Abstract
Tumor progression usually proceeds through several sequential stages, any of which could be targets for interrupting the progression process if one understood these steps at the molecular level. We extracted nascent plasma cell tumor (PCT) cells from within inflammatory oil granulomas (OG) isolated from IP pristane-injected BALB/c.iMycEμ mice at 5 different time points during tumor progression. We used laser capture microdissection to collect incipient PCT cells and analyzed their global gene expression on Affymetrix Mouse Genome 430A microarrays. Two independent studies were performed with different sets of mice. Analysis of the expression data used ANOVA and Bayesian estimation of temporal regulation. Genetic pathway analysis was performed using MetaCore (GeneGo) and IPA (Ingenuity). The gene expression profiles of PCT samples and those of undissected OG samples from adjacent sections showed that different genes and pathways were mobilized in the tumor cells during tumor progression, compared with their stroma. Our analysis implicated several genetic pathways in PCT progression, including biphasic (up- and then down-regulation) of the Spp1/osteopontin-dependent network and up-regulation of mRNA translation/protein synthesis. The latter led to a biologic validation study that showed that the AMPK-activating diabetes drug, metformin, was a potent specific PCT inhibitor in vitro.
Introduction
Carcinogenesis consists of a series of genetic events from initiation though progression to complete malignant transformation.1 Tumor progression could potentially be interrupted at any of its critical pathways if we understood this process at the molecular level. This understanding is difficult to achieve in studies of human cancer patients because of their different genetic backgrounds, lifestyles, and environment. In addition, it is rare to have the opportunity to follow the natural progression of an individual's malignancy by examining the tumor sequentially over a period of time without the effects of therapeutic interventions.
Plasma cell (PC) neoplasms, including multiple myeloma (MM), extraosseous plasmacytoma and monoclonal Ig deposition disease, occur in many mammalian species. MM is the chief PC malignancy in humans, and, despite recent advances in treatment, is extremely hard to cure. MM is always preceded by a precursor lesion designated monoclonal gammopathy of undetermined significance (MGUS).2,3 Global gene expression profiling (GEP) has identified major differences between MGUS and normal PCs, yet, no clear distinctions have emerged between MGUS and MM.4,5
Oil-induced peritoneal PC tumors (PCT) in BALB/c mice provide a valuable experimental model system for studying the progression of PC malignancies such as MM. First, both MM and PCT are PC neoplasms with considerable latent periods, and both acquire genetic aberrations during neoplastic cell transformation. Second, chromosomal translocations that deregulate oncogenes on juxtaposition to Ig heavy chain (Igh) enhancers are thought to initiate both human MM and mouse PCT. However, the translocations in MM are highly variable,6 whereas those in PCT are essentially uniform,7 a Myc-activating T(12;15)(Igh/Myc) in ∼ 85% of cases.8 Third, gene-targeted BALB/c.iMycEμ mice, which contain a mouse Myc (c-myc) cDNA gene inserted head-to-head into the Igh locus, recapitulate the naturally occurring T(12;15) initiation step, so that all B lymphocytes contain activated Myc and are setup for subsequent completion of the PCT transformation program.9,10 The iMycEμ transgene accelerates PCT formation, typically producing tumors with a 100% incidence within 3 months of pristane injection11 compared with a 65% tumor incidence and longer latency (mean tumor onset of 7 months) in parental BALB/c mice. Studying the genetic mechanisms involved in the progression of PCT in the BALB/c.iMycEμ mouse model could offer insights into ways to interrupt this process in both species.
Reasoning that the genetic events involved in the iMycEμ-driven progression of PCT are likely to be reflected in changes in the cell's transcriptome, we used GEP using microarray hybridization of RNA from peritoneal granulomas in BALB/c.iMycEμ mice isolated at 5 different times after IP pristane administration: early (7 days), 3 intermediary time points (17, 33 and 46 or 49 days) and late (104 or 105 days). The last is well beyond the 80-day postpristane time in which transplantable (ie, fully transformed) tumor cells have been found.11 Because pristane granulomas contain not only cancer cells but also large numbers of other types of cells, we used laser microdissection (LMD) to collect nascent tumor cells12 followed by isolation of RNA from these cells (generating PC samples) and analysis of their GEP.13 Because tumor development can be heavily influenced by the tumor microenvironment,14,15 we also prepared RNA from entire frozen sections of oil granulomas (OG) without performing LMD (generating OG samples, containing both tumor cells and surrounding stromal cells). Statistical analysis and interpretation of multiple changes in gene expression over time is challenging, so we supplemented traditional ANOVA with a tool specifically designed for analysis of time-dependent samples: Baysean estimation of temporal regulation (BETR).16 Genes identified as varying over the time course of our study were analyzed using MetaCore (GeneGo) and Ingenuity Pathway Analysis (IPA; Ingenuity Systems) to identify pathways that might be involved in tumor progression.
Methods
Mice, tumor induction, and tissue processing
The BALB/c.iMycEμ mouse was developed by gene targeting a his6-tagged murine Myc cDNA just 5′ of the Eμ enhancer of the Igh locus9 and backcrossing the transgene onto strain BALB/c. Two independent cohorts of BALB/c.iMycEμ mice were aged at least 8 weeks and injected IP with 0.5 mL of pristane (2,6,10,14-tetramethylpentadecane) oil. In the initial study (study 1), mice were maintained for 7, 17, 33, 49 and 104 days before collecting their mesenteric OGs. For the validation and confirmation study (study 2), the same timetable was used, except that granulomas were collected on days 46 and 105 instead of days 49 and 104, respectively. The mesenteric tissue was immediately frozen in OCT (Tissue-Tek), cryosectioned at 8 μm onto PEN membrane slides (Leica), and stored at −80° until use. Another portion of the OG tissue was fixed in formalin/acetic acid/ethanol and embedded in paraffin for H&E staining or immunolabeling using Avidin-biotin Vector Red techniques with an antiserum to immunoglobulin κ light chains (Southern Biotechnology Associates) to detect Ig production, a hallmark of PCs.8 All research involving mice was approved by the National Cancer Institute (NCI) Animal Care Committee.
Staining and LMD of tissue sections
Frozen sections were fixed in 70% ethanol for 20 seconds, stained to identify PCs using methyl green pyronin (MGP; Sigma-Aldrich)17 that also contained 300 U/mL of an RNase inhibitor (Superasin; Ambion) for 2 minutes with slight agitation, destained by a quick dip in RNAase-free water followed by 2 changes of 100% ethanol (3 and 30 seconds, respectively), dehydrated in 2 changes of xylene for 2 minutes each, and allowed to air dry. MGP-positive cells were microdissected on a Leica LMD6000 into 0.5 mL of RNase-free tube caps containing 50 μL of RLT extraction buffer (QIAGEN). Total RNA from both laser-microdissected MGP-positive cells (PC samples) and whole, undissected tissue (OG samples) sections was isolated using Allprep DNA/RNA Micro kits (QIAGEN) according to the manufacturer's protocol. RNA integrity was assessed via both NanoDrop 2000 Spectrophotometer (ThermoScientific) and Agilent's RNA 6000 Pico Labchip and 2100 Bioanalyzer. Samples showing degradation were not used in this investigation.
RNA amplification, hybridization, and microarray scanning
RNA samples underwent 2 rounds of linear amplification using Affymetrix's GeneChip 2-cycle cDNA Synthesis Kit according to the manufacturer's protocol. The quality of the amplified and fragmented RNAs was assessed by the RNA 6000 Nano Labchip/2100 Bioanalyzer (Agilent). Twenty to 50 ng of total intact RNA was labeled, fragmented, and biotin-labeled cDNA was hybridized to Affymetrix Mouse Genome 430A 2.0 arrays. Hybridized arrays were stained with PE-streptavidin (Molecular Probes), scanned with an Affymetrix Genechip 3000 Scanner at the default settings and analyzed with Affymetrix Microarray Analysis Suite Version 5.0. All microarray data are available at the Gene Expression Omnibus (GEO) database under accession number GSE34078.
Microarray analysis
Microarray data were normalized,18 visualized, and analyzed using the following methods:
ANOVA.
Affymetrix “.cel” files were imported into Partek GS Version 6.5 using the standard import tool with RMA normalization. Study 1 and study 2 files were imported and analyzed separately: categorized by date, and subjected to multi-way ANOVA. False discovery rate (FDR) correction was applied using the step-up method. Probe sets that met a FDR of ≤ 0.05 were selected for further study.
BETR.
BETR16 analysis was done in R using the BETR package from Bioconductor. The samples were imported using ReadAffy from the Affymetrix package and normalized using RMA. Replicate and day information was applied to the set, and BETR analysis was performed with an α cutoff of 0.01.
Visualization.
Probe sets that met the criteria above were hierarchically clustered with the gene mean values shifted to zero and the standard deviation adjusted to 1, using Partek GS Version 6.5, which was also used to create Venn diagrams.
Pathway analysis.
Genetic pathways were evaluated using the MetaCore Analytical Suite (GeneGo Inc). Enrichment analysis consisted of mapping gene IDs of the dataset onto gene IDs in entities of built-in functional ontologies represented in MetaCore by pathway maps and networks. Additional network associations were generated using IPA (Ingenuity Systems), which links specific genes to a database of gene functions gleaned from the biomedical research literature.
Cell culture and cell proliferation assay
Mouse PCT cell lines, XRPC24, TEPC2027, and ABPC20,8 were grown in RPMI 1640 (Gibco; Invitrogen) plus 10% FBS (Benchmark), 2mM l-glutamine (Biosource), 1mM sodium pyruvate (Gibco; Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco; Invitrogen), and 55μM β-mercaptoethanol (Gibco; Invitrogen). The mouse BM-derived stromal line S1019 was kindly provided by Kenneth Dorshkind (University of California–Los Angeles) and grown in DMEM (Gibco; Invitrogen), supplemented as in the previous sentence. Inhibition of cell proliferation by metformin (Sigma-Aldrich) was assayed over a 4-day period on an initial suspension of 6000 to 10 000 cells in 96-well plates. Cell proliferation was determined by measuring fluorescence of Cell Titer-Blue (Promega) at a wavelength of 590 nm using a FLUOstar Omega (BMG LABTECH) plate reader.
Results
LMD affords in situ collection of fresh tumor precursors to study PCT progression
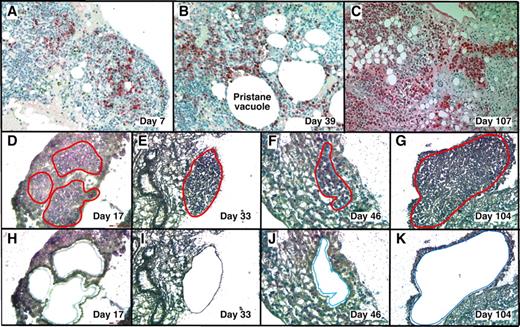
With LMD we harvested PCs undergoing malignant transformation in their native microenvironment (Figure 1) from cryosections of OCT-embedded pristane granulomas. MGP staining detected RNA-rich cells such as PCs12 in frozen sections of OG. This rapid method minimized the likelihood of mRNA loss by continued processing or RNAase damage, and it correlated well with the widely used method of identifying PCs in formalin-fixed paraffin-embedded tissue sections using Ab to Ig (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
LMD isolatesPCs undergoing tumor progression in inflammatory peritoneal granulomas. (A-C) Low-power images (originally using 10× objective) of peritoneal OGs containing PCs analyzed at early (day 7), middle (day 39), and late (day 107) times after IP injection of 0.5 mL of pristane oil. Paraffin-embedded formalin-fixed tissues were sectioned and stained with Ab against mouse Ig κ light chains to identify PCs (red), and then counterstained with hematoxylin to identify stromal cells (blue). (D-K) Microscopic images of MGP-stained frozen sections of pristane granulomas at early (day 17), middle (days 33 and 46), and late (day 104) times after IP pristane before (D-G) and after (H-K) collection of PCs using LMD. The contours of the tissue fragments targeted by the laser beam are indicated by red lines in panels D through G. The actual tissue fragments collected by LMD came from the holes seen in panels H through K. Microscopic slides were read using an Olympus BX-51 Light Microscope equipped with UPLSAPO objectives (Olympus) of the following magnifications and numerical apertures: 10×, 0.4 (panels A-C); 20×, 0.75 (panels D,H,F,J); 40×, 0.95 (panels E,I,G,K). The imaging medium was air. The light temperature of the microscope bulb varied between 3000 and 3400 K. Images were acquired with the help of a DP2 digital camera (Olympus) and DP2-BSW imaging software (Olympus), saved as TIF (tagged image file) data files, and enhanced—with respect to brightness, contrast, and color balance—using the Adobe Photoshop CS2 Version 9.0.2 software (Adobe Systems Inc).
LMD isolatesPCs undergoing tumor progression in inflammatory peritoneal granulomas. (A-C) Low-power images (originally using 10× objective) of peritoneal OGs containing PCs analyzed at early (day 7), middle (day 39), and late (day 107) times after IP injection of 0.5 mL of pristane oil. Paraffin-embedded formalin-fixed tissues were sectioned and stained with Ab against mouse Ig κ light chains to identify PCs (red), and then counterstained with hematoxylin to identify stromal cells (blue). (D-K) Microscopic images of MGP-stained frozen sections of pristane granulomas at early (day 17), middle (days 33 and 46), and late (day 104) times after IP pristane before (D-G) and after (H-K) collection of PCs using LMD. The contours of the tissue fragments targeted by the laser beam are indicated by red lines in panels D through G. The actual tissue fragments collected by LMD came from the holes seen in panels H through K. Microscopic slides were read using an Olympus BX-51 Light Microscope equipped with UPLSAPO objectives (Olympus) of the following magnifications and numerical apertures: 10×, 0.4 (panels A-C); 20×, 0.75 (panels D,H,F,J); 40×, 0.95 (panels E,I,G,K). The imaging medium was air. The light temperature of the microscope bulb varied between 3000 and 3400 K. Images were acquired with the help of a DP2 digital camera (Olympus) and DP2-BSW imaging software (Olympus), saved as TIF (tagged image file) data files, and enhanced—with respect to brightness, contrast, and color balance—using the Adobe Photoshop CS2 Version 9.0.2 software (Adobe Systems Inc).
Analysis of gene expression changes in the initial study (study 1)
The GEP of sections of undissected OGs and laser microdissected PCs undergoing tumor formation were first evaluated in study 1 by unsupervised cluster analysis. We used statistical criteria that were rather permissive (ie, initial variance filtration that considers only the genes with the top 25% variance, having at least 1 sample > 4-fold higher expressed than average expression), to yield a broad, inclusive overview of differentially expressed genes in the PC (cancer) cells and the stroma (OG). We found very different expression profiles for PC and OG RNAs (supplemental Figure 2). Regardless of sampling time points, the OG samples clustered together, separately from the clustered PC samples. This finding demonstrates the usefulness of dissecting the tumor precursor cells away from the stroma to determine the genes involved in tumor progression, both in nascent PCT cells and in their microenvironment.
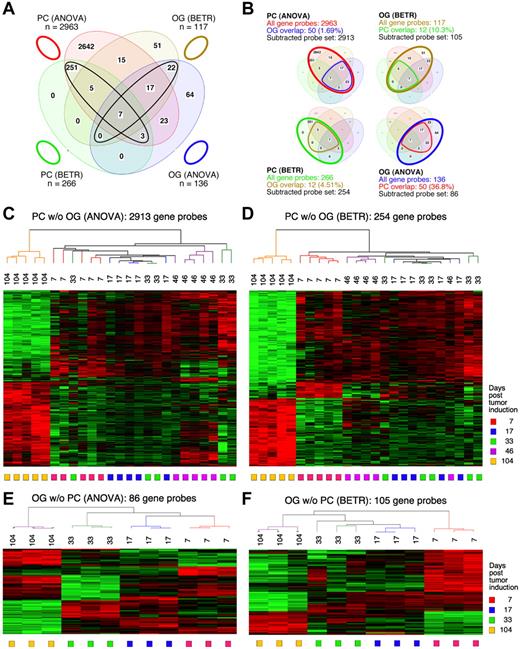
To identify the genes that showed significant differences of expression in the cancer cells themselves (the PC samples) during tumor progression, we performed both a 5-group ANOVA and a 5-group BETR analysis of the PC samples. The ANOVA analysis revealed 2963 Affymetrix gene sets (∼ 2516 unique genes, because several genes are represented by more than one gene set) that showed significant differences at a 1% FDR (P < .01; supplemental Table 1). BETR analysis revealed 266 genes with significant variability within the samples at a P < .05 (supplemental Table 2).
For the analysis of GEP in the OG samples, we performed analogous 5-group multi-way ANOVA and BETR analyses. When we applied the same cutoff value (P < .01), only 136 gene sets showed significant differences via ANOVA (supplemental Table 3) and 117 via BETR (supplemental Table 4). These findings are summarized in the Venn diagram in Figure 2A. They suggest that tumor progression involves fewer changes in gene expression in the cells surrounding the tumor cells than in the tumor cells themselves. Note that 100% of the 266 BETR-revealed gene probes in the PC sample are included in the ANOVA gene set (indicated by a black ellipse pointing up to the left), whereas only 46 of 117 (39.3%) BETR gene probes in the OG sample are included in the ANOVA gene set (black ellipse pointing up to the right). This suggests that BETR is better suited for the analysis of LMD samples (PC) that are more uniformly PCs than for analysis of OG samples because BETR requires a degree of cell uniformity at all time points, which may not be satisfied in OG samples, being composed of a varying mix of reactive cells in the inflammatory granulomas.
Gene expression changes during tumor progression instudy1 (initial study). (A) A Venn diagram showing the number of gene sets found to be significantly variable by ANOVA and BETR analysis of PC and OG samples. Overlap of genes from the different analyses and different samples is indicated. The list of gene sets revealed by ANOVA and BETR analysis of both gene sets, before removal of overlapping genes, can be found in supplemental Tables 1 through 4. (B) Venn diagrams that illustrate the gene subtraction approach used in this study. The ANOVA-revealed PC gene set (top left) consists of 2963 gene probes (red oval), 50 of which also occurred in the ANOVA-revealed OG gene set (blue cone). Thus, the corrected or subtracted ANOVA-PC gene set consists of 2913 gene probes. The same algorithm applies to the BETR-revealed PC gene set (bottom left), the BETR-revealed OG gene set (top right) and the ANOVA-revealed OG gene set (bottom right). (C-F) Heat maps of changing genes during tumor progression in LMD-isolated PC samples or nondissected, whole-tissue OG samples after removal of gene sets, as described in panel B. Differentially regulated genes identified by either ANOVA or BETR analyses were subjected to unsupervised hierarchical clustering. Up- and down-regulated genes are indicated by red and green, respectively. Time of tumor induction is indicated by colored squares and dendrograms below and above heat maps, respectively. N.B.: The day 46 OG samples are not included in the analyses shown here, because only 2 day 46 samples proved to be of suitable quality for analysis, and BETR analysis requires all groups to contain the same number of samples (3, in the case of samples for days 7, 17, 33 and 104).
Gene expression changes during tumor progression instudy1 (initial study). (A) A Venn diagram showing the number of gene sets found to be significantly variable by ANOVA and BETR analysis of PC and OG samples. Overlap of genes from the different analyses and different samples is indicated. The list of gene sets revealed by ANOVA and BETR analysis of both gene sets, before removal of overlapping genes, can be found in supplemental Tables 1 through 4. (B) Venn diagrams that illustrate the gene subtraction approach used in this study. The ANOVA-revealed PC gene set (top left) consists of 2963 gene probes (red oval), 50 of which also occurred in the ANOVA-revealed OG gene set (blue cone). Thus, the corrected or subtracted ANOVA-PC gene set consists of 2913 gene probes. The same algorithm applies to the BETR-revealed PC gene set (bottom left), the BETR-revealed OG gene set (top right) and the ANOVA-revealed OG gene set (bottom right). (C-F) Heat maps of changing genes during tumor progression in LMD-isolated PC samples or nondissected, whole-tissue OG samples after removal of gene sets, as described in panel B. Differentially regulated genes identified by either ANOVA or BETR analyses were subjected to unsupervised hierarchical clustering. Up- and down-regulated genes are indicated by red and green, respectively. Time of tumor induction is indicated by colored squares and dendrograms below and above heat maps, respectively. N.B.: The day 46 OG samples are not included in the analyses shown here, because only 2 day 46 samples proved to be of suitable quality for analysis, and BETR analysis requires all groups to contain the same number of samples (3, in the case of samples for days 7, 17, 33 and 104).
Differentially expressed genes in PC and OG samples were not exclusive but exhibited partial overlap. This is depicted in the Venn diagrams presented in Figure 2B, which indicate the degree of overlap between genes that changed their expression in nascent tumor cells (PC samples) and those that changed in the surrounding stroma (OG samples). These overlapping genes are graphically marked by the cone-shaped lines within the ovals that represent the total numbers of gene sets. Specifically, the PC-ANOVA gene probe set (n = 2963) contained 50 OG-ANOVA gene probes, an overlap of 1.69% (Figure 2B top left); the PC-BETR set (n = 266) contained 12 (4.51%) OG-BETR probes (Figure 2B bottom left); the OG-ANOVA set (n = 136) included 50 (36.8%) PC-ANOVA probes (Figure 2B bottom right); and the OG-BETR gene probe set (n = 117) included 12 (10.3%) PC-BETR probes (Figure 2B top right). Because we were interested in distinguishing tumor progression–dependent changes in gene expression in tumor precursor from those in bystander cells, we removed the genes that overlapped between analyses of the PC and OG samples before subjecting the samples to hierarchical cluster analysis.
Unsupervised cluster analysis of PC genes identified by ANOVA or BETR resulted in the dendrograms and heat maps presented in Figure 2C and D, respectively. Gradual changes in gene expression were evident in both heat maps, with the most dramatic difference appearing in the day 104 samples. These figures show that a large number of genes that are expressed during tumor progression are down-regulated in the fully transformed tumor cells abundant at day 104. A few of these genes can already be seen to be down-regulated in the intermediate time points, especially day 46. The heat map also shows that several genes are up-regulated in the day 104 tumor cells, some already up-regulated at day 46. Similar cluster analysis of the OG samples (Figure 2E-F) also revealed a unique heat map for the day 104 samples, but the day 7 samples, especially evident in the BETR analysis, also appeared unique. Compared with the PC samples, there was, perhaps, a more gradual transition from the early to the later pattern of gene expression evident in the intermediate time points of the OG samples.
Analysis of gene expression changes in the validation study (study 2)
Because 2-fold amplification of the tiny amounts of RNA available in each sample rendered them unsuitable for RT-PCR validation, we decided to validate the results from the initial study biologically by carrying out a completely new study. We subjected new sets of BALB/c.iMycEμ mice to the same PCT induction regimen as the mice from the first study, however, we paid greater attention to accelerating RNA processing and microarray hybridization. Specifically, OCT tissue blocks were stored at −80° for the shortest time possible; LMD was performed no more than one to 2 days after frozen sectioning; RNA was prepared immediately after sectioning; and all sample amplifications and microarray hybridizations were performed on the same day using master mixes for every reagent to ensure uniformity.
Supplemental Figure 3A shows heat maps derived from unsupervised cluster analysis of combined PC and OG samples from both studies, using ANOVA-selected genes at P < .01 after the same variance filtration. Inspection of the heat map reveals that cluster formation is driven in part by the difference of the overall GEP between the PC and OG samples from both studies. Another strong driver of sample clustering was study-specific gene expression patterns. This resulted in 2 subclusters (study 1 and study 2) within the PC samples from both studies and 2 similar subclusters of samples within the OG samples. Thus, PC and OG samples from studies 1 and 2 produced 2 nearly perfect superclusters, indicating that, regardless of sampling times and batches of mouse samples, the major differences come from the 2 different types of samples: PC versus OG. When we aligned the 2 independent PC datasets according to time points in a supervised manner (supplemental Figure 3B), the similarity of the changes in gene expression over time in both studies became apparent. We think that the 2 studies varied, in part, because of differences in the inbred BALB/c.iMycEμ mice that were still being introgressively inbred in the period between first and second experiments. Nevertheless, the 2 independent studies produced a highly comparable overall result, and the second study validated the PCT progression results of the initial study.
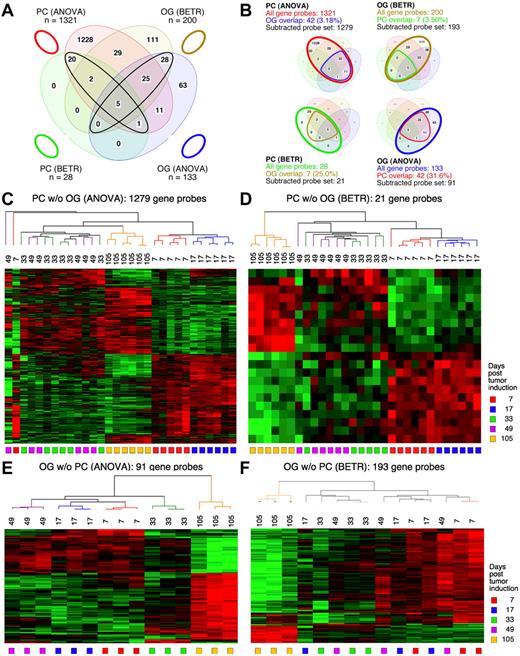
Figure 3 displays Venn diagrams, dendrograms, and heat maps of the results from study 2, analogous to those shown in Figure 2 for study 1. The improved LMD collection and handling of the PC samples resulted in a tighter analysis, with many fewer gene sets that vary among the LMD-collected samples in study 2 compared with study 1: 1321 via ANOVA (supplemental Table 5) and 28 via BETR (supplemental Table 6). The OG samples once again revealed fewer varying genes than the PC samples in ANOVA (supplemental Table 7), but not in BETR analysis (supplemental Table 8). Supplemental Table 9 summarizes the number of significantly varying genes in the 2 studies. As in study 1, all BETR-revealed gene probes in study 2 PC sample (n = 28) are also included in the ANOVA gene set, whereas only 58 of 193 (30.1%) BETR gene probes in the OG sample are included in the OG-ANOVA gene set. Likewise, as observed in study 1, the Venn diagrams that illustrate the “gene subtraction” approach for study 2 (Figure 3B) show that the highest overlap among gene probes was found in the OG-ANOVA sample: 31.6%. The results of unsupervised cluster analysis of all 4 “subtracted” gene probe sets are displayed in Figure 3C through F.
Gene expression changes during tumor progression in study 2 (validation study). Samples were analyzed as in study 1. Venn diagram showing the number of gene sets found to be significantly variable by ANOVA and BETR analysis of PC (plasma cell) and oil granuloma (OG) samples. Overlap of genes from the different analyses and different samples is indicated. The list of gene sets revealed by ANOVA and BETR analysis of both gene sets, before removal of overlapping genes, can be found in supplemental Tables 5 through 8. (B) Venn diagrams that illustrate the gene subtraction approach used in this study. The ANOVA-revealed PC gene set (top left) consists of 1321 gene probes (red oval), 42 of which also occurred in the ANOVA-revealed OG gene set (blue cone). Thus, the corrected or subtracted ANOVA-PC-gene set consists of 1279 gene probes. The same algorithm applies to the BETR-revealed PC gene set (bottom left), the BETR-revealed OG gene set (top right) and the ANOVA-revealed OG gene set (bottom right). Panels C, D, E, and F present heat maps of changing genes during tumor progression in LMD-isolated PC samples or non-dissected, whole-tissue OG samples after removal of gene sets, as described in panel B. Differently regulated genes identified by either ANOVA or BETR analyses were subjected to unsupervised hierarchical clustering. Up- and down-regulated genes are indicated by red and green, respectively. Time of tumor induction is indicated by colored squares and dendrograms below and above heat maps, respectively.
Gene expression changes during tumor progression in study 2 (validation study). Samples were analyzed as in study 1. Venn diagram showing the number of gene sets found to be significantly variable by ANOVA and BETR analysis of PC (plasma cell) and oil granuloma (OG) samples. Overlap of genes from the different analyses and different samples is indicated. The list of gene sets revealed by ANOVA and BETR analysis of both gene sets, before removal of overlapping genes, can be found in supplemental Tables 5 through 8. (B) Venn diagrams that illustrate the gene subtraction approach used in this study. The ANOVA-revealed PC gene set (top left) consists of 1321 gene probes (red oval), 42 of which also occurred in the ANOVA-revealed OG gene set (blue cone). Thus, the corrected or subtracted ANOVA-PC-gene set consists of 1279 gene probes. The same algorithm applies to the BETR-revealed PC gene set (bottom left), the BETR-revealed OG gene set (top right) and the ANOVA-revealed OG gene set (bottom right). Panels C, D, E, and F present heat maps of changing genes during tumor progression in LMD-isolated PC samples or non-dissected, whole-tissue OG samples after removal of gene sets, as described in panel B. Differently regulated genes identified by either ANOVA or BETR analyses were subjected to unsupervised hierarchical clustering. Up- and down-regulated genes are indicated by red and green, respectively. Time of tumor induction is indicated by colored squares and dendrograms below and above heat maps, respectively.
Candidate genes for intervention in tumor progression
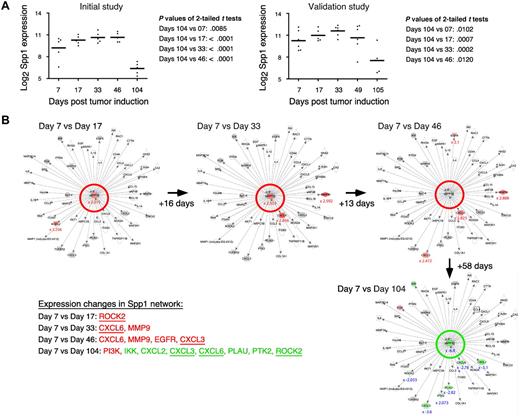
Table 1 shows a selection of PC and OG genes identified in the analyses presented in the preceding two sections. As expected for a B-lineage neoplasm that arises in a proinflammatory tissue environment, the table contains many B lymphocyte–associated genes (Ig genes, Ly6d, etc) and cancer genes involved in regulating programmed cell death (Bmi1, Bcl1a, Bcl2l1), chemokine signaling (Ccl8, Ccl9, Ccl24, Cxcl1), cell-cycle progression (Ccnb1, Ccnb2, Cdkn1b), growth factors (Igf1), inflammation (Cat, Cd163), and signal transduction (Mapk8, Socs2). Next, we asked whether the differentially expressed genes listed in Table 1 might include novel targets for drug intervention during PCT progression, for example, protein-encoding genes that are up-regulated in the course of tumor development and potentially drug-inhibitable. The time-course plots of 5 individual genes whose expression increased in PC samples (but not in OG samples) when PCs completed malignant transformation (day 104 or 105) are shown in Figure 4. These genes provide leads for new interventions specifically aimed at tumor cells. Csk encodes a tyrosine kinase that may play a role in mediating G-protein signals in the reorganization of the actin cytoskeleton in PCs,20 and, like Abl and many other kinases, Csk may be targeted by small drug inhibitors. Similarly attractive candidates are Bmi1 and Bcl-XL (encoded by Bcl2l1), for which an extensive drug development pipeline exists, and surface proteins (eg, Ly6d) that may be targeted by therapeutic Ab. The time-course plot of a gene differentially up-regulated in OG but not PC samples, Eif2ak3, suggests an intervention that may complement the genes found overexpressed in PCTs, described above, by targeting the tumor's microenvironment (supplemental Figure 4 top). Less appealing from a cancer intervention perspective, yet highly relevant for increased understanding of cancer biology, may be genes that become down-regulated when tumor precursors transition to frank neoplasia. Examples along this line are: Ccl11, Cd36, Ear1 and Rab34 in OG samples (supplemental Figure 4) and Spp1 in PC samples (Figure 5A). We focused on Spp1, because it appears in the center of the most prominent genetic network uncovered by IPA of differentially expressed PC genes, that is, “cancer-related genes,” and because Spp1 is a well-known metastatic factor.21 Spp1, also called osteopontin,22 expression increases throughout tumor progression but is dramatically down-regulated in the day 104/105 frank PCT samples. Consistent with this, several Spp1-controlled target genes exhibited high expression levels on days 7-49 but low levels on day 104/105 (Figure 5B).
Categories of genes differentially expressed
| Category . | Gene* . |
|---|---|
| Apoptosis/antiapoptosis | Bmi1,†‡Bcl1a,†‡Bcl2l1,†Dap,†Tlr4,†Tradd,†Xiap† |
| Cancer | Dpt,†Spp1,§Tgfb2,†Malat1,†Mta3† |
| Cell adhesion | Emb,†Enah,†Postn†‡ |
| Chemokines/receptors | Ccl2,†‡Ccl6,†Ccl8,†Ccl9,†Ccl11,†‡Ccl24,‡Cxcl1,†‡Cxcl5,†Cxcl12,†Cxxc5† |
| Collagens | Col3a1,†Col6a1† |
| Cyclins | Ccnb1,†Ccnb2,†Ccnd2,†Ccng2,†Cdkn1b† |
| Cytoskeleton | Gsn,†S100a10† |
| Growth factors/receptors | Csf1r,†Gpr137,†‡Igf1,†Igf2,†Igf2r,†Il2rg,†‡Jund,†Kitl,†Lifr,†‡Ndn,†Notch2,†Notch3,†Pdgfra,†Trf,†Trfc†‡ |
| Immune cells | Fcgr3,†IgA,†‡Igj,†‡IgG,†‡Igh6, Igk,†Igl,†‡Klra17,†‡Lcp1,†Ly6d,†Mpeg1,†Slamf9,†‡Tmpa,†‡Vpreb3† |
| Inflammation/macrophages | Cat,†‡Cd163,†‡Fcgr3,†‡Il1l1,†‡Ptger4,†‡Ptgs2,‡Sod1,†‡ |
| Oncogenes/tumor suppressors | Brca1,†‡Cebpd,†‡Crk,†Csk,†Dek,†Extl2,†‡Fos,‡Jund,†Mela,†‡Serpini1† |
| Proteases/inhibitors | Ctsc,†‡Klk1,†Serpini1,†Timp2† |
| Signal transduction | Dkk1,†‡Grb2z,†‡Mapk8,†‡Map3k7,†Map4k4,†Mxi1,†Ppap2a,†Ppap2b,†Prkcz,†Rb,†Smad 6,†Smad7,†Smoc1,†Socs2,†‡Tgfb2,†Tgfbr22 |
| Transcription/translation | Cited2,†‡Cebpd,†‡Egr1,†‡Eif2ak3,†Eif4e3,†Prpf19,†‡Sdpr,†Sp12 |
| Category . | Gene* . |
|---|---|
| Apoptosis/antiapoptosis | Bmi1,†‡Bcl1a,†‡Bcl2l1,†Dap,†Tlr4,†Tradd,†Xiap† |
| Cancer | Dpt,†Spp1,§Tgfb2,†Malat1,†Mta3† |
| Cell adhesion | Emb,†Enah,†Postn†‡ |
| Chemokines/receptors | Ccl2,†‡Ccl6,†Ccl8,†Ccl9,†Ccl11,†‡Ccl24,‡Cxcl1,†‡Cxcl5,†Cxcl12,†Cxxc5† |
| Collagens | Col3a1,†Col6a1† |
| Cyclins | Ccnb1,†Ccnb2,†Ccnd2,†Ccng2,†Cdkn1b† |
| Cytoskeleton | Gsn,†S100a10† |
| Growth factors/receptors | Csf1r,†Gpr137,†‡Igf1,†Igf2,†Igf2r,†Il2rg,†‡Jund,†Kitl,†Lifr,†‡Ndn,†Notch2,†Notch3,†Pdgfra,†Trf,†Trfc†‡ |
| Immune cells | Fcgr3,†IgA,†‡Igj,†‡IgG,†‡Igh6, Igk,†Igl,†‡Klra17,†‡Lcp1,†Ly6d,†Mpeg1,†Slamf9,†‡Tmpa,†‡Vpreb3† |
| Inflammation/macrophages | Cat,†‡Cd163,†‡Fcgr3,†‡Il1l1,†‡Ptger4,†‡Ptgs2,‡Sod1,†‡ |
| Oncogenes/tumor suppressors | Brca1,†‡Cebpd,†‡Crk,†Csk,†Dek,†Extl2,†‡Fos,‡Jund,†Mela,†‡Serpini1† |
| Proteases/inhibitors | Ctsc,†‡Klk1,†Serpini1,†Timp2† |
| Signal transduction | Dkk1,†‡Grb2z,†‡Mapk8,†‡Map3k7,†Map4k4,†Mxi1,†Ppap2a,†Ppap2b,†Prkcz,†Rb,†Smad 6,†Smad7,†Smoc1,†Socs2,†‡Tgfb2,†Tgfbr22 |
| Transcription/translation | Cited2,†‡Cebpd,†‡Egr1,†‡Eif2ak3,†Eif4e3,†Prpf19,†‡Sdpr,†Sp12 |
Bold type indicates genes found in study 1 or 2, using both ANOVA and BETR analysis. BETR indicates Baysean estimation of temporal regulation; PC, plasma cell; and OG, oil granuloma.
Light, normal type indicates genes found in study 1 or 2, using ANOVA.
Gene was detected in the PC samples.
Gene was detected in the OG samples.
Spp1 was detected in studies 1 and 2, using both ANOVA and BETR analysis.
Expression levels of 5 selected ANOVA-revealed PC genes are up-regulated in frank tumor cells compared with premalignant tumor precursors. Log2-transformed mean gene expression levels (horizontal lines) and individual gene expression levels (dots) at indicated times posttumor induction with pristane are plotted for study 1 (initial study) in the left column and study 2 (validation study) in the center column. The results of statistical comparisons (2-tailed t test) of mean expression levels of combined tumor precursor samples (□; days 7-46 in case of study 1 [n = 20]; days 7-49 in case of study 2 [n = 24]) and tumor samples (■; day 104 in case of study 1 [n = 5]; day 105 in case of study 2 [n = 6]) are indicated as P values in the right column. Bcl2l1 indicates BCL-2-like 1; Bmi, Bmi polycomb ring finger oncogene; Cxxc5, CXXC finger 5; Csk, c-src tyrosine kinase; and Ly6 days, lymphocyte Ag 6 complex, locus D.
Expression levels of 5 selected ANOVA-revealed PC genes are up-regulated in frank tumor cells compared with premalignant tumor precursors. Log2-transformed mean gene expression levels (horizontal lines) and individual gene expression levels (dots) at indicated times posttumor induction with pristane are plotted for study 1 (initial study) in the left column and study 2 (validation study) in the center column. The results of statistical comparisons (2-tailed t test) of mean expression levels of combined tumor precursor samples (□; days 7-46 in case of study 1 [n = 20]; days 7-49 in case of study 2 [n = 24]) and tumor samples (■; day 104 in case of study 1 [n = 5]; day 105 in case of study 2 [n = 6]) are indicated as P values in the right column. Bcl2l1 indicates BCL-2-like 1; Bmi, Bmi polycomb ring finger oncogene; Cxxc5, CXXC finger 5; Csk, c-src tyrosine kinase; and Ly6 days, lymphocyte Ag 6 complex, locus D.
Tumor progression–dependent changes are seen in Spp1 expression and in other genes in the Spp1 network. (A) The time course of the mean expression level of Spp1, secreted phosphoprotein 1 (better known as osteopontin) in the PC samples of study 1 (left) and study 2 (right). In both studies, the expression of Spp1 in frank tumor cells (days 104/105) was significantly lower (P < .05) than in tumor precursors, regardless which time of tumor induction (days 7-49) was used for t test analysis. (B) Ingenuity pathways analysis (IPA) comparisons of the Spp1-dependent gene network from study 1 on days 7 vs 17 (top row, left), days 7 vs 33 (center), days 7 vs 46 (right) and days 7 vs 104 (bottom row, right). Spp1 and Spp1-regulated target genes are in the center and periphery of network diagrams, respectively. Spp1 targets that were significantly up-regulated when the relatively high Spp1 level on day 7 exhibited further incremental increases (days 17-46) are highlighted in red to the bottom left: Rock2, Cxcl3, Cxcl6, EGFR, MMP9 (P < .001, > 3-fold differences). Spp1 targets that were significantly down-regulated in accordance with the drop in Spp1 levels on day 104 are highlighted in green (P < .001, > 3-fold differences). Note that 3 genes found to be up-regulated in earlier comparisons (Cxcl3, Cxcl6, Rock2) are now found to be significantly down-regulated. Similar results were obtained when the Spp1 network in study 2 was analyzed (not shown).
Tumor progression–dependent changes are seen in Spp1 expression and in other genes in the Spp1 network. (A) The time course of the mean expression level of Spp1, secreted phosphoprotein 1 (better known as osteopontin) in the PC samples of study 1 (left) and study 2 (right). In both studies, the expression of Spp1 in frank tumor cells (days 104/105) was significantly lower (P < .05) than in tumor precursors, regardless which time of tumor induction (days 7-49) was used for t test analysis. (B) Ingenuity pathways analysis (IPA) comparisons of the Spp1-dependent gene network from study 1 on days 7 vs 17 (top row, left), days 7 vs 33 (center), days 7 vs 46 (right) and days 7 vs 104 (bottom row, right). Spp1 and Spp1-regulated target genes are in the center and periphery of network diagrams, respectively. Spp1 targets that were significantly up-regulated when the relatively high Spp1 level on day 7 exhibited further incremental increases (days 17-46) are highlighted in red to the bottom left: Rock2, Cxcl3, Cxcl6, EGFR, MMP9 (P < .001, > 3-fold differences). Spp1 targets that were significantly down-regulated in accordance with the drop in Spp1 levels on day 104 are highlighted in green (P < .001, > 3-fold differences). Note that 3 genes found to be up-regulated in earlier comparisons (Cxcl3, Cxcl6, Rock2) are now found to be significantly down-regulated. Similar results were obtained when the Spp1 network in study 2 was analyzed (not shown).
Pathway prediction reveals PCT-intrinsic pathways of tumor progression
We used the MetaCore package to analyze the differentially expressed genes included in supplemental Tables 1 through 8. A 5% FDR (P < .05) analysis produced many pathway maps for the PC samples (n = 336) but only 7 pathway maps for the OG samples. We decided to continue the MetaCore analysis only with the PC samples, LMD-enriched for tumor precursor cells. To discover only the most significant gene maps, we increased the stringency of analysis by tightening FDR from 5% to 0.01% (P < 10−4). At this cutoff level, we detected 7 gene maps in the PC/ANOVA set of the initial study and 26 gene maps in the corresponding PC/ANOVA set of the validation study. These 33 gene maps are summarized in Table 2 and assembled according to significance levels in supplemental Figure 5. Overrepresented among these pathway maps are “Development” (n = 11), “Cell adhesion and Cytoskeletal remodeling” (n = 8), and “Immune Response” (n = 7). The PC-associated gene map with the highest P value (4.22 × 10−10) detected in the initial study, was “Development–Regulation of epithelial to mesenchymal transition (EMT)” (supplemental Figure 5B, no. 1), and a similar EMT pathway was also shown to be involved in the validation study (supplemental Figure 5A, no. 16; P = 1.74 × 10−6). EMT is often a hallmark of neoplastic transformation.23 Additional pathway maps identified were IGF-1, TGFβ, and NFκB signaling, which have previously been identified as contributors in the pathophysiology of PCTs.24-26 Novel pathways and genes heretofore not associated with PCT development in mouse, but known to play a role in other forms of tumorigenesis,27 include regulation of EIF4F and EIF2 activity during protein translation and ephrin signaling (supplemental Figure 5A, no. 12). One salient ephrin-regulated pathway involves VEGFR2 and angiogenesis and is known to drive the development of other tumors.28 Cell adhesion and cytoskeletal remodeling are understudied fields in plasma-cell neoplasia and have great potential relevance for tumor invasion and dissemination.
Summary of statistically significant GeneGo pathway maps (FDR 0.0001) discovered in ANOVA-revealed, differentially regulated genes in PC samples from study 1 and/or study 2
| Unique pathways included* . | P† . | Hits‡ . | Ratio, %§ . | SF5‖ . |
|---|---|---|---|---|
| GO category: development, n = 11#¶ | ||||
| Regulation of EMT* | 4.22 × 10−10 | 25/64 | 39.1 | B1 |
| IGF-1 receptor signaling | 2.83 × 10−8 | 14/51 | 27.4 | A5 |
| Flt3 signaling | 3.06 × 10−7 | 12/44 | 27.3 | A11 |
| FGFR signaling pathway | 2.69 × 10−6 | 12/53 | 22.6 | A22 |
| EGFR signaling pathway | 3.14 × 10−6 | 13/63 | 20.6 | A23 |
| GO category: cell adhesion and cytoskeleton remodeling, n = 8#¶ | ||||
| TGF, WNT and cytoskeletal remodeling | 4.47 × 10−8 | 31/111 | 27.9 | B2 |
| TGF, WNT and cytoskeletal remodeling | 7.69 × 10−8 | 20/111 | 18.0 | A8 |
| ECM remodeling | 1.89 × 10−7 | 19/52 | 36.5 | B3 |
| Chemokines and cell adhesion | 1.89 × 10−7 | 28/100 | 28.0 | B5 |
| Cytoskeleton remodeling | 2.98 × 10−7 | 28/102 | 27.5 | B6 |
| Cytoskeleton remodeling | 4.81 × 10−7 | 18/102 | 17.6 | A12 |
| PLAU signaling | 6.62 × 10−7 | 11/39 | 28.2 | A14 |
| GO category: immune response, n = 7#¶ | ||||
| Gastrin in inflammatory response | 5.54 × 10−9 | 17/69 | 24.6 | A2 |
| Classical complement pathway | 1.89 × 10−7 | 19/52 | 36.5 | B4 |
| Lectin-induced complement pathway | 3.62 × 10−7 | 18/49 | 36.7 | B7 |
| CD40 signaling | 6.16 × 10−7 | 14/64 | 21.9 | A13 |
| IL-7 signaling in B lymphocytes | 1.93 × 10−6 | 11/43 | 25.6 | A17 |
| GO category: others, n = 7††#¶ | ||||
| Apoptosis and survival–BAD phosphorylation | 1.80 × 10−8 | 13/42 | 31.0 | A3 |
| Translation–regulation of EIF4F activity | 4.85 × 10−8 | 14/53 | 26.4 | A6 |
| Translation–regulation of EIF2 activity | 6.94 × 10−8 | 12/39 | 30.8 | A7 |
| Some pathways of EMT in cancer cells | 1.73 × 10−6 | 12/51 | 23.5 | A16 |
| Signal transduction–AKT signaling | 1.93 × 10−6 | 11/43 | 25.6 | A18 |
| Unique pathways included* . | P† . | Hits‡ . | Ratio, %§ . | SF5‖ . |
|---|---|---|---|---|
| GO category: development, n = 11#¶ | ||||
| Regulation of EMT* | 4.22 × 10−10 | 25/64 | 39.1 | B1 |
| IGF-1 receptor signaling | 2.83 × 10−8 | 14/51 | 27.4 | A5 |
| Flt3 signaling | 3.06 × 10−7 | 12/44 | 27.3 | A11 |
| FGFR signaling pathway | 2.69 × 10−6 | 12/53 | 22.6 | A22 |
| EGFR signaling pathway | 3.14 × 10−6 | 13/63 | 20.6 | A23 |
| GO category: cell adhesion and cytoskeleton remodeling, n = 8#¶ | ||||
| TGF, WNT and cytoskeletal remodeling | 4.47 × 10−8 | 31/111 | 27.9 | B2 |
| TGF, WNT and cytoskeletal remodeling | 7.69 × 10−8 | 20/111 | 18.0 | A8 |
| ECM remodeling | 1.89 × 10−7 | 19/52 | 36.5 | B3 |
| Chemokines and cell adhesion | 1.89 × 10−7 | 28/100 | 28.0 | B5 |
| Cytoskeleton remodeling | 2.98 × 10−7 | 28/102 | 27.5 | B6 |
| Cytoskeleton remodeling | 4.81 × 10−7 | 18/102 | 17.6 | A12 |
| PLAU signaling | 6.62 × 10−7 | 11/39 | 28.2 | A14 |
| GO category: immune response, n = 7#¶ | ||||
| Gastrin in inflammatory response | 5.54 × 10−9 | 17/69 | 24.6 | A2 |
| Classical complement pathway | 1.89 × 10−7 | 19/52 | 36.5 | B4 |
| Lectin-induced complement pathway | 3.62 × 10−7 | 18/49 | 36.7 | B7 |
| CD40 signaling | 6.16 × 10−7 | 14/64 | 21.9 | A13 |
| IL-7 signaling in B lymphocytes | 1.93 × 10−6 | 11/43 | 25.6 | A17 |
| GO category: others, n = 7††#¶ | ||||
| Apoptosis and survival–BAD phosphorylation | 1.80 × 10−8 | 13/42 | 31.0 | A3 |
| Translation–regulation of EIF4F activity | 4.85 × 10−8 | 14/53 | 26.4 | A6 |
| Translation–regulation of EIF2 activity | 6.94 × 10−8 | 12/39 | 30.8 | A7 |
| Some pathways of EMT in cancer cells | 1.73 × 10−6 | 12/51 | 23.5 | A16 |
| Signal transduction–AKT signaling | 1.93 × 10−6 | 11/43 | 25.6 | A18 |
FDR indicates false discovery rate; PC, plasma cell; GO, GeneGo; EMT, epithelial to mesenchymal transition; IGF, insulin-like growth factor 1; FGFR, fibroblast growth factor receptor; EGFR, epidermal growth factor receptor; WNT, wingless-type; ECM, extracellular matrix; PLAU, plasminge activator, uroinase; BAD, BCL2-associated agonist of cell death; and NGF, naive grwoth factor.
Designation of GeneGo pathway.
Log (P), indicating the probability that the postulated GeneGo pathway is in fact not involved in the tumor sample.
Number of differentially regulated pathway genes versus total number of genes comprising the pathway.
Ratio of differentially regulated pathway genes to total number of pathway genes.
Number of pathway in panel A or panel B of supplemental Figure 5.
GeneGo category of functionally related, genetic pathway maps. Enrichment analysis scores and ranks the most relevant GeneGo pathways in a given category.
Number of unique GeneGo pathway maps discovered in a given GeneGo category.
Epithelial to mesenchymal transition.
Includes the following GO categories: Apoptosis and survival, NGF activation of NF-κB, G-protein signaling, Some pathways of EMT in cancer cells and signal transduction.
Comparison with human PC neoplasia
In a pilot study designed to relate our results for mouse PCT progression to that of human disease, we performed several computer-based comparisons among the mouse PCT and a human MM dataset from the Mayo Clinic that was publically available (http://www.broadinstitute.org/mmgp/). Our ANOVA analysis of the 18 737 genes from this dataset that have a mouse counterpart with the identical gene name revealed 2292 unique, annotated genes that varied significantly (FDR 5%) among the MGUS, smoldering multiple mycloma (SMM), and MM groups. Our heat map of an unsupervised hierarchical cluster analysis of these genes is shown in supplemental Figure 6A. The mouse ANOVA-PC gene probe sets from study 1 and study 2 (corrected for genes also found in OG stroma) were merged to identify probes that varied significantly during PCT progression in either study 1 or study 2 (“combined” probe set of 2218 mouse genes, supplemental Figure 6B-C) or in both study 1 and study 2 (“in-common” probe set of 315 mouse genes, supplemental Figure 6B,D). The “in-common” mouse genes were used for the statistical analysis of the Mayo Clinic Patient Dataset as depicted in supplemental Figure 6D and displayed as a heat map in supplemental Figure 6E, in which heretofore unknown genetic associations within the MGUS, SMM, and MM samples can be seen. Two examples of apparent cluster enrichment are boxed in supplemental Figure 6E and enlarged on the right: MGUS enrichment, in the top enlargement, and SMM/MM enrichment, in the bottom. These results, though encouraging, are not conclusive and need confirmation with other MM datasets and more refined analyses to fully plumb any possible biologic significance for progression of PC neoplasia in the 2 species.
Metformin is a potent inhibitor of PCT in vitro
To begin testing the biologic relevance of the pathways described above, and to initiate translation of our new findings into potential new interventions for PCT, we pursued the MetaCore lead that implicated the regulation of the translation initiation factors, Eif4f and Eif2, in tumor development. The importance of these proteins was re-enforced by the result of 2 additional MetaCore analyses using the Process Networks and Map Folder tools (not shown). The former revealed “Translation–regulation of initiation” as the topmost significant pathway in the ANOVA-PC validation gene set [P = 2.49 × 10−10; 47 hits among 127 pathway genes (37%)]. The latter tool scored “Protein synthesis” at rank 7 of coregulated pathways in both studies: P = 4.95 × 10−4 for the initial study and 5.31 × 10−12 for the validation study. The “Translation” pathway was of further interest because it can be readily inhibited by a Food and Drug Administration–approved drug, metformin. This drug activates AMPK (AMP-activated protein kinase), but it also has pleiotropic effects in many types of cancer, on cell cycle progression, growth factor signaling, regulation of apoptosis and, importantly, protein synthesis.29 On this backdrop, we decided to test, for the first time, metformin as a potential PCT drug and see whether metformin could inhibit the growth of malignant PCs in vitro.
Figure 6A, top panel, shows that the XRPC24 cell line exhibited a significant dose-dependent decrease in proliferation with as little as 1mM metformin. Treatment with 5mM metformin for 48 hours resulted in ∼ 50% inhibition of proliferation. The metformin effect on cell growth and proliferation was not because of general toxicity, for even 10mM metformin did not cause significant inhibition of proliferation of the mouse BM stromal cell line, S10 (Figure 6A bottom). When we exposed 3 additional PCT lines (ABPC20, AH2D11, and TEPC2027) to the same concentrations of metformin for 48 hours, these lines also showed marked inhibition of proliferation, ∼ 40% at 5mM drug (Figure 6B), similar to XRPC24 cells. These findings indicate that the sensitivity to metformin may be a general characteristic for PCTs, and it may be selective, because metformin has little inhibitory effect on stromal cells.
Metformin inhibits proliferation of PCT cells in vitro. (A) Top panel displays the dose-dependent decrease in the number of viable cells of the PC tumor line, XRPC24, after treatment with metformin. Cells were grown in vitro in presence of 3 different concentrations of metformin (1mM, 5mM, and 10mM) for 1, 2, or 3 days; then the number of cells was determined using the Cell Titer Blue assay. Statistical comparison of biologic replicates (n = 6) using a 2-tailed t test demonstrated the growth-inhibiting effect of metformin at all concentrations and time points compared with cells grown in absence of metformin (*P < .05). Metformin did not inhibit BM stroma cells, S-10 (bottom). (B) Inhibition of proliferation of PCT by metformin was confirmed in 3 additional lines of PCTs (ABPC20, AH2D11, TEPC2077) after 2 days in cell culture. Same conditions were used in all experiments.
Metformin inhibits proliferation of PCT cells in vitro. (A) Top panel displays the dose-dependent decrease in the number of viable cells of the PC tumor line, XRPC24, after treatment with metformin. Cells were grown in vitro in presence of 3 different concentrations of metformin (1mM, 5mM, and 10mM) for 1, 2, or 3 days; then the number of cells was determined using the Cell Titer Blue assay. Statistical comparison of biologic replicates (n = 6) using a 2-tailed t test demonstrated the growth-inhibiting effect of metformin at all concentrations and time points compared with cells grown in absence of metformin (*P < .05). Metformin did not inhibit BM stroma cells, S-10 (bottom). (B) Inhibition of proliferation of PCT by metformin was confirmed in 3 additional lines of PCTs (ABPC20, AH2D11, TEPC2077) after 2 days in cell culture. Same conditions were used in all experiments.
Discussion
Even though there have been many reports of gene expression changes at different stages of human cancers,4,5,30-34 the amount of information on changes in a primary tumor's transcriptome during the tumor's progression process is still very limited. Mouse model systems of human cancer, including MM, offer particular advantages to the study of tumor progression: opportunities for harvesting fresh tissue samples of different stages of tumor from identical genetic backgrounds and environmentally controlled conditions in a rapid, unprocessed, systematic, reproducible fashion. In the study presented here, we froze the OGs immediately on explanting to minimize postmortem changes in mRNAs from processing and/or degradation. We used LMD to collect PCs from IP OGs in BALB/c.iMycEμ mice, with minimal contamination by stromal cells, at different times of tumor progression to evaluate changes in gene expression that drive and/or accompany malignant PC transformation.
We found that genetic pathways related to development, cell adhesion, remodeling of the cytoskeleton, and regulation of the immune response appear to be involved in PCT progression, some particularly in the tumor cells themselves and others principally in the OG. Among the many genes shown to change expression during PCT progression in this study is Spp1, also known as osteopontin. Spp1 is involved in many steps of tumor development, progression, metastasis and angiogenesis35 in cancers of breast,36,37 stomach,38 lung,39,40 prostate,41 liver,42 colon,22 and, of particular import for this study, MM.43 While plasma concentrations of osteopontin are significantly increased in patients with MM,44 we found that Spp1 is strongly expressed at early stages of PCT progression but drops precipitously at day 104/105. It is possible that the observed up-regulation of Spp1 and its gene network is related to increased angiogenesis in early stages of MM and PCT. Osteopontin is a well-established proangiogenic factor in human myeloma,45 and blood vessel formation is a crucial process for formation of pristane-induced peritoneal granulomas.8 Indeed, our unpublished findings suggest that elevated angiogenesis is a common feature of mouse PCT, with some tumors exhibiting dramatic levels of vasculature. The reason why Spp1 expression drops precipitously in frank PCT in mouse, but remains high in human MM, is not known. It might reflect the additional, osteoclastogenic and bone-resorbing activity of osteopontin in myeloma46 that plays no role in extraosseous PCT. Inhibition of osteopontin, which offers promise in myeloma therapy,45 should be tested for PCT prevention in pristane-treated mice, and, if successful, such an approach may be of great relevance for preventing MGUS to MM progression.
Clearly, there is much more to do in this investigation of tumor progression, for example, overexpression or inhibition of individual genes or pathways are warranted to confirm their importance for the survival of iMycEμ-expressing tumor precursors and their maturation into fully transformed PCT cells. In addition, investigating epigenetic changes and changes in gene copy number over a similar time course of PCT progression would be appropriate, and initial stages of these studies are already underway. Going forward, high priority will be given to establishing the relevance of the mouse studies to human disease progression. Our initial studies along this line indicated a significant genetic overlap with progression of human MM, using as a paradigm 115 patients from the Mayo Clinic Dataset. Although initial results have been encouraging, they need to be refined and validated with additional MM datasets before we can report with confidence the degree of similarity in the genetics of PC neoplasia in mouse and man.
Follow-up studies on the report47 that diabetics who took metformin showed a 23% lower chance of developing cancer than diabetics who received insulin treatment have shown that metformin had pleiotropic effects in many types of cancer, for example, cell-cycle arrest, apoptosis and/or reduced growth factor signaling.48 Our experiments showed that PCTs in culture were very sensitive to metformin. The main downstream effect of AMPK activation on cell signaling is inhibition of mTOR, which is important for PCT development.49 However, one study using rapamycin to inhibit mTOR did not inhibit PCT growth in vitro.50 This result raises the interesting possibility, supported by findings in this study, that indirect, upstream targeting of multiple mTOR-controlling pathways by metformin may be more effective in inhibiting PCT. Although the mechanism by which metformin impedes the proliferation of PCT in vitro has not been determined, our result on the differential sensitivity of tumor versus stromal cells suggests that PCT progression could be specifically targeted in vivo in BALB/c.iMycEμ mice by treatment with metformin, the next step envisioned for this study. If this proves successful, targeting AMPK with metformin might be considered for use in treatment of patients with high-risk MGUS and/or SMM.34
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Kenneth Dorshkind for the S10 stromal cell line, and Dr Steven Bauer and Eva Rudikoff for their suggestions concerning its husbandry. They thank Dr Mark Simpson and Shelly Hooper for assistance with LMD, and Dr Michael Potter and Elizabeth Mushinski for some of the histology and IHC images. They thank Dr Beverly Mock for her careful reading of the manuscript.
This work was supported in part by the Intramural Research Program of the National Institutes of Health and the National Cancer Institute (NCI). This work was also supported, in part, by research awards from the Multiple Myeloma Research and International Waldenström's Macroglobulinemia Foundations (S.J.) and R01CA151354 from the NCI (S.J.).
National Institutes of Health
Authorship
Contribution: J.L. performed most of the research; E.S.P. performed data analysis, interpreted data, and wrote the manuscript; H.W. performed part of the research; S.G. performed part of the research; J.D.O. performed part of the research and edited the manuscript; P.J.N. performed part of the research; W.D. performed part of the research; T.B. performed statistical analysis; S.J. analyzed and interpreted data and wrote the manuscript; and J.F.M. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Siegfried Janz, Department of Pathology, The University of Iowa, Carver College of Medicine, 500 Newton Rd, 1030 ML, Iowa City, IA 52242; e-mail: siegfried-janz@uiowa.edu, or J. Frederic Mushinski, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 37 Convent Dr, Bldg 37, Rm 3146, Bethesda, MD 20892; e-mail: mushinsj@gmail.com.
References
Author notes
J.L. and E.S.P. contributed equally.
S.J. and J.F.M. are co–senior authors.

![Figure 4. Expression levels of 5 selected ANOVA-revealed PC genes are up-regulated in frank tumor cells compared with premalignant tumor precursors. Log2-transformed mean gene expression levels (horizontal lines) and individual gene expression levels (dots) at indicated times posttumor induction with pristane are plotted for study 1 (initial study) in the left column and study 2 (validation study) in the center column. The results of statistical comparisons (2-tailed t test) of mean expression levels of combined tumor precursor samples (□; days 7-46 in case of study 1 [n = 20]; days 7-49 in case of study 2 [n = 24]) and tumor samples (■; day 104 in case of study 1 [n = 5]; day 105 in case of study 2 [n = 6]) are indicated as P values in the right column. Bcl2l1 indicates BCL-2-like 1; Bmi, Bmi polycomb ring finger oncogene; Cxxc5, CXXC finger 5; Csk, c-src tyrosine kinase; and Ly6 days, lymphocyte Ag 6 complex, locus D.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/4/10.1182_blood-2011-06-363887/4/m_zh89991285420004.jpeg?Expires=1769084785&Signature=KLTmhyjz-8JTFEgjABHoPsvV3aaCpeudPD~JP~5vTfrHgWG3j-f7ZQzUUuupmh0lmjnjXGTMUQphUJFzsbYyMJgYENK0sGkEZxfg3VeHrServ274hWeCJb2KR-NAd1aP-NVQZYktn30dXtp~tPYTsqm4WCvhbpSVPu~lhJjHGOzl10aoc0dz8aPJ07Cx6IYQ2we-3JeJF18j1XcIyrRNUydBc1kJiRj0CcZtjYj3TZe6V74Pci0wKd8kWdGb2mYRgz3rTxVE7rd~OOQntxiZ8tgxokBCkDSqfSrA9Io63Ndn0SgVDBK9BHF6I3vXKmaMRrGjsDAARej48fwpUrkepA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal