of BDCA3 versus CD11b (top panel) among the GFP+ cells. Two populations were identified: gate I, BDCA3+, CD11b−; and gate II, BDCA3−, CD11b+. Numbers indicate percentage of cells in each of the indicated gates. (B) Frequency of BDCA3+ GFP+ (top panel) and of CD11b+ GFP+ cells (bottom panel) is shown. (C) Normalized expression of Batf3 within Lin− (CD3/14/16/19/20/56) HLA-DR+ GFP+ live cells, purified from the spleen of the humanized mice. (A,C) Data are representative of 2 independent experiments, with 4 mice for each sh-RNA. (B) Data are mean ± SEM of 2 independent experiments. *P < .05 (Mann-Whitney test). sh-CTRL indicates control; Sp, spleen; Lg, lung; and K, kidney.
Batf3 silencing is not sufficient to impair DNGR-1+ BDCA3+ DC development in humanized mice. (A) CD11c+ HLA-DR+ live cells (same as in Figure 3) were analyzed for the expression of BDCA3 versus CD11b (top panel) among the GFP+ cells. Two populations were identified: gate I, BDCA3+, CD11b−; and gate II, BDCA3−, CD11b+. Numbers indicate percentage of cells in each of the indicated gates. (B) Frequency of BDCA3+ GFP+ (top panel) and of CD11b+ GFP+ cells (bottom panel) is shown. (C) Normalized expression of Batf3 within Lin− (CD3/14/16/19/20/56) HLA-DR+ GFP+ live cells, purified from the spleen of the humanized mice. (A,C) Data are representative of 2 independent experiments, with 4 mice for each sh-RNA. (B) Data are mean ± SEM of 2 independent experiments. *P < .05 (Mann-Whitney test). sh-CTRL indicates control; Sp, spleen; Lg, lung; and K, kidney.
Discussion
DCs are potent antigen-presenting cells involved in priming and regulating adaptive immune responses.36 In addition, DCs can also play a key role in innate immunity, acting as a potent source of cytokines, such as IFN-α/β or IL-12.11,30 In recent years, the heterogeneity of the DC system has become increasingly apparent.2,6,37 An attempt to make sense of DC heterogeneity and to map equivalent DC subsets across species has led to efforts to define the ontogeny and transcriptome of phenotypically distinct DC types in the hope that they might help define related DC subtypes.5,7,8,10 This approach has led to the definition of a Batf3-dependent CD8α+-like DC family in mouse, which encompasses the CD8α+ DCs of lymphoid tissues and the CD103+ CD11b− DCs found in nonlymphoid organs. An XCR1+ DNGR-1+ BDCA-3+ subset of DCs in human blood, spleen, lymph nodes, and tonsil was recently described as the putative equivalent of mouse lymphoid tissue CD8α+ DCs on the basis of phenotypic and functional similarity.23-26 However, whether related human DCs also exist in nonlymphoid organs has not been established, and the actual equivalence of DNGR-1+ human DCs and mouse CD8α+ DCs awaits demonstration that the former, like the latter, depend on Batf3 for their development. Furthermore, a common marker for this lineage across species has not been reported, leading to the ambiguous use of “CD8α+-like DCs” to refer to the global Batf3-dependent family of mouse DCs, as well as their putative counterparts in human lymphoid organs and blood. Here, we demonstrate that high DNGR-1 expression identifies the “CD8α+-like DC” family in both lymphoid and nonlymphoid tissues of mouse, human, and humanized mouse and show that human DNGR-1+ DCs require Batf3 for their development in vitro. We propose that high expression of DNGR-1 uniquely marks a Batf3-dependent DC lineage across species, which might therefore be referred to as “DNGR-1hi DCs.”
Expression of DNGR-1 in mouse spleen has previously been shown to be restricted to CD8α+ DCs and pDCs.31,33 The latter express lower levels of the receptor.31,33 Expression of DNGR-1 on splenic DCs is stable, and the gene is not induced on other splenocytes, even after LPS injection into mice.38 Here we extended those observations to show that lymph node CD8α+ DCs also express high levels of DNGR-1 but that, in addition, the receptor also marks the Batf3-dependent extended CD8α+ DC family, including the CD103+ CD11b− DCs in nonlymphoid tissues and their progeny among lymph node immigrant DCs. As such, DNGR-1 appears similar to XCR-1, which was very recently also shown to selectively identify the Batf3-dependent DC lineage in mice.13
We previously exploited DNGR-1 as a marker to identify human DCs in human spleen and in the spleens of humanized mice that resemble mouse CD8α+ DCs in phenotype and function.26 Here, we show that DNGR-1 additionally marks a CD11c+ HLA-DR+ cell population in human lymph nodes and Peyer patches. These cells represent conventional DCs, as human pDCs do not express DNGR-126,31,33,39 or CD11c.40,41 In Peyer patches, which have no afferent lymph supply, DNGR-1+ DCs probably correspond to the CD11b− resident DCs in the sub-dome area that were first reported in the mouse and are thought to represent the equivalent of spleen CD8α+ DCs.42 In contrast, in MLN, DNGR-1 staining is expected to mark the human equivalents of both the resident and the migratory CD11b− DCs. The origin of the latter remains unclear. It has been argued that CD11b− DCs identified by flow cytometry in murine intestinal lamina propria cell preparations arise from contaminating gut-associated lymphoid tissues, including Peyer patches and isolated lymphoid follicles.43 In turn, this would suggest that DNGR-1+ CD11b− mig-DCs found in MLN immigrate from these sites. However, it remains possible that some CD11b− DCs reside in the intestinal lamina propria, an issue that cannot be resolved by flow cytometry alone. Our immunohistochemical analysis demonstrated that DNGR-1+ DCs are readily identified within the intestinal lamina propria, situated within the villous core. Thus, similar to other nonlymphoid tissues, gut lamina propria contains a discrete population of DNGR-1+ CD11b− DCs. These DCs may serve to sample luminal antigens or apoptotic epithelial cells and may give rise to the migratory CD8α+-like DCs that subsequently drain through the afferent lymph to the MLN.44
The presence of mouse or human DNGR-1+ DCs in multiple lymphoid and nonlymphoid organs of mice, humans, and humanized mice suggests that DNGR-1 marks a discrete population of DCs that is conserved across species and tissues. Notably, we were able to show that Batf3 is required for growing human DNGR-1+ DC populations in vitro, formally establishing that Batf3 is required in human as in mouse. This finding helps cement the equivalence between DNGR-1+ human DCs and mouse CD8α+ DCs. However, Batf3 dependence could not be established in vivo as DNGR-1+ DCs were seen to develop normally in humanized mice from progenitors transduced with Batf3 shRNA LV. Because of scarcity of material, we did not test the functionality of the cells and therefore cannot establish whether they were functionally impaired, for example, in their cross-presenting activity. Nevertheless, their presence in normal frequencies may indicate that the level of transcription factor knockdown achieved with shRNA LV is not sufficient to block development of human DNGR-1+ DCs in vivo, even though it suffices in vitro. An alternative explanation is that Batf3 dependence may be reduced or bypassed altogether in humanized mice under the influence of factors absent from our DC differentiation cultures. It is notable that, even in mouse, CD8α+ DCs in some organs can develop independently of Batf3, a fact that became especially apparent when Batf3-deficient mice, originally made in a 129/SvEv background, were backcrossed to the C57BL/6 strain.18 In addition to Batf3, CD8α+ DC development in the mouse depends on NFIL3/E4BP4,45 a transcription factor that acts upstream of Batf3, as well as on Id215 and IRF8.14 Interestingly, BDCA-3+ DCs are still found in the blood of 2 patients bearing an autosomal dominant mutation in IRF8.29 Thus, it is possible that partial abrogation of IRF8 function caused by the human mutation, which probably generates a dominant negative IRF8, as well as partial Batf3 knockdown, as shown here, is not sufficient to abrogate development of DNGR-1+ DCs in vivo, even if it does so under more stringent conditions in vitro. It would be interesting to combine knockdown of Batf3 with knockdown of NFIL3/E4BP4, IRF8, and Id2 to see if any such combination abrogates DNGR-1+ DC development in humanized mice.
Humanized mice offer an unprecedented tool for studying the human immune system in vivo. However, the development of human DCs in such mice may be limited by particular mouse factors that do not cross-react with human receptors. For example, human GM-CSF receptor cannot respond to mouse GM-CSF.46 As GM-CSF induces CD103 expression by mouse CD8α+-like DCs,19-21 this could explain why we do not detect CD103 on DNGR-1+ human DCs in humanized mice. Flow cytometric analysis of the same cells isolated from human tissues will be necessary to establish whether CD103 can act as a marker for the CD8α+-like DC lineage in humans. Similarly, whether DNGR-1+ human DCs can express CD8α in some situations remains to be determined. Independently of additional markers, our current data clearly identify DNGR-1 as a specific and universal identifier of the Batf3-dependent CD8α+ DC family across mouse and human. The ability to use anti–DNGR-1 to identify these cells in situ thus opens the door to further characterizing these cells in different human tissues, both in normal and pathologic conditions. Finally, the identification of a common marker and the ontogenetic verification of a common DC lineage in both mice and humans underscores the usefulness of mouse models of DC function and reinforces the notion that antigen targeting to DNGR-1, which has been so successful in mice,31,33,38,47,48 may yet serve as a useful strategy for immune modulation in humans.49
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the FACS Laboratory and Equipment Park of the London Research Institute for technical support and Biologic Resources for care of the animals used in these studies; Drs S. Gudjonsson, G. Baseckas, and O. Patchan (Skåne University Hospital, Malmö, Sweden) for collecting human ileum; and members of the Immunobiology Laboratory, Cancer Research UK for advice and discussions.
This work was supported by Cancer Research UK (core London Research Institute support, C.R.e.S. and D.B.) and the Swedish National Health Service (clinical research grant, W.W.A.). L.F.P. was supported by a Marie-Curie Intra-European Fellowship. Y.R. was supported by a clinical Leukemia Lymphoma Research Fund fellowship. C.R.e.S. was additionally supported by Fondation Bettencourt-Schueller and an ERC Advanced Researcher grant.
Authorship
Contribution: L.F.P., Y.R., H.U.-H., and B.U.S. performed the experiments and analyzed data; L.F.P., Y.R., H.U.-H., D.S., W.W.A., D.B., and C.R.e.S. designed the experiments and analyzed data; L.F.P., Y.R., H.U.-H., W.W.A., D.B., and C.R.e.S. wrote the paper; and U.K.H., L.F.M., and K.M.M. provided materials and reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.F.P. is Center for Infection and Immunity of Lille, Pasteur Institute of Lille, Lille, France.
Correspondence: Caetano Reis e Sousa, Immunobiology Laboratory, Cancer Research United Kingdom, London Research Institute, Lincoln's Inn Fields Laboratories, 44 Lincoln's Inn Fields, London WC2A 3LY, United Kingdom; e-mail: caetano@cancer.org.uk.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal